Abstract
A high frequency of mtDNA somatic mutation has been observed in many tumors as well as in aging tissues. In this study, we analyzed the mtDNA control region sequence variation in 3534 single normal cells and individual blasts from 18 patients with leukemia and 10 healthy donors, to address the mutation process in leukemic cells. We found significant differences in mtDNA sequence, as represented by the number of haplotypes and the mean number of cells with each nonaggregate haplotype in a population of cells, in patients compared to controls. Patients with similar clinical leukemia types, particularly acute myeloid leukemia (AML), did not show a uniform pattern of sequence variation in single blasts. Some patients at relapse presented a complex shift of major haplotypes in single cells. Four patients showed high frequencies of cells containing mutations 189, 260, 16150, and 16488, respectively, as a result of clonal expansion and could be considered as potential markers for their respective disease progression. To our knowledge, this is the first large-scale study of mtDNA variation in single malignant cells. Our results suggest that the somatic mutation process in leukemia is complex, leading to diverse levels of genetic alterations due to either intrinsic aspects of leukemia pathophysiology or chemotherapy effects.
Introduction
Since the recent discovery of a high frequency of mtDNA somatic mutations in cancer,1,2 a putative role of mtDNA has been sought in tumorigenesis. Two recent studies showed a potential role for pathogenic mtDNA mutations in the promotion of tumor by increasing reactive oxygen species (ROS)3 and preventing apoptosis.4 There is a proposal for adding cancer to the list of mitochondrial diseases.3 Despite the enormous number of mtDNA somatic mutations identified in cancer cells,5,6 not yet fully resolved are the questions of how mutations arise and become fixed in tumors, and whether the mutation rate is accelerated or slowed at different stages of malignant progression. A selective mechanism leading to a replicative advantage has been suggested to explain an apparently high rate of somatic mutation in tumors,1,2 but later studies and computer modeling indicated that random processes are sufficient to explain the high frequency of homoplasmic somatic mutations in both tumors and aging tissues.7–11
Several laboratories have reported unexpectedly large number of somatic mutations in leukemia. Ivanova et al12 compared the restriction endonuclease digestion pattern of mtDNA in patients with acute lymphoblastic leukemia (ALL) and in healthy controls. Carew et al13 determined mtDNA mutations in 6 short fragments from the control and coding regions in patients with chronic lymphocytic leukemia (CLL) with and without prior chemotherapy. Grist et al14 analyzed partial control region sequence variation in patients with acute myeloid leukemia (AML) or ALL. He et al15 sequenced the entire mtDNA genome in both normal and leukemic cells from each of the 24 patients. Finally, Linnartz et al16 analyzed the mtDNA alterations in 10 cases of acute leukemia evolving from myelodysplastic syndrome. There are also reports of change of mtDNA copy number in leukemic cells.15,17,18 None of these studies has traced somatic mutations in leukemia at the level of a single cell.
We have recently described age-dependent accumulation of mtDNA mutations in single hematopoietic cells from the bone marrow and peripheral blood (PB) of healthy donors19–21 and compared the mutation spectra of differentiated blood cells from the same donors by using an optimized method for analyzing mtDNA variation in single flow cytometry-sorted cells.21 In the present study, we used single-cell analysis to detect low-frequency individual somatic mutations that are not well represented in bulk tissue to quantify a somatic mutation process in leukemia, as well as to determine whether mtDNA could be used as a marker for monitoring disease progression. We analyzed mtDNA control region sequence variation among single cells obtained from patients with leukemia and healthy controls. Our results indicate that the mutation process in leukemia is complex, leading to diverse levels of genetic sequence alterations due to either intrinsic aspects of leukemia pathophysiology or chemotherapy effects. Shifts in major mtDNA haplotypes in single cells can be observed during different stages of leukemia in some individual patients.
Patients, materials, and methods
Patients
Eighteen patients with leukemia were recruited in this study with informed consent provided under protocols approved by the Institutional Review Boards of the National Heart, Lung, and Blood Institute (NHLBI, Bethesda, MD), M. D. Anderson Cancer Center, and University of São Paulo at Ribeirão Preto Medical School (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). PB from 10 patients (UPN1-3, UPN16-22) was collected at the M. D. Anderson Cancer Center and was transferred to Bethesda. Leukemic blasts, mononuclear cells, and granulocytes from PB were separated by Ficoll density-gradient centrifugation on arrival and were frozen in liquid nitrogen. Frozen PB cells from 8 patients with AML from Brazil were shipped to Bethesda on dry ice. We also obtained a PB sample from patient OAM at the time of relapse. The recently available data of mtDNA variation in single PB hematopoietic cells from 5 healthy donors,21 together with data from PB CD34+ single cells from 5 newly analyzed healthy volunteers in the current study, served as healthy controls.
Single-cell sorting
We sorted for blasts in each patient according to specific cell surface markers for the malignant cell population as determined in a clinical laboratory during diagnostic evaluation (Table S1). For some patients, we also sorted “normal” cells, such as granulocytes, and used them as controls. We failed to successfully sort the blast cells of patient UPN22 due to poor survival after shipping, freezing, and thawing, and possibly also chemotherapy effects, and for this case we only analyzed CD34+ cells and granulocytes. We followed the staining procedures for each antibody provided by the manufacturer (BD Biosciences, San Jose, CA) and the procedures described in our recent publication.21 Briefly, 1 × 106 cells suspended in 100 μL PBS containing 0.5% bovine serum albumin (BSA) were incubated with respective antibodies (anti-CD34 phycoerythrin [PE]–conjugated monoclonal antibody; anti-CD33 allophycocyanin [APC]–conjugated monoclonal antibody or anti-CD33 PE-conjugated monoclonal antibody; anti-CD13 PE-conjugated monoclonal antibody; anti-CD5 fluorescein isothiocyanate [FITC]–conjugated monoclonal antibody) or a combination of 2 antibodies (Table S1) for 30 minutes at 4°C. Cells were then washed and resuspended in 600 μL PBS supplemented with 0.5% BSA.
