Abstract
Epigenetic regulation in eukaryotes is executed by a complex set of signaling interactions among small RNA species and chromatin marks, including histone modification and DNA methylation. We identified vim1 (VARIANT IN METHYLATION 1), an Arabidopsis mutation causing cytosine hypomethylation and decondensation of centromeres in interphase. VIM1 is a member of a small gene family, encoding proteins containing PHD, RING, and SRA (SET- and RING-associated) domains, which are found together in mammalian proteins implicated in regulation of chromatin modification, transcription, and the cell cycle. VIM1 is an unconventional methylcytosine-binding protein that interacts in vitro with 5mCpG- and 5mCpHpG-modified DNA (via its SRA domain), as well as recombinant histones (H2B, H3, H4, and HTR12) in plant extracts. VIM1 associates with methylated genomic loci in vivo and is enriched in chromocenters. Our findings suggest that VIM1 acts at the DNA methylation–histone interface to maintain centromeric heterochromatin.
Keywords: Cytosine methylation, methylcytosine-binding protein, centromere, epigenetic, SRA domain, heterochromatin
Large regions of higher eukaryotic genomes, particularly the domains surrounding and encompassing centromeres, are composed of repetitive elements, which are preferential targets for heterochromatin assembly. Heterochromatic regions of the genome are characterized by increased chromatin condensation and decreased or differential accessibility to regulatory proteins (Craig 2005; Huisinga et al. 2006). Most forms of heterochromatin are stably inherited and contain one or more epigenetic marks that direct its maintenance during cell division. Specification and maintenance of heterochromatin and other functionally distinct chromatin domains rely on complex interactions among cytosine methylation, histone modification, and RNA interference (RNAi) pathways (Tamaru and Selker 2001; Jaenisch and Bird 2003; Chan et al. 2005; Matzke and Birchler 2005).
Cytosine methylation is an important mechanism for establishing stable heritable epigenetic marks, thereby linking primary nucleotide sequence to chromatin organization (Jaenisch and Bird 2003). The cytosine methylation reaction is catalyzed by enzymes known as cytosine-DNA-methyltransferases (DNMTs) (Goll and Bestor 2005). Notably, mammalian cytosine methylation is mostly restricted to symmetrical CpG sequences, although plant cytosine methylation occurs at CpG, CpHpG, and CpHpH sequences (where H = A, C, T). Once established, cytosine methylation can be inherited through mitosis, and sometimes through meiosis, providing a stable epigenetic mark (Genereux et al. 2005; Richards 2006). Cytosine methylation cooperates with histone modification to generate a self-reinforcing cycle of epigenetic events that lead to long-term transcriptional repression (Jenuwein and Allis 2001; Nightingale et al. 2006). The regulatory information encoded by cytosine methylation is deciphered by a family of proteins that, through a conserved methyl-CpG-binding domain (MBD), selectively binds methylated CpG dinucleotides regardless of the sequence context (Klose and Bird 2006). Some MBD proteins can recruit histone deacetylase (HDAC) or histone methyltransferase (HMT) complexes as well as chromatin remodeling factors (Klose and Bird 2006). Chromatin modification can also influence cytosine methylation. For example, methylation at histone H3K9 creates binding sites for adaptor molecules, such as HP1 (Heterochromatin Protein 1) and related chromo-domain proteins, that ultimately recruit DNMTs (Fuks et al. 2003; Lehnertz et al. 2003). Through these types of interactions, covalent modifications of DNA and core histones play important roles in heterochromatin formation in mammals and plants.
The last several years have witnessed tremendous advances in the genetic analysis of eukaryotic DNA methylation, resulting in the identification of proteins and pathways necessary for specification and maintenance of genomic cytosine methylation patterns (Jaenisch and Bird 2003; Rangwala and Richards 2004; Chan et al. 2005). Two major protein categories important for genomic cytosine methylation, identified through both forward and reverse genetic approaches, are DNMTs and chromatin modification enzymes (e.g., HDACs, HMTs, nucleosome remodelers). Post-transcriptional silencing/RNAi pathways mutants also have cytosine hypomethylation phenotypes, highlighting the importance of RNA-directed DNA methylation mechanisms guided by small RNA species. However, only a handful of these mutations affect centromere DNA methylation, despite the fact that the centromere is a principle target for cytosine methylation and heterochromatin formation in the cell. In an effort to identify novel alleles or new loci affecting centromere DNA methylation, we chose an alternative approach of screening natural strains of the flowering plant Arabidopsis thaliana for changes in genomic methylation patterns. Through this approach, we identified a novel genetic locus, VIM1 (VARIANT IN METHYLATION 1), encoding a non-MBD class methylcytosine-binding protein that is required for full centromere DNA methylation. In addition, loss of VIM1 function leads to decondensation of the centromere repeat sequence in interphase and an alteration in the localization pattern of the centromere-specific histone H3 variant, HTR12. Our results suggest that VIM1 acts at the interface between DNA methylation and chromatin to maintain centromere heterochromatin.
