Abstract
Eukaryotic cells distinguish their chromosome ends from accidental DNA double-strand breaks by packaging them in a protective structure referred to as the telomere “cap.” Here we investigate the nature of the telomere cap by examining events at DNA breaks generated adjacent to either natural telomeric sequences (TG repeats) or arrays of Rap1-binding sites that vary in length. Although DNA breaks adjacent to either short or long telomeric sequences are efficiently converted into stable telomeres, they elicit very different initial responses. Short telomeric sequences (80 base pair [bp]) are avidly bound by Mre11, as well as the telomere capping protein Cdc13 and telomerase enzyme, consistent with their rapid telomerase-dependent elongation. Surprisingly, little or no Mre11 binding is detected at long telomere tracts (250 bp), and this is correlated with reduced Cdc13 and telomerase binding. Consistent with these observations, ends with long telomere tracts undergo strongly reduced exonucleolytic resection and display limited binding by both Rpa1 and Mec1, suggesting that they fail to elicit a checkpoint response. Rap1 binding is required for end concealment at long tracts, but Rif proteins, yKu, and Cdc13 are not. These results shed light on the nature of the telomere cap and mechanisms that regulate telomerase access at chromosome ends.
Keywords: DNA double-strand break, telomere capping, telomerase, DNA damage checkpoint, telomere length regulation
The ends of linear eukaryotic chromosomes require special mechanisms to assure their complete replication (Lingner et al. 1995) and to prevent them from being either degraded or joined with other telomeres or accidental DNA breaks (for review, see Smogorzewska and De Lange 2004). An elaborate and dynamic protein–DNA complex referred to as the telomere has thus evolved to cope with these problems. In most eukaryotes, chromosome end replication is assisted by the telomerase enzyme, which can synthesize de novo TG-rich repeat sequences at chromosome 3′ ends (Greider and Blackburn 1985), thus reversing the loss of information resulting from the 5′ exonucleolytic degradation (5′ end resection) observed at telomeres in all organisms examined to date. How telomerase action is regulated so as to bring about a fixed average TG-tract length at telomeres is still unknown. The second problem, end protection or “capping,” is resolved by proteins that bind to telomeres through mechanisms that are still poorly understood. In mammalian cells, capping requires a protein complex called “shelterin” (de Lange 2005), which may act by helping to form a protective structure, the t-loop, wherein the chromosome 3′ single-stranded terminus is buried in a more internal telomere repeat sequence (Griffith et al. 1999). In budding yeast, capping requires a protein complex consisting of Cdc13, Stn1, and Ten1 that localizes to the 3′ TG-rich single-stranded DNA at telomeres by virtue of Cdc13 DNA binding (Bertuch and Lundblad 2006). The loss of Cdc13 function in the temperature-sensitive cdc13-1 mutant results in rapid 5′ end resection, activation of the DNA damage checkpoint, and a disruption of telomere structure (Weinert and Hartwell 1993; Garvik et al. 1995).
Both telomerase action and telomere capping are tightly linked to the length of the TG-repeat tract at individual telomeres. [In budding yeast, telomeric repeats take the form G2–3(TG)1–6, but will be referred to hereafter simply as “TG repeats.”] Duplex TG-repeat sequences are bound by specific proteins (Rap1 in budding yeast, Taz1 in fission yeast, and Trf1/Trf2 in mammals) whose amount at the telomere is thought to increase with TG length. In several systems evidence has been presented indicating the involvement of TG-repeat-binding proteins in a negative feedback mechanism controlling telomerase recruitment and/or action in cis at the telomere (Marcand et al. 1997; van Steensel and de Lange 1997; Ray and Runge 1999). Consistent with this model, a recent study that examined individual telomere elongation events provided direct evidence that telomerase acts preferentially at short telomeric tracts (Teixeira et al. 2004). Interestingly, the block to telomerase action at long ends requires the Rif1 protein (Teixeira et al. 2004), which, together with Rif2, is recruited to telomeres through an interaction with the Rap1 C terminus, where it acts as a negative regulator of telomerase action (Hardy et al. 1992; Wotton and Shore 1997). However, the mechanism(s) by which the Rap1 C terminus and its interacting Rif proteins regulate telomerase action is still unclear.
Studies employing chromatin immunoprecipitation (ChIP) have revealed that telomerase access to the chromosome end is indeed highly regulated, with simultaneous binding of telomerase enzyme (Est2) and the essential holoenzyme subunit Est1 occurring only in late S phase (Taggart et al. 2002; Schramke et al. 2004), when telomere replication is known to occur (Marcand et al. 2000). Est1/Est2 recruitment depends critically on an interaction between Est1 and Cdc13 (Evans and Lundblad 1999; Pennock et al. 2001; Bianchi et al. 2004), the latter also displaying an S-phase-specific peak of telomere binding (Taggart et al. 2002; Schramke et al. 2004). Moving one step further back, the resection of the 5′ end of the telomere required to generate a Cdc13-binding site is promoted by the Mre11/Rad50/Xrs2 (MRX) complex, which is recruited to telomeres in S phase, presumably following completion of conventional DNA replication (Larrivee et al. 2004; Takata et al. 2005). So far, however, ChIP experiments have been unable to reveal which of these steps in telomere replication, if any, are regulated by TG-tract length.
Short telomere tracts, generated either by the loss of telomerase or the loss of factors required for normal telomerase action, have reduced capping function and are thus more prone to engage in either homologous recombination (HR) or nonhomologous end-joining (NHEJ) reactions (DuBois et al. 2002; Mieczkowski et al. 2003). Similarly, inactivation of Cdc13 or its associated proteins Stn1 and Ten1 leads to a loss of capping (Garvik et al. 1995; Grandin et al. 1997; DuBois et al. 2002), as does inactivation of the duplex TG-repeat-binding protein Rap1 (Pardo and Marcand 2005) or the combined loss of Est2 and Tel1 (Chan and Blackburn 2003). Interestingly, in the case of Rap1 inactivation, telomere–telomere fusions occur even when TG-tract length is at or near normal levels (Pardo and Marcand 2005). Thus, several different factors contribute to generating a capped structure at telomeric TG-repeat tracts in yeast. At present, the precise nature of the cap structure is unknown, as is the minimal TG-repeat length required for its generation.
A paradoxical feature of telomeres is the presence of several proteins that are also found at accidental double-strand breaks (DSBs), where they promote recombinational repair (NHEJ or HR), processes that are blocked at capped telomeres. For example, the highly conserved MRX complex is rapidly recruited to DSBs in yeast (Lisby et al. 2004; Shroff et al. 2004), where it promotes either NHEJ, together with yKu70/80 and Lig4, or 5′ end resection and HR, depending on whether cyclin-dependent kinase (CDK) activity is low or high (Ira et al. 2004; Garber et al. 2005). How the action of MRX and yKu at telomeres is controlled so as to promote telomerase action when appropriate, and to prevent at all costs NHEJ and HR, is still a major outstanding question.
One impediment to understanding the molecular mechanisms underlying telomere capping and length regulation is the heterogeneous length of TG tracts at native telomeres. Hence, possible differences between telomeres with long or short TG tracts will be obscured by experiments that measure a population average. To bypass this problem, we have turned to a simplified system in which a telomere is generated by inducing a DSB, using a galactose-regulated version of the endogenous HO endonuclease gene, adjacent to a “seed” of native TG-repeat sequence or arrays of Rap1-binding sites (Diede and Gottschling 1999; Grossi et al. 2001). Because all ends in this case have a uniform TG-tract length (or fixed number of Rap1-binding sites) just after generation of the DSB, one can use this system to examine the effect of telomeric tract length on the processing of the break site. DSB sites with as few as approximately four Rap1-binding sites (80 base pair [bp] of native TG repeat) are very efficiently “healed” to produce novel telomeres (Diede and Gottschling 1999; Grossi et al. 2001), suggesting that this system might possess key features of native telomeres. Indeed, we show that short TG tracts are rapidly elongated by telomerase following cutting by HO, whereas tracts equivalent to average native telomere length are not—a property recently demonstrated for native telomeres (Teixeira et al. 2004). Using ChIP to examine the binding of various proteins at the break sites, we find remarkable differences between the molecular response to short versus long TG-tract or Rap1-binding site arrays. Whereas short telomeric tracts behave similarly to nontelomeric ends, apart from their ability to recruit Cdc13 and telomerase, long telomeric tracts are notably inert. These differences are discussed in relationship to possible mechanisms of telomere capping and telomere length regulation.