Cell sorting was performed on a MoFlo Cytometer (DakoCytomation, Ft Collins, CO), using 100 mW of the 488 nm line of an argon laser (I-90, Coherent, Palo Alto, CA) for excitation. Forward scatter was the triggering parameter. Single-cell deposition was accomplished using the CyClone automated cloner (DakoCytomation) in the 0.5 single-drop mode with gating based on forward scatter and fluorescence. Single blasts, CD34+ cells, and granulocytes were sorted into each well of a 96-well plate containing 50 μL lysis buffer (10 mM Tris-HCl, pH 8.0, 50 mM KCl, 100 μg/mL proteinase K, 1% Triton X-100). The whole plate was then incubated at 56°C for 15 minutes to liberate total DNA. Proteinase K in the lysate was inactivated by incubation at 96°C for 6 minutes.
DNA amplification and sequencing
To generate sufficient template from single cells for sequencing of the entire mtDNA control region sequence, 2-step nested polymerase chain reaction (PCR) amplification was used with the outer and inner primer pairs and conditions as described in our recent studies.19–21 In brief, the first PCR was performed in 30 μL of reaction mixture containing 400 μM of each dNTP, 1 U TaKaRa LA Taq, which has proof reading activity (Takara Bio, Otsu, Japan), 0.5 μM of each forward and reverse outer primer (L15594: 5′-CGCCTACACAATTCTCCGATC-3′ and H901: 5′-ACTTGGGTTAATCGTGTGACC-3′), and 5 μL cell lysates. The amplification was run on the GeneAmp PCR system 9700 (Applied Biosystems, Foster City, CA) with the following cycles: one cycle of 94°C for 3 minutes; then 35 cycles of 94°C for 30 seconds, 52°C for 40 seconds, and 72°C for 1 minute with a 5-second increase per cycle; and ending with a full extension cycle of 72°C for 10 minutes. The second PCR was performed in 50 μL reaction mixture containing 400 μM of each dNTP, 2 U TaKaRa LA Taq, 0.5 μM of each forward and reverse inner primer (L15990: 5′-TTAACTCCACCATTAGCACC-3′ and H650: 5′-GAAAGGCTAGGACCAAACCTA-3′), and 5 μL of first PCR product under the same amplification condition as the first PCR but with an extension time of 90 seconds at 72°C/cycle. Second PCR products were purified using the QIA quick PCR purification kit (Qiagen, Valencia, CA) and were directly sequenced by using the BigDye Terminator v3.1 Cycle Sequencing kit on a 3100 DNA sequencer (Applied Biosystems) according to the manufacturer's manual. We used the second PCR primers and the following primers to overlap sequence the entire mtDNA control region: L15996, 5′-CTCCACCATTAGCACCCAAAGC-3′; L16209, 5′-CCCCATGCTTACAAGCAAGT-3′; L16517, 5′-CATCTGGTTCCTACTTCAGG-3′; H26, 5′-GCATGGAGAGCTCCCGTGAGTGG-3′; L29, 5′-GGTCTATCACCCTATTAACCAC-3′; L332, 5′-CCCGCTTCTGGCCACAGCAC-3′.
Sequence analyses
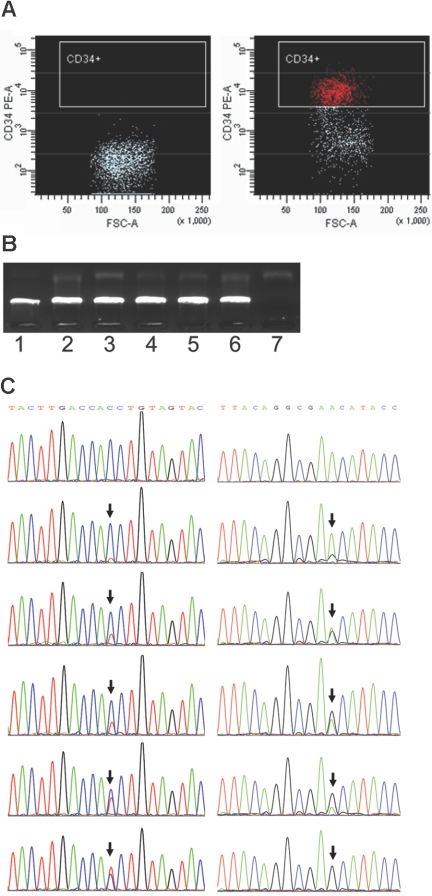
Sequences were aligned by the SeqMan program (DNASTAR, Madison, WI) and were proofread by eye. A mutation was scored relative to the revised Cambridge Reference Sequence (rCRS).22 Cells showing sequence difference at all the variable sites of the donor's consensus sequence, which suggest external mtDNA contamination, were discarded. Due to the limits of sequencing, mutation heteroplasmy (coexistence of wild-type and the mutant allele) was scored when a mutant allele was present at more than 10% level chromatography (Figure 1). The length mutations of C-tract in region 16184-16193 due to 16189T>C mutation were not scored in single cells because most of them were heteroplasmic of multiple polyC-tracts and could not be conservatively counted according to sequencing chromatographs. We scored the length mutation of C-tract in region 303-309 in the second hypervariable segment (HVS-II) of control region by direct counting of the base shift of T at site 310. The length mutation of the AC repeat in region 515-524 in the third hypervariable segment (HVS-III) was also based on the sequencing chromatographs.
Figure 1.
Single-cell sorting, amplification, and mutation scoring. (A) Representative isotype IgG (negative control) and CD34 stainings for patient UPN21. (B) Single-cell 2-step PCR results. Lanes 1-6, DNA from single cells as the template. Lane 7, negative control. (C) Different levels of heteroplasmy of mtDNA mutations at sites 16150 and 189 (arrows) in single cells from leukemia patients DC and UPN17, respectively.