Results
Identification of a natural variant with hypomethylated centromere DNA
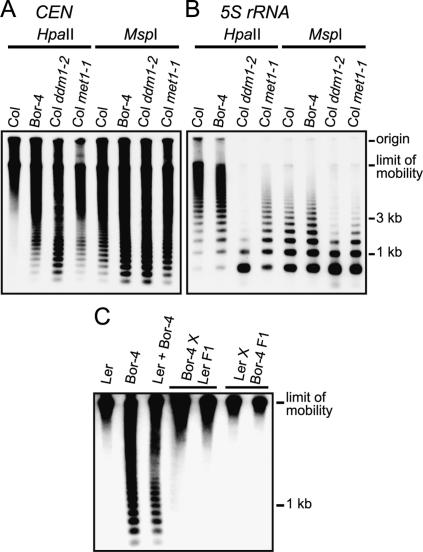
In a screen for natural variation in cytosine methylation among 89 different strains of A. thaliana, we identified one strain that contained hypomethylated centromeric 180-base-pair (bp) repeats (CEN). Centromere repeats from strain Borky-4 (Bor-4) showed an increased level of digestion with the restriction endonuclease HpaII (5′-CCGG-3′), which is inhibited by either 5mCpG or 5mCpHpG, relative to that seen for repeats in wild-type Columbia (Col) plants (Fig. 1A). A similar enhanced digestion was seen when the centromere repeats were cut with MspI, which cleaves the same site as HpaII but is inhibited only by 5mCpHpG. The effect of two previously characterized DNA hypomethylation mutations in strain Col are shown for comparison: ddm1, which reduces cytosine methylation in all sequence contexts (Vongs et al. 1993); and met1, which predominantly reduces 5mCpG (Kankel et al. 2003; Saze et al. 2003). These observations suggest that Bor-4 contains centromeric repeats that are hypomethylated at both CpG and CpHpG sites.
Figure 1.
Reduced cytosine methylation of centromeric repeats in Bor-4. (A) Genomic DNA samples from the indicated genotypes were digested with the isoschizomers HpaII or MspI, and DNA blot hybridization with a 180-bp centromere repeat probe (CEN) was performed. (B) The filter shown in A was rehybridized with a 5S rRNA probe (5S rRNA). (C) A DNA blot hybridization pattern with the CEN probe after HpaII digestion demonstrates that centromeric repeat arrays hypomethylated in Bor-4 were fully remethylated in F1 hybrids resulting from reciprocal crosses between strains Bor-4 and Ler. The lane labeled Ler + Bor-4 contains a 1:1 mixture of Ler and Bor-4 HpaII-digested genomic DNA and shows the hybridization pattern expected if no remethylation occurred in the F1 hybrids.
To determine whether reduced cytosine methylation in Bor-4 occurs at other genomic locations, we examined the extent of cytosine methylation at three additional loci that are heavily methylated in wild-type strains: the FWA gene (Soppe et al. 2000) and the pericentromeric tandemly repeated 5S rRNA genes (Campell et al. 1992) and Athila retrotransposable elements (Pelissier et al. 1995). Compared with wild-type Col, we did not find a significant change in Bor-4 cytosine methylation at the 5S rRNA genes (Fig. 1B), in the Athlia elements or at the FWA locus (data not shown). In agreement with previous reports, we observed a strong hypomethylation of all these loci in Col plants containing either ddm1-2 or met1-1 alleles. These results indicate that DNA hypomethylation in wild strain Bor-4 preferentially affects the 180-bp centromere repeats.