Results
Telomerase recruitment occurs preferentially at DSBs with short TG tracts
A current model for telomere length regulation postulates that telomerase action is controlled by a negative feedback mechanism that senses telomere (TG repeat) length (for review, see Shore 2001). According to one specific version of this model, telomerase recruitment is the key regulatory step that is favored at short telomeres, but reduced or eliminated at ends with TG repeats equal to or longer than the population average. To test this idea we turned to a simplified model system in which native telomeric TG-repeat sequences of different lengths are placed immediately adjacent to a recognition site for the endogenous HO endonuclease (Fig. 1A). As shown previously by us and others (Diede and Gottschling 1999; Bianchi et al. 2004), galactose induction of HO endonuclease and generation of a DNA DSB adjacent to a short (80-bp) tract of telomeric TG-repeat sequence (TG-80) results in rapid and efficient telomerase-dependent elongation of the break. Elongation of the TG-containing DSB is associated with Cdc13-dependent recruitment of the telomerase accessory factor Est1, as well as the telomerase enzyme itself, as measured by ChIP (Bianchi et al. 2004). In contrast, non-TG-containing breaks do not recruit telomerase efficiently and are not elongated, but are instead either repaired by recombination (NHEJ or HR) or break-induced replication (BIR) (McEachern and Haber 2006), or degraded (Sandell and Zakian 1993). These observations suggest that a DSB that terminates with a telomere repeat sequence is recognized by the cell as a telomere, and thus might serve as a model to understand how the cell regulates telomerase action as a function of TG repeat length.
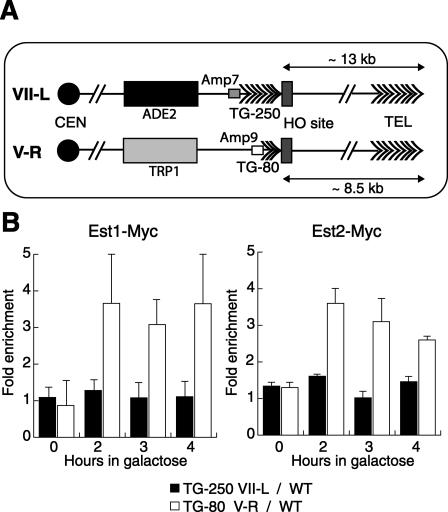
Figure 1.
Preferential telomerase recruitment at short TG tracts. (A) Schematic representation of the modified subtelomeric regions of Chr. VII-L and Chr. V-R. (B) Analysis by ChIP of the binding of Est1-Myc and Est2-Myc after galactose induction of a DSB. See Materials and Methods for details.
We began by comparing the behavior of a TG-80 break site to one containing 250 bp of telomere repeats (TG-250), which is essentially equivalent in length to a native telomere. The two TG tracts are present in the same cell, but on different chromosomes, as indicated in Figure 1A. This allows for a direct and quantitative comparison between the two DSBs by ChIP, where each break site is compared with the same internal control sequence. As shown previously (Bianchi et al. 2004), we detected specific recruitment of both Est1 and Est2 at the TG-80 break site by 2 h following HO induction (Fig. 1B). However, neither protein appeared to accumulate at the TG-250 break site at any point during the 4 h following HO induction (Fig. 1B). Consistent with the apparent absence of both telomerase subunits at the 250-bp tract, we observed no elongation (or shortening) of this end, as measured by Southern blot, during this 4-h period (Supplementary Figure S1, left panel). In contrast, and as reported previously, the TG-80-containing break was actively elongated during this time course (Supplementary Figure S1, middle panel). Although the TG-250 break appeared to be unreactive even by 4 h after HO induction, it later took on the appearance of a normal telomere, with a heterogeneous TG-repeat length centered ∼250–300 bp, as did the end beginning as an 80-bp TG-repeat tract (data not shown). Furthermore, cells containing TG-250 at a break site formed stable telomeres following HO induction at a similar frequency to TG-80 cells, as judged by a genetic assay that monitors this event (Supplementary Table S1). This suggests that the TG-250 break, like the TG-80 one, is recognized initially by the cell as a telomere. Significantly, however, the TG-250 break appears to actively block telomerase association, in comparison with the TG-80 site.
End processing and recruitment of Cdc13 and Mre11 are reduced at long TG tracts
We next considered the possibility that reduced telomerase association at the longer TG-250 site might be due to a block at some step prior to telomerase recruitment. As mentioned above, Cdc13 is required for normal telomerase recruitment both at native telomeres and at a TG-80 DSB site. We therefore used the ChIP assay to compare Cdc13 recruitment at short and long TG repeats. As shown in Figure 2A, there is indeed a dramatic (∼25-fold) reduction in Cdc13 recruitment at the long TG-250 site, compared with the active TG-80 array. Increased Cdc13 binding at the TG-80 site compared with the longer array is not a consequence of telomerase action at the former (which will create additional single-stranded TG-rich DNA for Cdc13 binding) since this difference is still largely maintained in cells carrying a point mutation in telomerase (est2-D670A) that abolishes its activity (Fig. 2A).
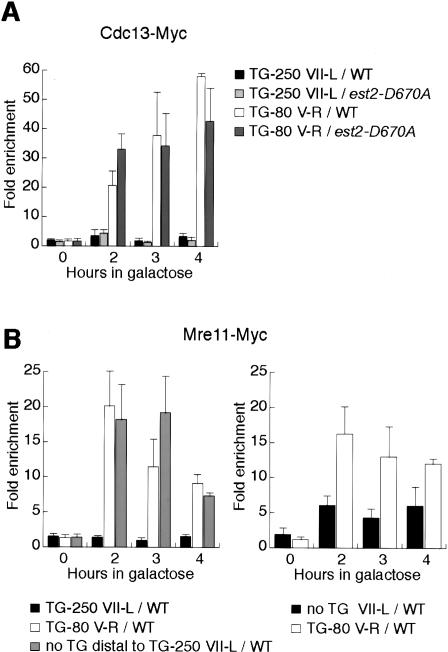
Figure 2.
Recruitment of Cdc13 and Mre11 is strongly reduced at long TG tracts. Analysis by ChIP of the binding of Cdc13-Myc in wild-type and est2-D670A strains (A) and of Mre11-Myc (B) in wild-type strains after galactose induction of a DSB.
Because the MRX complex is implicated in the generation of a 3′ TG-rich single-strand overhang at telomeres (Larrivee et al. 2004; Takata et al. 2005), and had previously been shown to be required for full Cdc13 binding at a TG-80-containing DSB (Diede and Gottschling 2001), we next compared Mre11 binding at the short and long TG-tract sites. Strikingly, we observed that the TG-250 break displayed markedly reduced Mre11 binding, compared with either TG-80 breaks (Fig. 2B, left panel) or breaks containing no TG at all (Fig. 2B, right panel). Again, the quantitative difference in Mre11 binding between short and long TG repeats, like that of Cdc13, was dramatic. Significantly, we found that the distal (non-TG) side of the TG-250 DSB contains normal amounts of Mre11 (Fig. 2B, left panel), comparable to a break lacking TG on either side, or to the TG-80 DSB.