Measurement of the number and size of mtDNA haplotypes in single-cell populations
We used 2 parameters to quantify the difference of the subclones marked by respective mtDNA haplotypes in healthy donors and leukemia patients. First, we determined the number of haplotypes identified in a population of cells from each sample; this index reflects the total number of mutations that have occurred within a given number of cells, which in turn reflects time over which the subclone has developed and the net mutation rate per unit time. Second, we calculated the mean number of cells bearing each nonaggregate haplotype; this parameter reflects the mean size of the subclones marked by the nonaggregate haplotypes, which correlates with factors such as the time that each mutation occurred, genetic drift, and the magnitude of selection or clonal expansion of the subclones harboring a particular nonaggregate haplotype. Both parameters have some limitations, but direct comparison of the leukemia and control samples based on either parameter should reveal differences between the 2 groups and may be correlated with distinct biologic processes.
The unpaired t test was used to compare differences between healthy controls and patients by using Prism 4 (GraphPad Software, San Diego, CA). F test was used to compare variances between the 2 groups; if variances were significantly different, the unpaired t test with the Welch correction was used to quantify the difference between the 2 groups. P values less than .05 were regarded as statistically significant.
Results and discussion
Data quality control
As has been reemphasized recently in a critical paper,5 analysis of mtDNA from minute amounts of DNA derived from minimal amounts of tissue makes contamination and PCR errors particularly problematic. Previous reports of somatic mutations in cancer tissues have not been free of laboratory error and misinterpretation of flawed data.5 In the current study, we were especially attentive to systematic errors that might introduce artifacts and we sought to maintain stringent quality control. First, a high-fidelity Taq polymerase with proof reading activity (with a relative error rate of 0.16 as compared with a regular Taq polymerase, according to the manufacturer), and negative controls were used during the amplification. Second, single nucleotide mutations were verified by independent amplification and sequencing using the first PCR product as template. Although recognizing that such a confirmation is not entirely independent because it does not use the original cell lysate, comparison of triplicates of independent amplifications derived from the original lysates followed by sequencing did indicate that 85% to 90% of mutations could be consistently reproduced from single CD34+ cells.21 In the current study, we compared substitutions among triplicates of independent amplifications for single cells from donor 6. Several differences were observed: the cell harboring mutation 16172 T>Y appeared as 16172T>C and 151T>C in one replicate; cell harboring mutation 16240G>R had one more mutation 106G>R in one replicate; mutations 16267C>C/A, 16039G>R, and 412G>R were observed in 2 of the 3 independent amplifications. Nucleotide substitutions in the other 5 cells could be confirmed in all the triplicates. The mutations that presented with marginal levels of heteroplasmy, less than about 20% as observed in the chromatographs, were not always reproducible. Finally, a phylogenetic approach that was described in previous studies5,23,24 was used for data quality control.
Aggregate mtDNA sequence and haplogroup status
We determined the aggregate mtDNA sequences of each patient and healthy control by taking the consensus sequence of the single cells and assigned each mtDNA to the nominated haplogroup system of world human mtDNA phylogeny.25–29 The resulting classification of each mtDNA revealed that 6 leukemia patients had matrilineal origin of Native Americans (haplogroups A2 and D125). Three patients and 2 healthy donors had matrilineal origin of Africans (haplogroups L2a, L3f, and L0a128). One healthy donor and one patient were of East Asian matrilineal origin (haplogroups M7a and B4b27). The remaining cases and controls were of European matrilineal origin26 (Table S2).
We tentatively grouped the samples according to mtDNA haplogroup background and compared the mutation level in a population of cells among samples with the same haplogroup status. Although the numbers of patients and controls were small, we did not observe any obvious correlation between haplogroup background and mtDNA variation in single cells (Table 1)
Table 1.
Summary of mtDNA sequence variation in single cells from the leukemia patients and healthy controls