Altered centromere organization in Bor-4 interphase nuclei
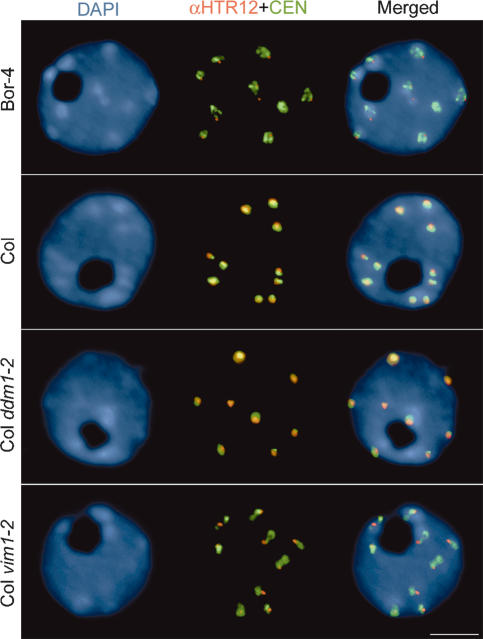
To investigate whether centromere heterochromatin was affected in Bor-4, we used fluorescence in situ hybridization (FISH) to detect the 180-bp centromere repeats in interphase nuclei. The spatial distribution of hybridization signal indicated that the centromere repeats in Bor-4 were decondensed relative to the repeats in Col nuclei (Fig. 2; Supplementary Table 1). We also stained fixed interphase nuclei with anti-HTR12 antibody (Talbert et al. 2002) to obtain a more comprehensive understanding of centromere heterochromatin in Bor-4. HTR12 is the Arabidopsis homolog of human CENP-A, Drosophila Cid, and yeast Cnp1, which are histone H3 variants that define the specialized chromatin structure associated with kinetochore assembly (Henikoff and Dalal 2005). We found that the area of the HTR12 immunostained signal was decreased in Bor-4 nuclei compared with the pattern observed in Col nuclei. We noted that neither the distribution of 180-bp repeats, nor the pattern of HTR12 staining is affected by the ddm1 mutation in the Col background (Fig. 2). Thus, Bor-4 exhibits not only abnormal centromere DNA methylation, but a unique centromere organization phenotype not exhibited by a well-characterized mutation with severe centromere DNA hypomethylation defects.
Figure 2.
Centromeric heterochromatin is altered in Bor-4. One-hundred-eighty-base-pair centromeric repeats (CEN) were detected by FISH, and HTR12 protein was immunolocalized in interphase nuclei from root tip cells of Bor-4, Col, Col ddm1-2, and Col vim1-2 (SALK_050903) plants. The DNA was counterstained with DAPI; chromocenters are more intensely stained. Bar, 5 μm.
A trans-acting mutation causes centromere DNA hypomethylation in Bor-4
To distinguish whether the Bor-4 centromere phenotypes result from the action of a trans-acting mutation or epigenetic inheritance of stochastic DNA hypomethylation, we outcrossed Bor-4 to standard laboratory strains Col and Ler and followed the segregation of the DNA hypomethylation phenotype in subsequent generations. In the F1 generation, regardless of the direction of the cross, the hypomethylated 180-bp repeat arrays originating from Bor-4 were fully remethylated, demonstrating that centromere DNA hypomethylation is reset efficiently (Fig. 1C; data not shown). This result contrasts with the persistence of hypomethylated genomes in F1 hybrids resulting from outcrossing ddm1 or met1 mutants (Vongs et al. 1993; Kakutani et al. 1999; Kankel et al. 2003), and argues against the possibility that Bor-4 centromere repeat hypomethylation is due strictly to epigenetic inheritance of a hypomethylated state. The hypomethylated centromere trait segregated as a monogenic recessive trait in two independent Ler X Bor-4 F2 families (normal:hypomethylated∷112:36). Recombinational mapping in Ler X Bor-4 F2 families indicated that the hypomethylated centromere phenotype of Bor-4 is caused by variation at a single trans-acting locus, VIM1, at a map position distinct from known Arabidopsis mutations affecting DNA methylation or chromatin modification (Supplementary Fig. 1).