To determine whether the reduced binding of both Cdc13 and Mre11 at the TG-250 end might be an artifact due to the greater distance between this end and the PCR amplicon (probe), compared with the TG-80 construct, we generated strains in which the probe-end distance for a TG-80 tract was increased by insertion of a 200-bp “spacer” sequence, and thus became slightly longer than that for the TG-250 construct. We then compared the binding of both Cdc13 and Mre11 at this modified TG-80 break on Chromosome (Chr.) VII-L to the original TG-80 break at Chr. V-R, in the same strain. Significantly, the quantitative binding of both proteins at the different ends was remarkably similar (Supplementary Figure S2), thus ruling out the possibility that the TG-80/TG-250 differences we observed were due to different probe-end distances. To test whether the TG-80 end might exert an effect in trans on binding at a TG-250 end, we examined both Cdc13 and Mre11 binding in cells carrying only a single break at a TG-250 site. Binding of both proteins was also low under these conditions (Supplementary Figure S3), thus ruling out an indirect effect of the TG-80 DSB on binding at the TG-250 break. Finally, we asked whether cell cycle- or checkpoint-dependent events, perhaps triggered by cleavage at one site prior to cleavage at the second, might underlie the differences we observed. Since in all the experiments described above cells display a uniform G2/M arrest ∼1–2 h following HO induction (data not shown), we addressed this issue by first blocking the cells in G2/M, by treatment with nocodozole, before HO induction. Even in cells blocked in G2/M throughout the course of the experiment, we observed the same large difference in both Cdc13 and Mre11 binding at the long versus short TG tracts (Supplementary Figure S4). Taken together, the observations described above suggest that some feature of the long TG tract is able to restrict the binding of Cdc13 and Mre11 to the break in cis, without affecting the other end and in the absence of any influence exerted by a second break site. The ChIP assay is unable to determine whether or not binding is completely restricted from the TG-250 tract, though the data do suggest that binding of both proteins is either severely reduced in amount, limited to a small fraction of the total number of ends, or both.
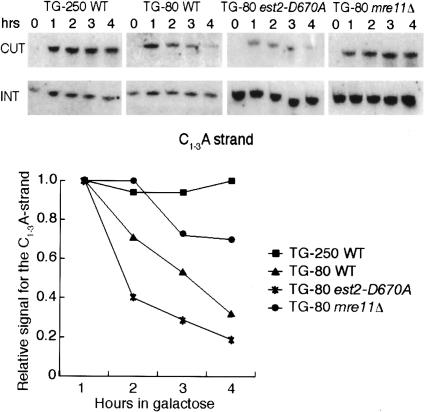
Reduced or abolished Mre11 binding at the long TG tract suggests that this end might not be subject to 5′ end resection. We tested this notion using a sensitive Southern blotting assay (Diede and Gottschling 2001) in which a riboprobe generated from the Dbp amplicon 7 was used as an indirect end label to follow the integrity of the TG strand (see Materials and Methods for details). In the 4 h following HO induction, the 5′ end of the TG-250 tract appears remarkably stable, whereas the same end at the TG-80 tract, as expected, disappears with a half-life of ∼2 h (Fig. 3). This disappearance occurs even in cells carrying a catalytically dead allele of telomerase (est2-D670A) and thus cannot be a consequence of telomerase addition and subsequent CA-strand fill-in (Fig. 3). We also confirmed that resection of the short TG tract is MRE11 dependent, since it is strongly reduced in an mre11Δ strain (Fig. 3). We conclude that the absence of resection at the long TG tract is consistent with, and likely a consequence of, its reduced Mre11 binding. It is worth noting that our experiments, as well as those reported previously (Diede and Gottschling 2001), fail to detect resection intermediates, presumably because they are too heterogeneous in length. Instead, we measure the initiation of resection, an event that is itself regulated at non-TG-containing breaks (Frank-Vaillant and Marcand 2002). Our data thus indicate that long TG tracts exert an additional block to the initiation of 5′ end resection.
Figure 3.
5′ end resection is inhibited at long TG arrays. (Top panel) Southern blots monitoring resection of the C1–3A strand at TG-250 in a wild-type strain and TG-80 repeats in wild-type, est2-D670A, and mre11-Δ strains. (INT) The internal loading control (band of 165 bp specific for a region on Chr. XV); (CUT) a band of 418 bp or 239 bp, corresponding to the C1–3A strand of the long or short telomeric tracts, respectively. (Bottom panel) Quantification of the relative signal for the C1–3A strand obtained by measuring the amount of the “CUT” band relative to the internal control “INT,” normalized to the efficiency of cleavage.
Inhibition of protein binding at long TG tracts requires Rap1 binding but not its C-terminal regulatory domain
The suppression of both Mre11 and Cdc13 binding at the TG-250 end would seem to explain why it fails to bind telomerase and elongate, and might therefore reflect a mechanism for length regulation of native telomeres. We therefore tested whether decreased Mre11 and Cdc13 binding at the TG-250 end is due to the Rap1 C terminus and/or the Rif1 and Rif2 proteins, which are known to act in cis as negative regulators of telomere elongation (Hardy et al. 1992; Wotton and Shore 1997). Cells carrying a deletion of the Rap1 C terminus, or null mutations in either RIF1 or RIF2, have overelongated telomeres, suggesting that the TG-250 array might be an active site for telomerase action (and presumably Mre11 and Cdc13 binding) in these cells.
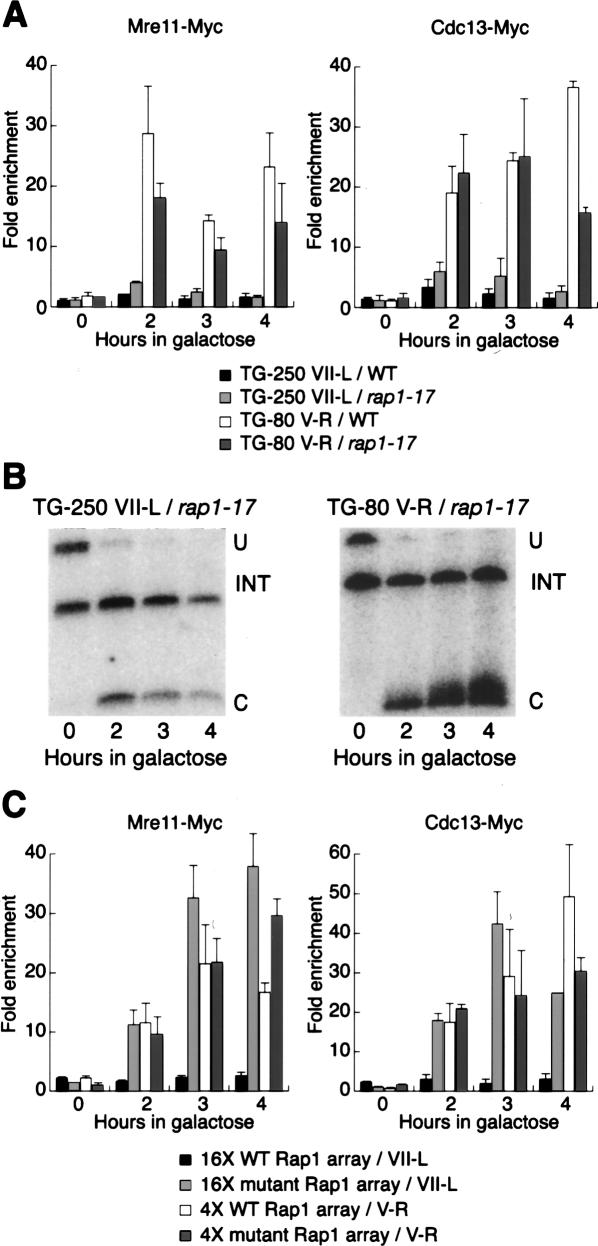
Unexpectedly, we found that recruitment of both Mre11 and Cdc13 is still strongly suppressed at the TG-250 break in a strain carrying a Rap1 C-terminal truncation mutation (rap1-17) (Fig. 4A). Similarly, deletion of neither RIF1 nor RIF2 had any significant effect on the behavior of the long TG tract (data not shown). Although the TG-250 tract eventually elongates due to the rap1-17 mutation (data not shown), we found no evidence for elongation during the first 4 h following induction of the break (Fig. 4B, left panel). Thus, the initial response to the exposure of the TG-250 tract does not appear to be regulated by either the Rap1 C terminus or the Rif proteins.
Figure 4.