| Sample | Leukemiatype | Age, y/sex | Haplogroup | No. of cells | No. ofhaplotypes, (%) | No. ofnonaggregate haploytpes by nucleotide substitutions (%) | Mean no. of cells with nonaggregate haplotype | Mean no. of cells with nonaggregate haplotype by nucleotide substitutions |
|---|---|---|---|---|---|---|---|---|
| CD34+ and blasts | ||||||||
| Patients | ||||||||
| LAA | AML M0 | 61/F | H | 95 | 16 (16.84) | 11 (11.58) | 2.58 | 1.05 |
| AGM | AML M1 | 62/M | B4b | 96 | 11 (11.46) | 7 (7.29) | 5.07 | 1.19 |
| MFSS | AML M1 | 36/F | L2a | 96 | 10 (10.42) | 8 (8.33) | 1.04 | 1.04 |
| JCS | AML M1 | 40/M | L3f | 96 | 15 (15.63) | 12 (12.50) | 3.67 | 1.04 |
| DC | AML M1 | 15/M | L2a | 96 | 18 (18.75) | 12 (12.50) | 3.25 | 3.27 |
| OAM at diagnosis | AML M1 | 33/M | D1 | 92 | 11 (11.96) | 9 (9.78) | 1.51 | 1.56 |
| EMB | AML M1 | 16/M | D1 | 95 | 34 (35.79) | 30 (31.58) | 1.05 | 1.05 |
| ERR | AML M1 | 53/F | A2 | 94 | 6 (6.38) | 5 (5.32) | 1.28 | 1.28 |
| UPN21 | AML | 16/F | D1 | 82 | 7 (8.54) | 3 (3.66) | 1.42 | 1.22 |
| UPN1 | AML M2 | 47/F | T2 | 146 | 10 (6.85) | 5 (3.42) | 1.73 | 0.68 |
| UPN16 | 2nd AML/MDS | 74/F | J1c | 92 | 5 (5.43) | 3 (3.26) | 2.42 | 1.09 |
| UPN22 | Relapsed AML | 11/F | H | 65 | 20 (30.77) | 11 (16.92) | 3.32 | 2.63 |
| OAM at relapse | Relapsed AML | 36/M | D1 | 96 | 6 (6.25) | 4 (4.17) | 5.61 | 6.78 |
| UPN18 | AML, prior NHL | 62/F | A2 | 92 | 8 (8.70) | 4 (4.35) | 2.30 | 1.09 |
| UPN20 | CML | 53/M | A2 | 85 | 3 (3.53) | 1 (1.18) | 6.90 | 1.18 |
| UPN17 | CML | 27/F | X2b | 80 | 17 (21.25) | 10 (12.50) | 3.97 | 4.42 |
| UPN2 | AML from CMML | 58/M | J1c | 96 | 14 (14.58) | 13 (13.54) | 2.31 | 2.31 |
| UPN3 | CLL | 64/F | H | 89 | 9 (10.11) | 4 (4.49) | 2.10 | 1.12 |
| UPN19, blasts | APML | 21/F | H | 94 | 22 (23.40) | 15 (15.96) | 2.60 | 1.06 |
| UPN19, normal CD 34+ cells | APML | 21/F | H | 96 | 10 (10.42) | 4 (4.17) | 4.76 | 1.04 |
| Grand mean ± SD | — | — | — | 94 ± 15 | 13.85 ± 8.56 | 9.33 ± 7.04 | 2.94 ± 1.63 | 1.81 ± 1.49 |
| Donors | ||||||||
| Donor 1 | — | 48/F | U5b | 85 | 14 (16.47) | 8 (9.41) | 2.74 | 1.47 |
| Donor 2 | — | 44/M | L2a | 92 | 16 (17.39) | 11 (11.96) | 2.27 | 1.09 |
| Donor 3 | — | 36/M | L0a1 | 93 | 12 (12.90) | 6 (6.45) | 3.19 | 2.13 |
| Donor 4 | — | 55/M | T2 | 93 | 14 (15.05) | 8 (8.60) | 3.32 | 3.02 |
| Donor 5 | — | 35/M | U5b | 93 | 13 (13.98) | 10 (10.75) | 2.92 | 1.18 |
| Donor 6 | — | 35/M | M7a1 | 95 | 15 (15.79) | 10 (10.53) | 2.91 | 1.05 |
| Donor 7 | — | 32/M | U6 | 93 | 40 (43.01) | 35 (37.63) | 1.45 | 1.08 |
| Donor 8 | — | 25/M | H | 93 | 38 (40.86) | 35 (37.63) | 1.58 | 1.08 |
| Donor 9 | — | 57/M | K1a | 93 | 31 (33.33) | 28 (30.11) | 1.36 | 1.34 |
| Donor 10 | — | 39/M | pre-V | 94 | 17 (18.09) | 12 (12.77) | 2.66 | 1.06 |
| Grand mean ± SD | — | — | — | 92 ± 3 | 22.69 ± 11.65 | 17.58 ± 12.40 | 2.44 ± 0.73 | 1.45 ± 0.64 |
| Granulocytes | ||||||||
| Patients | ||||||||
| UPN21 | AML | 16/F | D1 | 24 | 8 (33.33) | 6 (25.00) | 4.75 | 4.17 |
| UPN22 | Relapsed AML | 11/F | H | 70 | 42 (60.00) | 30 (42.86) | 2.00 | 1.89 |
| UPN20 | CML | 53/M | A2 | 92 | 17 (18.48) | 13 (14.13) | 1.80 | 1.09 |
| UPN19 | APML | 21/F | H | 79 | 23 (29.11) | 15 (18.99) | 2.47 | 1.27 |
| Grand mean ± SD | — | — | — | 66 ± 30 | 35.23 ± 17.66 | 25.25 ± 12.56 | 2.76 ± 1.36 | 2.11 ± 1.42 |
| Donors | ||||||||
| Donor 1 | — | 48/F | U5b | 64 | 28 (43.75) | 23 (35.94) | 2.12 | 1.56 |
| Donor 2, PB | — | 44/M | L2a | 82 | 27 (32.93) | 22 (26.83) | 1.77 | 1.33 |
| Donor 2, bone marrow | — | 45/M | L2a | 96 | 16 (16.67) | 12 (12.50) | 2.36 | 1.04 |
| Donor 3 | — | 36/M | L0a1 | 72 | 26 (36.11) | 23 (31.94) | 1.66 | 1.39 |
| Donor 4 | — | 55/M | T2 | 76 | 27 (35.53) | 19 (25.00) | 1.99 | 1.66 |
| Donor 5 | — | 35/M | U5b | 82 | 31 (37.80) | 26 (31.71) | 1.34 | 1.27 |
| Grand mean ± SD | — | — | — | 79 ± 11 | 33.80 ± 9.14 | 27.32 ± 8.25 | 1.87 ± 0.36 | 1.38 ± 0.22 |
The mtDNA haplogroup classification was based on sequence variation in the consensus sequence of the single cells from respective individuals (Table S2). We counted the number of haplotypes (including the aggregate mtDNA haplotype and the nonaggregate haplotypes that differed from aggregate sequence by nucleotide substitutions or indels or both) and the number of nonaggregate haplotypes that differed from the aggregate type only by nucleotide substitutions, based on the mutations detected in a population of cells from each sample (Table S3). The geometric mean number of cells harboring each nonaggregate haplotype or nonaggregate haplotype by nucleotide substitutions was computed as another parameter to show the difference between the leukemia patients and healthy donors. The data of peripheral blood CD34+ cells and granulocytes for healthy donors 1 to 5 are from Ogasawara et al.21 Single blasts at diagnosis and relapse were analyzed for patient OAM. Both blasts (CD33+ CD34−) and normal CD34+ cells were analyzed for patient UPN19.
Granulocytes from peripheral blood and bone marrow were analyzed for healthy donor 2.
MDS indicates myelodysplastic syndrome; NHL, non-Hodgkin lymphoma; CMML, chronic monomyelocytic leukemia; —, not applicable.
mtDNA variation in leukemic blasts versus normal cells from patients and healthy donors
Because the nuclear genetic background of each patient and healthy donor was different, comparison of mtDNA variation of “normal” single cells and single blasts from the same patient would be desirable. However, in our recent analysis of mtDNA mutation spectra of CD34+, T, and B cells, and granulocytes from the same healthy donors, we found varied levels of mtDNA alterations among the 4 cell types from the same individual, with generally greater variation among granulocytes.21 Therefore, we anticipated that such a comparison of “normal” cells and leukemic blasts from the same patient might be confounded by the higher level of mtDNA alterations in polymorphonuclear cells.
Consistent with the pattern in healthy donors, we found marked variance of mtDNA variation in different cell types from the same patient. For example, we observed more haplotypes among blasts (22 haplotypes in 94 blast cells) of acute promyelocytic leukemia (APML) UPN19 than in the same patient's presumably normal CD34+ cells (10 haplotypes in 96 cells). Granulocytes from this patient contained even more mutations (23 haplotypes in 79 granulocytes). In another 3 patients (UPN20-22), in which analyses of both granulocytes and blast cells (or CD34+ cells) were conducted, we found a similar trend of greater variation in granulocytes (Table 1). Thus, it appears that somatic mutations might accumulate at different rates or mechanisms in differentiated cells and progenitor cells from the same individual.
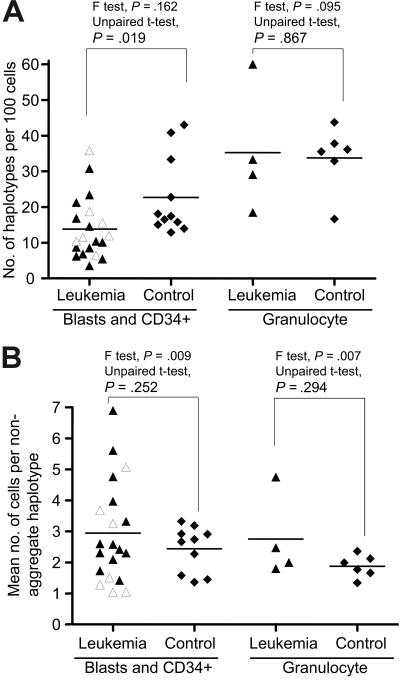
We next obtained the PB CD34+ cells from the healthy donors as controls for comparison with blasts in patients (Tables 1 and S3–S4). The mtDNA variation in single blast cells and CD34+ cells for each subject, as represented by the number of haplotypes and the mean number of cells bearing each nonaggregate haplotype, varied substantially among the patients compared with controls (Table 1; Figure 2). The overall difference in the number of haplotypes/100 cells between the patient and control groups was statistically significant (P = .02 < .05, 2 tailed unpaired t test); the leukemia group harbored fewer haplotypes than in controls. Among the AML patients who were sampled at the time of diagnosis and prior to any treatment, particularly subtype M1, we did not observe a uniform pattern of mtDNA variation in blast cells (Table 1; Figures 2 and S1). Patients with AML who had previously undergone medical treatment, including the relapsed AML patients and the AML patients in which leukemia was secondary to chemotherapy or evolved from another hematologic disease, did not generally show a higher level of variation compared to de novo leukemia patients (for both parameters, P > .05; 2-tailed unpaired t test). Based on the pattern of mtDNA variation in single cells, AML appeared heterogeneous despite its character as a clonal disease.
Figure 2.
mtDNA sequence variation pattern in single-cell populations from patients with leukemia and healthy controls. (A) The number of mtDNA haplotypes (including the aggregate haplotype and the nonaggregate haplotypes that differ from aggregate sequence by nucleotide substitutions or indels or both) observed in a population of single cells from each subject in the patient and control groups. (B) The mean number of cells bearing each nonaggregate haplotype in a population of cells from leukemia patients and healthy controls. The unpaired t test was used when the F test was not statistically significant; otherwise unpaired t test with the Welch correction was used. The 7 AML M1 patients at diagnosis and prior treatment are marked by open triangles. Horizontal lines refer to mean values.
mtDNA marker for leukemia progression and clonal proliferation?
Rapid expansion and other neoplastic features of blast cells in leukemia could affect the mutation process in mtDNA and be detectable during progression. These changes therefore might be used as markers for tracking individual clones. The C-tract alteration in region 303-309 in HVS-II has been identified as a major target for mtDNA alterations in cancer30 and related to premalignant lesions of the head and neck as a marker.31 In contrast, the current and other data21 indicate that this C-tract alteration is also a “hot spot” for somatic mutation in single cells in healthy individuals. The heteroplasmy of certain point mutations has been found to vary during diagnosis, remission, and relapse stages of the same leukemia patient.15,16 In the present study, besides the C-tract alteration, which occurred in 12 of 18 leukemia patients and in 9 of the 10 controls (albeit with different frequency in each sample), we also observed mtDNA substitution with different levels of heteroplasmy in many single cells from each of the 5 patients UPN2 (260G>R), UPN17 (189A>R), UPN22 (16320C>Y), DC (16150C>Y), and OAM (16488C>C/A). In patient UPN20, 12.9% (11 of 85) of blasts were distinguished by different levels of heteroplasmy of a dinucleotide AC repeat insertion at region 515-524 relative to its aggregate sequence (note that the aggregate sequence of UPN20 only had 4 AC repeats in region 515-524, as compared with the rCRS22). These different levels of heteroplasmy of certain mutation in different single cells from the same individual, as exemplified by mutations at sites 189 and 16150 in patients UPN17 and DC, respectively, in Figure 1, obviously resulted from clonal expansion and genetic drift due to the rapid proliferation of the blasts, which may ultimately result in a shift of the predominant allele in single cells. Coding region mutations may also exist in leukemic blasts and present a similar heteroplasmy pattern as observed here but sequencing was restricted to the control region in the present study. Potential damage to DNA in single blasts may also contribute to observed heteroplasmy owing to random statistical effects.