Multiple Rap1 binding suppresses Mre11 and Cdc13 recruitment at long TG tracts. (A) Analysis by ChIP of the binding of Mre11-Myc and Cdc13-Myc after galactose induction of a DSB in wild-type and rap1-17 strains. (B) Southern blots monitoring cleavage at the HO site in a rap1-17 strain. (INT) The internal loading control; (U) an uncut fragment; (C) the fragment resulting from “U” after induction of the HO cut. (C) Analysis by ChIP of the binding of Mre11-Myc and Cdc13-Myc after galactose induction of a DSB adjacent to 16× and 4× wild-type and mutant Rap1 site arrays.
We thus decided to ask whether Rap1 binding itself is required to inhibit both Mre11 and Cdc13 binding at the TG-250 end. Because RAP1 is an essential gene, we were unable to perform a straightforward test of a loss-of-function mutation. Instead, we used “synthetic” arrays of Rap1-binding sites adjacent to an HO cut site, and compared them with similar arrays in which a single critical base pair within each binding site was mutated (Grossi et al. 2001). We found that synthetic arrays of four and 16 Rap1-binding sites, corresponding roughly to the TG-80 and TG-250 constructs in terms of the number of Rap1-binding sites, responded similarly to their native counterparts with respect to Cdc13 and Mre11 binding (Fig. 4C). As expected, the 4× Rap1 site array showed robust elongation during the 4 h following HO induction, whereas the 16× Rap1 site array appeared unchanged during this period, as we had observed for the corresponding native arrays (data not shown). Whereas the single point mutation in each of the binding sites of the 4× Rap1 site array had relatively little effect on either Mre11 or Cdc13 binding, the same point mutations in the context of the 16× Rap1 site array completely relieved the inhibition of both Mre11 and Cdc13 binding, such that the 4× and 16× mutant arrays behaved similarly to each other and to the wild-type 4× array, in terms of both protein binding and elongation (Fig. 4C; data not shown). We confirmed by ChIP that the mutant arrays, as expected, bind little if any Rap1 (data not shown). These results suggest that the ability to bind Rap1 is an essential feature that allows the long TG arrays to suppress both Mre11 and Cdc13 binding.
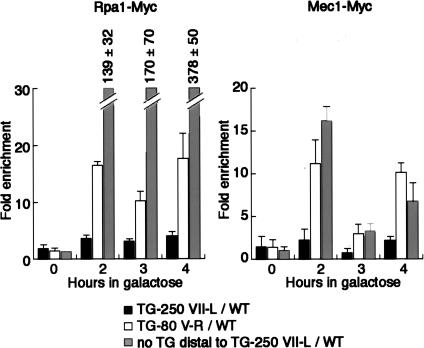
Long TG tracts fail to elicit a strong checkpoint response
DSBs initiate a checkpoint response through the activation of a defined signaling pathway that leads to cell cycle arrest and induction of genes involved in repair of the damage (Zhou and Elledge 2000). In yeast, the major checkpoint response to DSBs is initiated by the ATR-like kinase Mec1, which binds together with an associated protein (Ddc2) to 5′ resected, single-stranded DNA ends that are coated with RPA (Garber et al. 2005). Given the refractory nature of ends containing long TG tracts or Rap1 arrays documented above, we asked whether or not they are recognized as DNA damage by measuring their association with both Rpa1 and Mec1. As shown in Figure 5, we detected near-background levels of both Rpa1 and Mec1 at the Chr. VII-L TG-250 break site, whereas the distal end of this DSB, which does not contain TG repeats, was bound strongly by both proteins. We also detected robust binding of both Rpa1 and Mec1 at the Chr. V-R TG-80 break site, consistent with the binding of Mre11 to this site and its resection. These data suggest that breaks with long telomeric TG tracts or Rap1 site arrays may be unable to initiate a checkpoint response due to an inability to recruit the Mec1 kinase.
Figure 5.
Long TG tracts fail to induce a strong checkpoint response. Analysis by ChIP of the binding of Rpa1-Myc and Mec1-Myc after galactose induction of a DSB in wild-type strains.
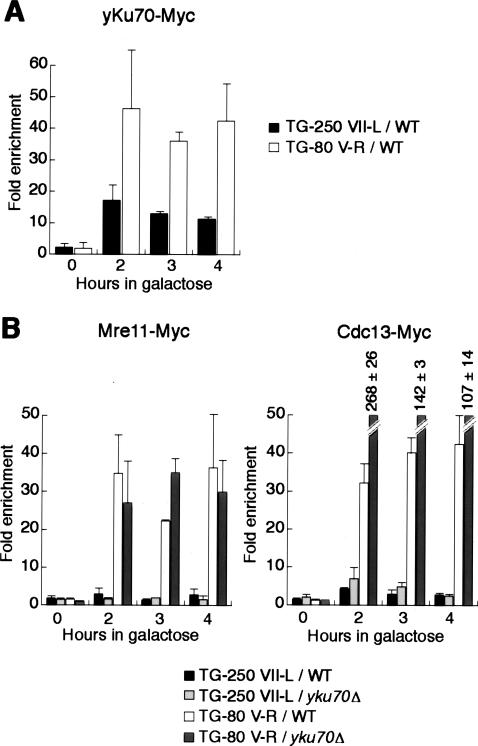
yKu binds at long TG breaks but does not block their resection
We next tested binding of yKu, a conserved heterodimeric protein known to bind to both DSBs and telomeres and to play a role in telomere capping (Fisher and Zakian 2005). Despite the absence or restriction to binding of all other proteins tested at long TG-tract break sites, we found clear evidence for yKu70 binding at such sites, at levels only slightly lower than those detected at short (TG-80) sites (Fig. 6A). To determine what function yKu might have at the longer TG-break sites, we next examined Cdc13 and Mre11 binding in yku70Δ cells. Somewhat surprisingly, we found no effect of the yku70Δ mutation on Mre11 binding at either long or short TG tracts (Fig. 6B, left panel). Furthermore, Cdc13 binding at a TG-250 site was also unaffected by the loss of yKu, even though the shorter TG-80 site experienced a dramatic increase in Cdc13 binding in the mutant (Fig. 6B, right panel), as reported previously (Fisher et al. 2004). These data indicate that although yKu is bound at long TG-tract sites, it is not required to prevent these ends from being recognized by Mre11. We also presume that yKu is not involved in the blockage of resection at these ends.
Figure 6.
Binding of yKu at long TG tracts does not inhibit resection. Analysis by ChIP after galactose induction of a DSB of the binding of yKu70-Myc in a wild-type strain (A) and of the binding of Mre11-Myc and Cdc13-Myc in wild-type and yku70-Δ strains (B).
A limited role for Cdc13 in ‘capping’ long TG tract ends
Although we clearly observed reduced binding of Cdc13 (and Mre11) at long TG or Rap1 site tracts compared with shorter ones, binding was consistently detected above background by the ChIP assay. It thus seems possible that some fraction of the long TG-tract ends are bound by at least one Cdc13 molecule, raising the possibility that Cdc13, in the long TG-tract context, is involved in masking these ends from resection and subsequent RPA binding and Mec1 recruitment.