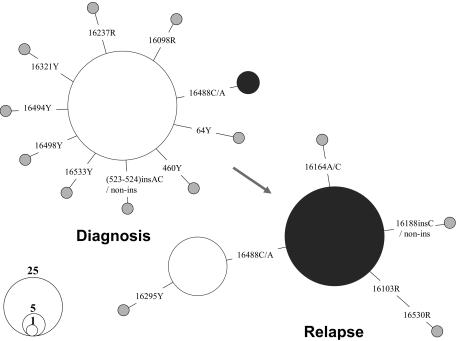
The comparison of the mtDNA mutation pattern in single blasts of one AML patient, OAM, at diagnosis and relapse provided direct evidence of the dynamics of different leukemic clones during disease progression. At first diagnosis in September 2002, only 5 of 92 blasts harbored a mtDNA haplotype, defined by the presence of an extra rare C to A transversion at site 16488 in a heteroplasmic status. However, 76.1% of single blasts (70 of 92) were found to bear this mutation at relapse in May 2005, suggesting its origin from the residual blast clone harboring mutation 16488C>C/A (Figure 3). This clone was apparently selected and then expanded after treatment. Although we were unable to analyze samples at different stages for all leukemia patients in the current study, this analysis of mtDNA sequence variation in single blasts suggests the feasibility of applying mtDNA somatic mutation to trace the progression of leukemia in individual cells. The feasibility of such an approach depends on the probability that cells that result from clonal expansion harbor the sentinel mutation; in most cases, however, there was little evidence for specific mutation markers for each leukemia patient or for different leukemia (sub)types.
Figure 3.
Shift of mtDNA major haplotypes in patient OAM with AML at diagnosis and relapse. The relationship between the haplotypes is presented in a network profile. The circle area is proportional to the haplotype frequency.
mtDNA mutation process in leukemia
Somatic mutations accumulated in tumors and aging tissues.1,2,5–11,19–21,32–34 Among the models proposed, random genetic drift provides the simplest explanation for this process and has been supported by mathematic simulations.7–11 Determining mtDNA variation in single cells could further test this hypothesis. Rapid clonal expansion of blasts (from a few founders) at an early stage of disease, together with altered intracellular mtDNA copy number in tumor cells,11,17,18 should enhance random drift and result in a pattern of a generally lower amount of mtDNA variation in patients' blast cells compared with controls. However, our analysis for mtDNA variation in single cells from leukemia patients revealed an unexpectedly complex pattern; the extent of mtDNA alteration varied greatly among the patients analyzed and was not generally increased in the later stages of leukemia or after medical treatment (Tables 1 and S3). Some leukemia patients at diagnosis presented levels of mtDNA alterations comparable to controls. Our previous studies had shown that mtDNA sequence is relatively homogenous in CD34+ cells from cord blood and much more heterogeneous in CD34+ cells from middle-aged adults.19 We thus anticipated homogeneity among acute leukemic blasts, which could be assumed to have arisen over a much shorter time period from limited number of founders. The observed complex pattern in leukemia patients, although on average showing fewer mutations than that in controls, suggests that mutations appear at a relatively high rate in leukemic cell mtDNA and that homoplasmy (uniformity of sequence within an individual cell) and clonal expansion (expression of the fixed mtDNA mutation in a substantial proportion of progeny) can be achieved over months rather than requiring decades.
Among those patients who presented a high level of mtDNA variation, we also observed different levels of heteroplasmy for certain mutation, such as variant at site 16150 in patient DC (Figure 1), which reflects the effect of random drift and clonal proliferation. In such cases, drift may play an active role in the fixation of mutations. The observed mtDNA variation pattern in these leukemia patients may reflect dynamic or counterbalanced processes of cumulative mutation induction by mutagens and mutation loss by genetic drift during clonal expansion, as well as effects of cytotoxic drug treatment. The mutation pattern observed in patient UPN22 may be an example; we observed several mutations with different levels of heteroplasmy, such as a length mutation in C-tract in HVS-II and variants C16320T and T195C in the majority of single cells that had a consensus sequence 16222-16519-263-319, whereas several cells bore the consensus sequence 16239-16519-263. The latter consensus sequence type was present at different frequency in CD34+ cells (4.6%, 3 of 65) and granulocytes (11.4%, 8 of 70; Table S3). It is possible that one of the 2 mtDNA consensus sequence types arose during chemotherapy. Grist et al14 also reported a shifting pattern of mtDNA types in a number of their leukemia patients and depicted a complex hypothetical evolutionary pathway for mtDNA haplotypes during diagnosis, remission, and relapse of one acute leukemia patient in their Figure 1. In the study by Carew et al,13 patients with CLL with chemotherapy had many heteroplasmic mutations in the short ND1 fragment (532 bp) and in other regions analyzed. These results favor an increased rate of mtDNA mutation during and after chemotherapy in some patients. Notable also is the presence of 3 mutations in the 2 consensus sequence types in UPN22, without evidence of intermediary types with single or double mutations; this patient had received a platelet transfusion proximate to sample acquisition, and contamination by donor blood cells cannot be excluded, as has been reported in a patient with ALL.35 In any case, mutations at sites 16320 and 195 and other mutations that stepped from the consensus sequence 16222-16519-263-319 in the majority of single cells in patient UPN22 were obviously the product of a quick mutation process.
About 60% of mutations (not including the length mutation in the C-tract and the dinucleotide AC repeat in HVS-III), which were observed in single cells from leukemia patients, are not listed on MITOMAP (www.mitomap.org) as reported polymorphisms; this database is derived mainly from population genetics, medical, and forensic studies. The percentage of such “novel” mutations was also very high (up to 55%) among the single cells from healthy donors (this study and Ogasawara et al21). Most of these mutations also are not in the list of phantom mutation hot spots that were detected in a massive screening of more than 5000 sequencing electropherograms,36 and thus are unlikely to be reading errors of sequencing chromatographs. Why these nucleotide changes have only occurred as somatic mutations and are not fixed in human populations as a germline substitution remains an open question. When we quantitated absolute numbers of mutations irrespective of their frequency, some mutations, such as 42T>Y, 146T>Y, 185G>R, 204T>Y, 378C>Y, 571C>Y, 16068T>Y, 16131T>Y, 16189T>Y, 16222C>Y, 16390G>R, and 16565C>Y, were present in single cells from 3 or more individuals. Among them, mutations 146, 185, 204, and 16189 have been scored as mutation hot spots in human mtDNA control region that was based on population data.23,24,37,38 It thus seems that somatic mutation events in single hematopoietic cells are also prone to occur at well-characterized mutational hot spots, as has been observed in patients with primary tumors of the central nervous system.33 Our observed mtDNA mutation spectrum of single cells from the normal and leukemic hematopoietic system also differs from the patterns that were described in aging tissues,39–41 Alzheimer disease brain,42 cardiomyocytes,43 and neurons and glia,44 suggesting tissue specificity and adding further complexity to our view of the mutation process in leukemia.
Supplementary Material
Acknowledgments
We thank Ms Leigh Samsel for technical assistance and Dr Rodrigo Calado for help in obtaining the samples. We are also grateful to the 2 anonymous reviewers for valuable comments.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
References
- 1.Polyak K, Li Y, Zhu H, et al. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat Genet. 1998;20:291–293. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- 2.Fliss MS, Usadel H, Caballero OL, et al. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000;287:2017–2019. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- 3.Petros JA, Baumann AK, Ruiz-Pesini E, et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci U S A. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shidara Y, Yamagata K, Kanamori T, et al. Positive contribution of pathogenic mutations in the mitochondrial genome to the promotion of cancer by prevention from apoptosis. Cancer Res. 2005;65:1655–1663. doi: 10.1158/0008-5472.CAN-04-2012. [DOI] [PubMed] [Google Scholar]
- 5.Salas A, Yao Y-G, Macaulay V, Vega A, Carracedo Á, Bandelt JH- A critical reassessment of the role of mitochondria in tumorigenesis. PLoS Med. 2005;2:e296. doi: 10.1371/journal.pmed.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salas A, Yao Y-G, Bandelt H-J. Mitochondria: more than mitochondrial DNA in cancer—author's reply. PLoS Med. 2006;3:e166. doi: 10.1371/journal.pmed.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chinnery PF, Samuels DC, Elson J, Turnbull DM. Accumulation of mitochondrial DNA mutations in ageing, cancer, and mitochondrial disease: is there a common mechanism? Lancet. 2002;360:1323–1325. doi: 10.1016/S0140-6736(02)11310-9. [DOI] [PubMed] [Google Scholar]
- 8.Elson JL, Samuels DC, Turnbull DM, Chinnery PF. Random intracellular drift explains the clonal expansion of mitochondrial DNA mutations with age. Am J Hum Genet. 2001;68:802–806. doi: 10.1086/318801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coller HA, Khrapko K, Bodyak ND, Nekhaeva E, Herrero-Jimenez P, Thilly WG. High frequency of homoplasmic mitochondrial DNA mutations in human tumors can be explained without selection. Nat Genet. 2001;28:147–150. doi: 10.1038/88859. [DOI] [PubMed] [Google Scholar]
- 10.Taylor RW, Barron MJ, Borthwick GM, et al. Mitochondrial DNA mutations in human colonic crypt stem cells. J Clin Invest. 2003;112:1351–1360. doi: 10.1172/JCI19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones JB, Song JJ, Hempen PM, Parmigiani G, Hruban RH, Kern SE. Detection of mitochondrial DNA mutations in pancreatic cancer offers a “mass”-ive advantage over detection of nuclear DNA mutations. Cancer Res. 2001;61:1299–1304. [PubMed] [Google Scholar]
- 12.Ivanova R, Lepage V, Loste MN, et al. Mitochondrial DNA sequence variation in human leukemic cells. Int J Cancer. 1998;76:495–498. doi: 10.1002/(sici)1097-0215(19980518)76:4<495::aid-ijc9>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 13.Carew JS, Zhou Y, Albitar M, Carew JD, Keating MJ, Huang P. Mitochondrial DNA mutations in primary leukemia cells after chemotherapy: clinical significance and therapeutic implications. Leukemia. 2003;17:1437–1447. doi: 10.1038/sj.leu.2403043. [DOI] [PubMed] [Google Scholar]
- 14.Grist SA, Lu XJ, Morley AA. Mitochondrial mutations in acute leukaemia. Leukemia. 2004;18:1313–1316. doi: 10.1038/sj.leu.2403380. [DOI] [PubMed] [Google Scholar]
- 15.He L, Luo L, Proctor SJ, et al. Somatic mitochondrial DNA mutations in adult-onset leukaemia. Leukemia. 2003;17:2487–2491. doi: 10.1038/sj.leu.2403146. [DOI] [PubMed] [Google Scholar]
- 16.Linnartz B, Anglmayer R, Zanssen S. Comprehensive scanning of somatic mitochondrial DNA alterations in acute leukemia developing from myelodysplastic syndromes. Cancer Res. 2004;64:1966–1971. doi: 10.1158/0008-5472.can-03-2956. [DOI] [PubMed] [Google Scholar]
- 17.Boultwood J, Fidler C, Mills KI, et al. Amplification of mitochondrial DNA in acute myeloid leukaemia. Br J Haematol. 1996;95:426–431. doi: 10.1046/j.1365-2141.1996.d01-1922.x. [DOI] [PubMed] [Google Scholar]
- 18.Jia L, Liu KZ, Newland AC, Mantsch HH, Kelsey SM. Pgp-positive leukaemic cells have increased mtDNA but no increased rate of proliferation. Br J Haematol. 1999;107:861–869. doi: 10.1046/j.1365-2141.1999.01771.x. [DOI] [PubMed] [Google Scholar]
- 19.Shin MG, Kajigaya S, McCoy JP, Jr, Levin BC, Young NS. Marked mitochondrial DNA sequence heterogeneity in single CD34+ cell clones from normal adult bone marrow. Blood. 2004;103:553–561. doi: 10.1182/blood-2003-05-1724. [DOI] [PubMed] [Google Scholar]
- 20.Shin MG, Kajigaya S, Tarnowka M, McCoy JP, Jr, Levin BC, Young NS. Mitochondrial DNA sequence heterogeneity in circulating normal human CD34 cells and granulocytes. Blood. 2004;103:4466–4477. doi: 10.1182/blood-2003-11-3949. [DOI] [PubMed] [Google Scholar]