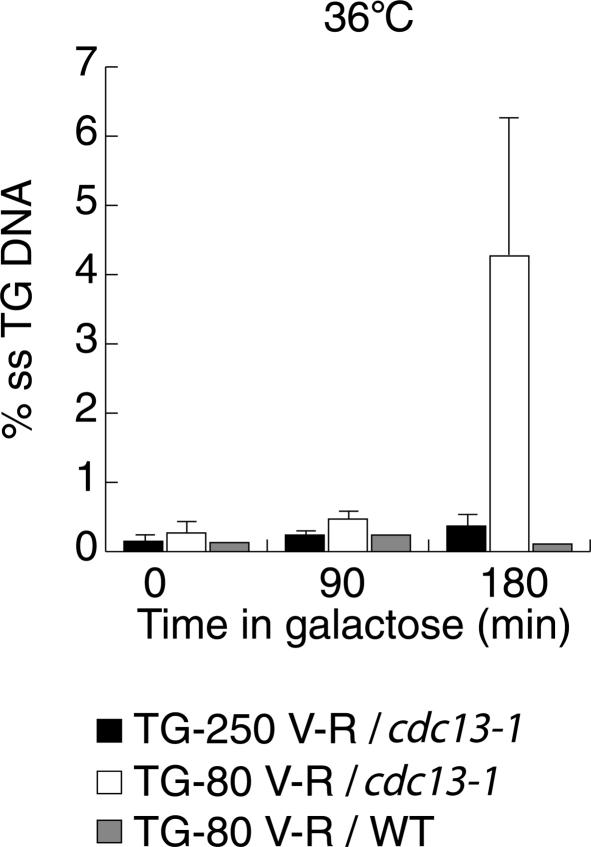
To test this idea, we directly monitored 5′ end resection, using the QAOS (quantitative amplification of single-stranded DNA) assay (Booth et al. 2001), at both TG-80 and TG-250 breaks, in cdc13-1 cells held at the nonpermissive temperature (36°C) (see Materials and Methods for details). As shown in Figure 7, the TG-80 break at Chr. V-R displays considerable single-stranded DNA (3%–6%) at a site >1.1 kb internal to the break. This value is similar to that observed at this site in native Chr. V-R under these conditions (Booth et al. 2001; A. Puglisi, unpubl.). Strikingly, however, single-stranded DNA internal to the TG-250 end is considerably (>10-fold) reduced compared with the TG-80 end (Fig. 7). As expected, no resection is detected at the TG-80 break in wild-type (CDC13+) cells at 36°C, indicating that the effect observed in mutant cells is in fact due to the loss of Cdc13 function.
Figure 7.
Limited role for Cdc13 in capping long TG-tract ends. Detection of single-stranded DNA at the YER188W locus in cdc13-1 mutant or CDC13+ cells following HO cutting adjacent to either TG-80 or TG-250 tracts (as indicated) at Chr. V-R. (YER188W is 1130 or 1309 bp centromere-proximal to HO cut site at the short or long TG tracts, respectively). Cells were first grown for 3 h at 24°C in YPLG, then transferred to fresh medium containing galactose (2%) at 36°C. The QAOS assay was used to measure single-stranded DNA levels on the TG strand at the indicated times following galactose induction of the DSB. See Materials and Methods for additional details.
We have extended these observations by comparing binding of Rpa1 at a Chr. VII-L TG-250 tract to that at a TG-80 break on Chr. V-R in cdc13-1 cells held at 36°C. As predicted by the resection results described above, Rpa1 binding at the TG-250 tract is consistently lower than that observed at the TG-80 tract or at the native Chr. VI-R telomere in the cdc13-1 mutant (Supplementary Fig. S5). We also found that the cdc13-1 mutation caused only a slight increase in Mre11 binding at the TG-250 tract (data not shown). Taken together, these data suggest that Cdc13 plays only a limited role in blocking resection and checkpoint activation at a TG-250 break site or that it may act at only at a small fraction of these ends. Ends with long arrays of Rap1-binding sites would thus appear to be protected by a Cdc13-independent mechanism.
Discussion
Here we examine the molecular response in cis to the generation of DNA DSBs in which one end at the break site contains a variable number of native telomeric TG repeats or Rap1-binding sites. Our results indicate that both long TG tracts (at or near the average length of native telomeres) or comparable arrays of Rap1-binding sites behave remarkably differently from shorter arrays of the same sequences or sites containing nontelomeric sequences. Specifically, long telomeric arrays are largely refractory to binding of both Mre11 and Cdc13, proteins implicated in telomere end processing and telomerase recruitment, respectively. Consistent with these observations, we fail to detect resection of these ends or significant telomerase binding. Similarly, the telomere-like DSBs appear to bind little if any Rpa1 or Mec1, suggesting that they fail to initiate a checkpoint response. Although Rap1 binding is required for the unusual behavior of long telomere-like ends, neither its C terminus nor the interacting Rif proteins, both of which have been previously implicated in telomere length regulation, are necessary. The mechanism underlying this effect of long arrays of bound Rap1 is still unclear.
Several lines of evidence suggest that the model system described here may provide useful insights into telomere function. To begin with, both the long and short TG tracts (or Rap1 site ends) are efficiently converted into stable telomeres, indicating that at some point following cutting by HO these ends are not different from native telomeres. In addition, the short tracts are actively and efficiently elongated by telomerase whereas the longer ones are not. This is precisely what was observed in a recent study in which the elongation of individual telomeres of variable length was measured following their introduction by mating into telomerase-positive cells (Teixeira et al. 2004). These authors found that the probability of elongation increases dramatically as TG-tract length decreases below 200 bp, consistent with our observation that TG-80 ends are efficiently and rapid elongated whereas TG-250 ends are not. Finally, we find that long TG tracts or Rap1 site arrays are unlikely to provoke a DNA-damage checkpoint based on their inability to recruit significant amounts of either Rpa1 or Mec1. This absence of checkpoint activation is a property shared by full-length native telomeres, though the mechanism underlying it is not known. In contrast, telomere shortening caused by telomerase loss leads to uncapping and induction of a checkpoint response (Hackett et al. 2001; Enomoto et al. 2002). Thus, by several criteria, the TG- or Rap1 site-containing ends display features in common with native full-length telomeres.
The results presented here indicate that telomerase-mediated elongation at a DSB is negatively regulated by increasing TG-tract length at a step upstream of telomerase recruitment itself, namely at the resection step that generates a single-stranded Cdc13-binding site. We have previously shown that mutants defective in a Cdc13–Est1 interaction (cdc13-2 and est1-60) both fail to accumulate telomerase enzyme (Est2) at a TG-80-containing DSB (Bianchi et al. 2004), so it follows that a break site bound by little or no Cdc13 will also fail to accumulate either Est1 or Est2. Since Cdc13 binding itself requires single-stranded DNA, its reduced levels (or absence) at long telomeric DSB ends can be explained by the failure of these ends to undergo resection, even 4 h following induction of the break. Likewise, this absence of resection can be at least partially ascribed to reduced Mre11 binding. Our data raise the interesting possibility that short telomeres are actively resected, whereas those at or near equilibrium length may undergo either limited resection or none at all.
Although the regulation of resection and Cdc13 binding might be an initial and primary step in telomere length regulation, it is clear that at least one other mechanism must function either in parallel with or downstream from these events. Thus, although both the Rap1 C terminus and the interacting factors Rif1 and Rif2 ultimately restrict the elongation of a TG-250 tract, they do not appear to play a major role in protecting this long telomeric tract from either Mre11 or Cdc13 binding in the first few hours following HO induction. These data suggest that the telomere length regulatory pathway defined by the Rap1 C terminus and Rif proteins might have a relatively minor influence on telomerase access. These factors might increase the efficiency of the pathway dependent on Rap1 DNA binding or act through another as yet unknown mechanism. An alternative explanation for these results is that the Rap1–Rif regulatory system does not efficiently assemble at the DSB. However, the observation that Rif1 is present at the TG-250 tract even before DSB formation (S. Negrini and D. Shore, unpubl.) would argue against this idea.
The block or severe restriction of Mre11 binding at ends bound by greater than four Rap1 molecules is particularly notable given the rapid binding of Mre11 at accidental (nontelomeric) DSBs (Lisby et al. 2004; Shroff et al. 2004) and the involvement of this protein as part of the MRX complex in both DSB repair and telomere replication (D’Amours and Jackson 2002). The failure of Mre11 to accumulate at long telomeric ends cannot be due to a different global cellular response to such breaks, since a short TG-tract break in the same cell is avidly bound by Mre11. In fact, the effect of long telomeric tracts is remarkably local, since even the distal (nontelomeric) side of the break binds Mre11 strongly. The same considerations apply to both RPA and Mec1 binding at long TG tracts and contrast with the recent finding that an elongating TG-80 tract generates a signal that can counteract the DNA damage checkpoint, even at a second break site located 0.6 kb away (Michelson et al. 2005). Taken together, our results raise the possibility that native telomeres whose TG tract length is at or near the population average fail to elicit a checkpoint response because of a strict limit on their resection. This may reflect an inherent resistance of these ends to exonuclease attack, or a block to the binding of an unidentified factor that promotes resection. Yku can be ruled out as such a factor, since the long TG tracts still appear to be resistant to resection in a yku70 mutant, as judged by their failure to recruit Rpa1. This observation is also consistent with the finding that Yku inhibits, rather than promotes, resection at DSBs (Lee et al. 1998).
The absence or strong reduction of resection and Cdc13 binding at long telomeric tracts adjacent to a DSB points to a Cdc13-independent mechanism that “caps” these ends, and by extension a similar mechanism at native telomeres. Using a temperature-sensitive allele of CDC13, we showed that Cdc13 activity plays only a minor role in the blockage of 5′ exonucleolytic attack at a TG-250 break. Unless the ends generated by HO cleavage possess a capping mechanism unavailable to native telomeres, these data would suggest that the latter also can be protected by a Cdc13-independent mechanism, at least when their duplex TG-tract length is relatively long. At present it is unclear what fraction of native telomeres in any given cell become uncapped when Cdc13 is inactivated, and it is known that only one uncapped telomere is sufficient to engage the DNA damage checkpoint (Sandell and Zakian 1993).
The unusual effect of long Rap1 arrays described here points to a highly cooperative process, rather than simple steric hinderance by the Rap1 molecule bound closest to the DSB end. Several explanations can be considered. One possibility is that DNA bending elicited by Rap1 binding (Muller et al. 1994) promotes a looped structure at long telomeric tracts. Interestingly, in vitro studies indicate that Rap1 binding to duplex DNA can promote its association with single-stranded telomeric DNA (Gilson et al. 1994). Taken together with our results, these observations raise the possibility that long arrays of Rap1 favor the formation of a t-loop structure similar to that observed in many other organisms (de Lange 2005) that may physically hide the DSB end from MRX and/or exonucleases. The apparent absence of resection at the long end, though, would seem to argue against this model. Alternatively, multiple Rap1 arrays might lead to rapid sequestration of the end at telomere clusters, perhaps through redundant cooperative interactions with Sir, Rif, and/or Ku proteins, where Mre11 access might be limited. Finally, we consider the possibility that long Rap1 arrays may affect DNA replication fork progression at the break site in such a way that leads to their protection following replication. Telomeric DNA sequences, whether present at their normal location or at internal sites, are known to cause replication fork blockage (Ivessa et al. 2002; Makovets et al. 2004), the strength of which is proportional to tract length. Although protein binding at telomeric tracts has been considered an impediment to replication fork passage, a recent report indicates just the opposite in fission yeast, where the Taz1 protein was shown to promote fork passage through telomeric repeats (Miller et al. 2006). Whether or not this turns out to be the case in budding yeast, it points to the possibility that replication fork passage at long TG tracts is a highly regulated process that could influence processing at nearby DSBs. However, the fact that long TG tracts are protected in cells blocked in G2/M before (and after) the break is induced (Supplementary Fig. S4) would argue that replication fork passage is not required for the effect.
In summary, we have shown that DSBs formed adjacent to telomeric tracts (or Rap1-binding site arrays) comparable in length to native telomeres appear to be capped by a mechanism that blocks access to the MRX complex, 5′ exonucleolytic resection, Cdc13 binding, and checkpoint activation. The unique protective property of these ends depends on Rap1 binding, but does not require either the Rif proteins, known to play a role in telomere length regulation through an interaction with the Rap1 C terminus, nor the end-binding protein Cdc13. Although the precise molecular nature of this capped structure is unknown, we suggest that it operates at native telomeres where it provides the first line of defense to guard against nucleolytic attack and activation of the DNA damage checkpoint. Because this concealment mechanism also inhibits telomerase access, and thus leads to TG-tract shortening, it is inherently unstable and requires intermittent Cdc13 and telomerase action as a reinforcing mechanism. A challenge for future studies will be to test various features of this model at native telomeres.
Materials and methods
Strains and plasmids
All strains constructed in this study (see Supplementary Table S2) are derived from a W303 strain lacking the HO site at the MAT locus (mat∷loxP) and carrying a galactose-inducible copy of the HO endonuclease gene at the LEU2 locus (Bianchi et al. 2004). This strain also contains a copy of the LYS2 gene at the MNT2 locus. For ChIP experiments, the subtelomeric region of Chr. VII-L was modified by the insertion, between the ADH4 and MNT2 genes, of a cassette containing the ADE2 gene, a sequence from the mouse Dbp gene (amplicon 7); either a 250-bp TG-repeat sequence from a native telomere, 80 bp of phage λ DNA, or an array of 16 Rap1-binding sites (either wild-type or mutated; Grossi et al. 2001); and the HO cut site. In the same strain, the subtelomeric region of Chr. V-R was modified by inserting a cassette at the YER188W locus containing the TRP1 gene, a second sequence from the mouse Dbp gene (amplicon 9); either 80 bp TG or an array of four Rap1-binding sites (wild-type or mutated); and the HO cut site (Fig. 1A). For resection assays, the subtelomeric region of Chr. VII-L was modified with the same cassette described above containing either TG-250 or TG-80 tracts.
The MRE11 and YKU70 deletion mutants (mre11∷kanMX and yku70∷kanMX) were generated by transformation of PCR products obtained from amplification of pUG6 (Guldener et al. 1996). The cdc13-1 allele was introduced at the endogenous CDC13 locus by transformation with XhoI-digested pVL451 (a gift from V. Lundblad, Salk Institute, CA) and selection on SC-Ura followed by selection on 5-FOA and screening for growth at 37°C. The rap1-17 allele was introduced by transformation with SnaBI-cleaved pAB357. The est2-D670A allele was introduced by allele replacement with pVL726 (Bianchi et al. 2004). Strains carrying the mutation were maintained through the presence of URA3 2-μm plasmid expressing wild-type EST2 (pAB370), which was eliminated before performing the experiment by growth on FOA-containing medium. The Myc epitope-tagged (13xMyc) versions of EST1, CDC13, MRE11, RPA1, and YKU70 (all C-terminal tags) were obtained by transformation of PCR products derived from pFA6a-13Myc-HIS3MX6 (Longtine et al. 1998). An N-terminal Myc tag on EST2 was obtained using pAB296 (Bianchi et al. 2004). The MEC1-18Myc allele (N-terminal tag) was constructed by transformation with StuI-linearized pAB378; this plasmid was derived from pML191.17 (a gift from M.P. Longhese, Milan, Italy; Paciotti et al. 2000) by subcloning into pRS303. None of the above tagged alleles had any detectable effect on telomere length.
ChIP
ChIP assays were performed as described previously (Bianchi et al. 2004). Quantification of immunoprecipitated DNA was achieved by real-time PCR on an Applied Biosystem ABI Prism 7700. Amplicon 7 is located adjacent to either TG-250, an array of 16 Rap1-binding sites, or 80 bp of phage λ DNA on Chr. VII-L; amplicon 9 is located adjacent to either TG-80 or an array of four Rap1-binding sites on Chr. V-R; amplicon 14 is located distal to the HO site on Chr. VII-L; and the internal control is located within the PDI1 gene (50 kb from left telomere of Chr. III). Enrichment of amplicons 7, 9, and 14 (AMP), over an internal control (INT) was determined after normalization with values obtained for mock-treated samples (no Myc antibody, Ab−) and with the efficiency of HO cutting (%HO cut) at Chr. VII-L and Chr. V-R. The equation used to determine fold enrichment was as follows:
For each strain, results were obtained from at least two experiments, and in some cases three or four. Data are reported as averages (bars), with standard deviations indicated by lines above.
Resection assay
EagI-digested genomic DNA was run in denaturing polyacrylamide gel (6%) and electroblotted. DNA was then probed with both a single-stranded riboprobe hybridizing to the Dbp amplicon 7, in order to measure resection of the C1–3A-rich strand, and a single-stranded riboprobe specific for a genomic sequence on Chr. XV that was used as internal loading control. Details of the plasmids used to generate these probes are available on request. The Dbp amplicon 7 (73 bp) is located 37 bp upstream of the TG repeats in both TG-80 and TG-250 constructs. Bands were detected using a PhosphorImager and quantified using Quantity One Software (Bio-Rad). The signal for the C1–3A strand relative to the internal control was obtained following normalization by the efficiency of HO cleavage.