- 21.Ogasawara Y, Nakayama K, Tarnowka M, et al. Mitochondrial DNA spectra in single CD34+ cells, T-cells, B-cells and granulocytes. Blood. 2005;106:3271–3284. doi: 10.1182/blood-2005-01-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA [letter]. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 23.Yao Y-G, Bravi CM, Bandelt H-J. A call for mtDNA data quality control in forensic science. Forensic Sci Int. 2004;141:1–6. doi: 10.1016/j.forsciint.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Bandelt HJ, Quintana-Murci L, Salas A, Macaulay V. The fingerprint of phantom mutations in mitochondrial DNA data. Am J Hum Genet. 2002;71:1150–1160. doi: 10.1086/344397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bandelt H-J, Herrnstadt C, Yao Y-G, et al. Identification of Native American founder mtDNAs through the analysis of complete mtDNA sequences: some caveats. Ann Hum Genet. 2003;67:512–524. doi: 10.1046/j.1469-1809.2003.00049.x. [DOI] [PubMed] [Google Scholar]
- 26.Palanichamy MG, Sun C, Agrawal S, et al. Phylogeny of mitochondrial DNA macrohaplogroup N in India, based on complete sequencing: implications for the peopling of South Asia. Am J Hum Genet. 2004;75:966–978. doi: 10.1086/425871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong Q-P, Yao Y-G, Sun C, Bandelt H-J, Zhu C-L, Zhang Y-P. Phylogeny of east Asian mitochondrial DNA lineages inferred from complete sequences. Am J Hum Genet. 2003;73:671–676. doi: 10.1086/377718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salas A, Richards M, Lareu MV, et al. The African diaspora: mitochondrial DNA and the Atlantic slave trade. Am J Hum Genet. 2004;74:454–465. doi: 10.1086/382194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reidla M, Kivisild T, Metspalu E, et al. Origin and diffusion of mtDNA haplogroup X. Am J Hum Genet. 2003;73:1178–1190. doi: 10.1086/379380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez-Cespedes M, Parrella P, Nomoto S, et al. Identification of a mononucleotide repeat as a major target for mitochondrial DNA alterations in human tumors. Cancer Res. 2001;61:7015–7019. [PubMed] [Google Scholar]
- 31.Ha PK, Tong BC, Westra WH, et al. Mitochondrial C-tract alteration in premalignant lesions of the head and neck: a marker for progression and clonal proliferation. Clin Cancer Res. 2002;8:2260–2265. [PubMed] [Google Scholar]
- 32.Gattermann N. Mitochondrial DNA mutations in the hematopoietic system. Leukemia. 2004;18:18–22. doi: 10.1038/sj.leu.2403209. [DOI] [PubMed] [Google Scholar]
- 33.Vega A, Salas A, Gamborino E, Sobrido MJ, Macaulay V, Carracedo Á. mtDNA mutations in tumors of the central nervous system reflect the neutral evolution of mtDNA in populations. Oncogene. 2004;23:1314–1320. doi: 10.1038/sj.onc.1207214. [DOI] [PubMed] [Google Scholar]
- 34.Greaves LC, Preston SL, Tadrous PJ, et al. Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc Natl Acad Sci U S A. 2006;103:714–719. doi: 10.1073/pnas.0505903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meierhofer D, Ebner S, Mayr JA, Jones ND, Kofler B, Sperl W. Platelet transfusion can mimic somatic mtDNA mutations. Leukemia. 2006;20:362–363. doi: 10.1038/sj.leu.2404070. [DOI] [PubMed] [Google Scholar]
- 36.Brandstätter A, Sanger T, Lutz-Bonengel S, et al. Phantom mutation hotspots in human mitochondrial DNA. Electrophoresis. 2005;26:3414–3429. doi: 10.1002/elps.200500307. [DOI] [PubMed] [Google Scholar]
- 37.Malyarchuk BA, Rogozin IB, Berikov VB, Derenko MV. Analysis of phylogenetically reconstructed mutational spectra in human mitochondrial DNA control region. Hum Genet. 2002;111:46–53. doi: 10.1007/s00439-002-0740-4. [DOI] [PubMed] [Google Scholar]
- 38.Stoneking M. Hypervariable sites in the mtDNA control region are mutational hotspots. Am J Hum Genet. 2000;67:1029–1032. doi: 10.1086/303092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michikawa Y, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G. Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science. 1999;286:774–779. doi: 10.1126/science.286.5440.774. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Michikawa Y, Mallidis C, et al. Muscle-specific mutations accumulate with aging in critical human mtDNA control sites for replication. Proc Natl Acad Sci U S A. 2001;98:4022–4027. doi: 10.1073/pnas.061013598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Asin-Cayuela J, Fish J, et al. Strikingly higher frequency in centenarians and twins of mtDNA mutation causing remodeling of replication origin in leukocytes. Proc Natl Acad Sci U S A. 2003;100:1116–1121. doi: 10.1073/pnas.242719399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coskun PE, Beal MF, Wallace DC. Alzheimer's brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc Natl Acad Sci U S A. 2004;101:10726–10731. doi: 10.1073/pnas.0403649101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nekhaeva E, Bodyak ND, Kraytsberg Y, et al. Clonally expanded mtDNA point mutations are abundant in individual cells of human tissues. Proc Natl Acad Sci U S A. 2002;99:5521–5526. doi: 10.1073/pnas.072670199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cantuti-Castelvetri I, Lin MT, Zheng K, et al. Somatic mitochondrial DNA mutations in single neurons and glia. Neurobiol Aging. 2005;26:1343–1355. doi: 10.1016/j.neurobiolaging.2004.11.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.