Southern blotting
Genomic DNA digested with either EcoRV or NdeI and AvaI was separated in a 0.7% agarose gel and transferred to Hybond N+ nylon membrane (Amersham Biosciences). EcoRV-digested DNA was then probed with both a 32P-radiolabeled ADE2 DNA fragment and a 32P-radiolabeled NMD5 fragment, while NdeI/AvaI-digested DNA was probed with both a 32P-radiolabeled TRP1 fragment and a 32P-radiolabeled CDC24 fragment. Quantification of HO cleavage was obtained by measuring the amount of the uncut band relative to the internal loading control, normalized to the uninduced (t = 0) sample.
QAOS assay
QAOS analysis was performed as described (Booth et al. 2001), except for the following modifications. Overnight cultures were diluted in YPLG (YEP + 2% lactic acid, 2% glycerol) and grown at 23°C for 3 h. Before galactose, addition cultures were shifted at 36°C. Cells were cross-linked at time 0 min, 90 min, and 180 min after galactose addition. DNA was sonicated and a reverse cross-link was performed overnight at 65°C. DNA was purified using a Qiagen QIAquick purification column. Primers and probes for the YER188W locus (1130 and 1309 bp centromere-proximal to the short and long TG tracts, respectively) were used for single-stranded DNA detection. PCR conditions were as follows. Step 1: one cycle; 5 min at 40°C, ramp to 72°C, at 2°C per minute. Step 2: one cycle; 10 min at 72°C. Step 3: one cycle; 5 min at 95°C. Step 4: 40 cycles; 15 sec at 95°C, 30 sec at 67°C, and 30 sec at 72°C.
Acknowledgments
This work is dedicated to the memory of Stephan B. Schawalder, a passionate scientist and a dear friend. We thank A. Puglisi for help and advice on the QAOS assay, V. Lundblad and M.P. Longhese for plasmids, S. Schawalder for Rap1 antibody, and S. Citi for providing the 9E10 antibody. We also thank A. Puglisi and all the members of the Shore laboratory for helpful discussions during the course of this work and insightful comments on the manuscript. We are grateful to N. Roggli for expert assistance with artwork. This work was supported by the Swiss National Science Foundation, the NCCR program “Frontiers in Genetics” (Swiss National Science Foundation), the Swiss Cancer League (OncoSuisse), and the Canton of Geneva.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.400907
References
- Bertuch A.A., Lundblad V., Lundblad V. The maintenance and masking of chromosome termini. Curr. Opin. Cell Biol. 2006;18:247–253. doi: 10.1016/j.ceb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Bianchi A., Negrini S., Shore D., Negrini S., Shore D., Shore D. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol. Cell. 2004;16:139–146. doi: 10.1016/j.molcel.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Booth C., Griffith E., Brady G., Lydall D., Griffith E., Brady G., Lydall D., Brady G., Lydall D., Lydall D. Quantitative amplification of single-stranded DNA (QAOS) demonstrates that cdc13-1 mutants generate ssDNA in a telomere to centromere direction. Nucleic Acids Res. 2001;29:4414–4422. doi: 10.1093/nar/29.21.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.W., Blackburn E.H., Blackburn E.H. Telomerase and ATM/Tel1p protect telomeres from nonhomologous end joining. Mol. Cell. 2003;11:1379–1387. doi: 10.1016/s1097-2765(03)00174-6. [DOI] [PubMed] [Google Scholar]
- D’Amours D., Jackson S.P., Jackson S.P. The Mre11 complex: At the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- de Lange T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes & Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Diede S.J., Gottschling D.E., Gottschling D.E. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases α and δ. Cell. 1999;99:723–733. doi: 10.1016/s0092-8674(00)81670-0. [DOI] [PubMed] [Google Scholar]
- Diede S.J., Gottschling D.E., Gottschling D.E. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr. Biol. 2001;11:1336–1340. doi: 10.1016/s0960-9822(01)00400-6. [DOI] [PubMed] [Google Scholar]
- DuBois M.L., Haimberger Z.W., McIntosh M.W., Gottschling D.E., Haimberger Z.W., McIntosh M.W., Gottschling D.E., McIntosh M.W., Gottschling D.E., Gottschling D.E. A quantitative assay for telomere protection in Saccharomyces cerevisiae. Genetics. 2002;161:995–1013. doi: 10.1093/genetics/161.3.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto S., Glowczewski L., Berman J., Glowczewski L., Berman J., Berman J. MEC3, MEC1, and DDC2 are essential components of a telomere checkpoint pathway required for cell cycle arrest during senescence in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:2626–2638. doi: 10.1091/mbc.02-02-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S.K., Lundblad V., Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- Fisher T.S., Zakian V.A., Zakian V.A. Ku: A multifunctional protein involved in telomere maintenance. DNA Repair (Amst.) 2005;4:1215–1226. doi: 10.1016/j.dnarep.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Fisher T.S., Taggart A.K., Zakian V.A., Taggart A.K., Zakian V.A., Zakian V.A. Cell cycle-dependent regulation of yeast telomerase by Ku. Nat. Struct. Mol. Biol. 2004;11:1198–1205. doi: 10.1038/nsmb854. [DOI] [PubMed] [Google Scholar]
- Frank-Vaillant M., Marcand S., Marcand S. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol. Cell. 2002;10:1189–1199. doi: 10.1016/s1097-2765(02)00705-0. [DOI] [PubMed] [Google Scholar]
- Garber P.M., Vidanes G.M., Toczyski D.P., Vidanes G.M., Toczyski D.P., Toczyski D.P. Damage in transition. Trends Biochem. Sci. 2005;30:63–66. doi: 10.1016/j.tibs.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Garvik B., Carson M., Hartwell L., Carson M., Hartwell L., Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E., Muller T., Sogo J., Laroche T., Gasser S.M., Muller T., Sogo J., Laroche T., Gasser S.M., Sogo J., Laroche T., Gasser S.M., Laroche T., Gasser S.M., Gasser S.M. RAP1 stimulates single- to double-strand association of yeast telomeric DNA: Implications for telomere–telomere interactions. Nucleic Acids Res. 1994;22:5310–5320. doi: 10.1093/nar/22.24.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N., Reed S.I., Charbonneau M., Reed S.I., Charbonneau M., Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes & Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- Greider C.W., Blackburn E.H., Blackburn E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H., de Lange T., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H., de Lange T., Rosenfield S., Stansel R.M., Bianchi A., Moss H., de Lange T., Stansel R.M., Bianchi A., Moss H., de Lange T., Bianchi A., Moss H., de Lange T., Moss H., de Lange T., de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- Grossi S., Bianchi A., Damay P., Shore D., Bianchi A., Damay P., Shore D., Damay P., Shore D., Shore D. Telomere formation by rap1p binding site arrays reveals end-specific length regulation requirements and active telomeric recombination. Mol. Cell. Biol. 2001;21:8117–8128. doi: 10.1128/MCB.21.23.8117-8128.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldener U., Heck S., Fielder T., Beinhauer J., Hegemann J.H., Heck S., Fielder T., Beinhauer J., Hegemann J.H., Fielder T., Beinhauer J., Hegemann J.H., Beinhauer J., Hegemann J.H., Hegemann J.H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett J.A., Feldser D.M., Greider C.W., Feldser D.M., Greider C.W., Greider C.W. Telomere dysfunction increases mutation rate and genomic instability. Cell. 2001;106:275–286. doi: 10.1016/s0092-8674(01)00457-3. [DOI] [PubMed] [Google Scholar]
- Hardy C.F.J., Sussel L., Shore D., Sussel L., Shore D., Shore D. A RAP1-interacting protein involved in silencing and telomere length regulation. Genes & Dev. 1992;6:801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- Ira G., Pellicioli A., Balijja A., Wang X., Fiorani S., Carotenuto W., Liberi G., Bressan D., Wan L., Hollingsworth N.M., Pellicioli A., Balijja A., Wang X., Fiorani S., Carotenuto W., Liberi G., Bressan D., Wan L., Hollingsworth N.M., Balijja A., Wang X., Fiorani S., Carotenuto W., Liberi G., Bressan D., Wan L., Hollingsworth N.M., Wang X., Fiorani S., Carotenuto W., Liberi G., Bressan D., Wan L., Hollingsworth N.M., Fiorani S., Carotenuto W., Liberi G., Bressan D., Wan L., Hollingsworth N.M., Carotenuto W., Liberi G., Bressan D., Wan L., Hollingsworth N.M., Liberi G., Bressan D., Wan L., Hollingsworth N.M., Bressan D., Wan L., Hollingsworth N.M., Wan L., Hollingsworth N.M., Hollingsworth N.M., et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa A.S., Zhou J.Q., Schulz V.P., Monson E.K., Zakian V.A., Zhou J.Q., Schulz V.P., Monson E.K., Zakian V.A., Schulz V.P., Monson E.K., Zakian V.A., Monson E.K., Zakian V.A., Zakian V.A. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes & Dev. 2002;16:1383–1396. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrivee M., LeBel C., Wellinger R.J., LeBel C., Wellinger R.J., Wellinger R.J. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes & Dev. 2004;18:1391–1396. doi: 10.1101/gad.1199404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.E., Moore J.K., Holmes A., Umezu K., Kolodner R.D., Haber J.E., Moore J.K., Holmes A., Umezu K., Kolodner R.D., Haber J.E., Holmes A., Umezu K., Kolodner R.D., Haber J.E., Umezu K., Kolodner R.D., Haber J.E., Kolodner R.D., Haber J.E., Haber J.E. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Lingner J., Cooper J.P., Cech T.R., Cooper J.P., Cech T.R., Cech T.R. Telomerase and DNA: No longer a lagging strand problem? Science. 1995;269:1533–1534. doi: 10.1126/science.7545310. [DOI] [PubMed] [Google Scholar]
- Lisby M., Barlow J.H., Burgess R.C., Rothstein R., Barlow J.H., Burgess R.C., Rothstein R., Burgess R.C., Rothstein R., Rothstein R. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R., Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R., Wach A., Brachat A., Philippsen P., Pringle J.R., Brachat A., Philippsen P., Pringle J.R., Philippsen P., Pringle J.R., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Makovets S., Herskowitz I., Blackburn E.H., Herskowitz I., Blackburn E.H., Blackburn E.H. Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol. Cell. Biol. 2004;24:4019–4031. doi: 10.1128/MCB.24.9.4019-4031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S., Gilson E., Shore D., Gilson E., Shore D., Shore D. A protein-counting mechanism for telomere length regulation in yeast. Science. 1997;275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- Marcand S., Brevet V., Mann C., Gilson E., Brevet V., Mann C., Gilson E., Mann C., Gilson E., Gilson E. Cell cycle restriction of telomere elongation. Curr. Biol. 2000;10:487–490. doi: 10.1016/s0960-9822(00)00450-4. [DOI] [PubMed] [Google Scholar]
- McEachern M.J., Haber J.E., Haber J.E. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- Michelson R.J., Rosenstein S., Weinert T., Rosenstein S., Weinert T., Weinert T. A telomeric repeat sequence adjacent to a DNA double-stranded break produces an anticheckpoint. Genes & Dev. 2005;19:2546–2559. doi: 10.1101/gad.1293805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski P.A., Mieczkowska J.O., Dominska M., Petes T.D., Mieczkowska J.O., Dominska M., Petes T.D., Dominska M., Petes T.D., Petes T.D. Genetic regulation of telomere–telomere fusions in the yeast Saccharomyces cerevisae. Proc. Natl. Acad. Sci. 2003;100:10854–10859. doi: 10.1073/pnas.1934561100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K.M., Rog O., Cooper J.P., Rog O., Cooper J.P., Cooper J.P. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- Muller T., Gilson E., Schmidt R., Giraldo R., Sogo J., Gross H., Gasser S.M., Gilson E., Schmidt R., Giraldo R., Sogo J., Gross H., Gasser S.M., Schmidt R., Giraldo R., Sogo J., Gross H., Gasser S.M., Giraldo R., Sogo J., Gross H., Gasser S.M., Sogo J., Gross H., Gasser S.M., Gross H., Gasser S.M., Gasser S.M. Imaging the asymmetrical DNA bend induced by repressor activator protein 1 with scanning tunneling microscopy. J. Struct. Biol. 1994;113:1–12. doi: 10.1006/jsbi.1994.1027. [DOI] [PubMed] [Google Scholar]
- Paciotti V., Clerici M., Lucchini G., Longhese M.P., Clerici M., Lucchini G., Longhese M.P., Lucchini G., Longhese M.P., Longhese M.P. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes & Dev. 2000;14:2046–2059. [PMC free article] [PubMed] [Google Scholar]
- Pardo B., Marcand S., Marcand S. Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J. 2005;24:3117–3127. doi: 10.1038/sj.emboj.7600778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennock E., Buckley K., Lundblad V., Buckley K., Lundblad V., Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- Ray A., Runge K.W., Runge K.W. The yeast telomere length counting machinery is sensitive to sequences at the telomere–nontelomere junction. Mol. Cell. Biol. 1999;19:31–45. doi: 10.1128/mcb.19.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell L.L., Zakian V.A., Zakian V.A. Loss of a yeast telomere: Arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- Schramke V., Luciano P., Brevet V., Guillot S., Corda Y., Longhese M.P., Gilson E., Geli V., Luciano P., Brevet V., Guillot S., Corda Y., Longhese M.P., Gilson E., Geli V., Brevet V., Guillot S., Corda Y., Longhese M.P., Gilson E., Geli V., Guillot S., Corda Y., Longhese M.P., Gilson E., Geli V., Corda Y., Longhese M.P., Gilson E., Geli V., Longhese M.P., Gilson E., Geli V., Gilson E., Geli V., Geli V. RPA regulates telomerase action by providing Est1p access to chromosome ends. Nat. Genet. 2004;36:46–54. doi: 10.1038/ng1284. [DOI] [PubMed] [Google Scholar]
- Shore D. Telomeric chromatin: Replicating and wrapping up chromosome ends. Curr. Opin. Genet. Dev. 2001;11:189–198. doi: 10.1016/s0959-437x(00)00178-7. [DOI] [PubMed] [Google Scholar]
- Shroff R., Arbel-Eden A., Pilch D., Ira G., Bonner W.M., Petrini J.H., Haber J.E., Lichten M., Arbel-Eden A., Pilch D., Ira G., Bonner W.M., Petrini J.H., Haber J.E., Lichten M., Pilch D., Ira G., Bonner W.M., Petrini J.H., Haber J.E., Lichten M., Ira G., Bonner W.M., Petrini J.H., Haber J.E., Lichten M., Bonner W.M., Petrini J.H., Haber J.E., Lichten M., Petrini J.H., Haber J.E., Lichten M., Haber J.E., Lichten M., Lichten M. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A., De Lange T., De Lange T. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- Taggart A.K., Teng S.C., Zakian V.A., Teng S.C., Zakian V.A., Zakian V.A. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]
- Takata H., Tanaka Y., Matsuura A., Tanaka Y., Matsuura A., Matsuura A. Late S phase-specific recruitment of Mre11 complex triggers hierarchical assembly of telomere replication proteins in Saccharomyces cerevisiae. Mol. Cell. 2005;17:573–583. doi: 10.1016/j.molcel.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Teixeira M.T., Arneric M., Sperisen P., Lingner J., Arneric M., Sperisen P., Lingner J., Sperisen P., Lingner J., Lingner J. Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- van Steensel B., de Lange T., de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- Weinert T.A., Hartwell L.H., Hartwell L.H. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics. 1993;134:63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton D., Shore D., Shore D. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes & Dev. 1997;11:748–760. doi: 10.1101/gad.11.6.748. [DOI] [PubMed] [Google Scholar]
- Zhou B.B., Elledge S.J., Elledge S.J. The DNA damage response: Putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]