Identification of VIM1
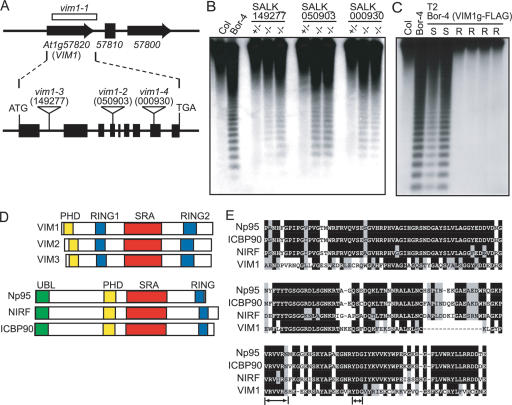
Using a Ler X Bor-4 F2 mapping population, we narrowed the region containing the vim1-1 allele from Bor-4 to an interval corresponding to a 113-kb region on the lower arm of chromosome 1. In Col, this window contains 30 annotated genes (Supplementary Fig. 1). The Bor-4 allele of one gene in this interval, At1g57820, carries a large deletion. The missing segment spans a 3.2-kb region from just downstream from the first intron donor site to the middle of the intergenic region between At1g57820 and At1g57810 (Fig. 3A; Supplementary Fig. 2). We could not detect At1g57820 transcript in Bor-4 using RT–PCR analysis; however, the predicted transcript was observed in Col and Ler (data not shown). To test whether mutation of At1g57820 causes hypomethylation of the 180-bp centromere repeats, we analyzed the effect of Agrobacterium T-DNA insertion alleles in this gene in the Col background. The T-DNA inserts in At1g57820 disrupt the first intron, the fourth exon, and the eighth exon, respectively (Fig. 3A). We could not detect full-length transcripts from any Col At1g57820 homozygous T-DNA mutant (data not shown). All three T-DNA insertion mutants in Col showed increased HpaII cleavage of the 180-bp centromere repeats (Fig. 3B), although the diagnostic ladder-like hybridization pattern was weaker than that seen in Bor-4 (Fig. 1A). The muted effect of the T-DNA insertion alleles in At1g57820 may be due to the action of strain-specific modifiers that partially cover for At1g57820 loss of function in Col. However, the vim1-2 allele (SALK_050903) in the Col strain caused distinct centromere defects similar to those seen in Bor-4 (Fig. 2), arguing that disruption of VIM1 function is responsible for both the altered centromere heterochromatin and centromere DNA hypomethylation phenotypes.
Figure 3.
Identification of the VIM1 gene. (A) Diagram of the intron–exon structure of the VIM1 gene and positions of T-DNA insertions in At1g57820 with allele designations. At1g57810 (reverse transcriptase pseudogene) is located between At1g57800 and At1g57820. The deletion in Bor-4 is shown as an open box (vim1-1 allele). Filled boxes represent exons and thin lines represent introns. The positions of the start and stop codon are indicated, as are the T-DNA insertion sites in At1g57820. (B) DNA blot hybridization (HpaII digest, CEN probe) analysis of T-DNA insertion mutants in At1g57820. Genomic DNA was isolated from independent plants in segregation populations. (+/+) Homozygous wild-type allele; (+/−) heterozygous for mutant allele; (−/−) homozygous for mutant allele. (C) DNA blot hybridization (HpaII digest, CEN probe) analysis demonstrating complementation of the hypomethylation phenotype of Bor-4 with a transgene expressing At1g57820 (from Col strain, coding sequence fused to a C-terminal Flag epitope tag). Analysis of six individuals in the T2 generation segregating the transgene (carrying a Basta herbicide resistance marker: [R] resistant; [S] sensitive) is shown. (D) Schematic representation of possible homologs of VIM1. The RING (Pfam PF00097), PHD (Pfam PF000628), and SRA (Pfam PF02182) domains are labeled. The mammalian proteins contain an N-terminal ubiquitin-like domain (UBL), which is absent in the VIM1-like Arabidopsis proteins, and only one RING domain. (E) Sequence alignment of SRA domains from VIM1 and three related mammalian proteins. Black shading shows identical residues and gray shading shows similar residues. Arrows denote the conserved VRV(I/V)RG and YDG motifs in SRA domains.
We confirmed that At1g57820 corresponds to the VIM1 locus by two independent genetic tests. Crosses between Bor-4 and Col plants homozygous for an At1g57820 T-DNA insertion allele demonstrated a lack of complementation for the centromere DNA methylation phenotype in the resulting F1 individuals (Supplementary Fig. 3). Furthermore, a 5.5-kb genomic fragment containing the Col sequence of At1g57820 was sufficient to complement the hypomethylation phenotype of Bor-4 (Fig. 3C). The VIM1 locus encodes a 645-amino-acid protein with a PHD domain, two RING finger domains, and an SRA (SET- and RING-associated) domain (Fig. 3D; Supplementary Fig. 4A). Four additional genes encoding related proteins (≥68% amino acid identity to VIM1) with a similar domain organization exist in the Arabidopsis genome: At1g57800, At1g66040, At1g66050, and At5g39550 (Fig. 3D; Supplementary Fig. 4B). Only three of these genes (At1g57820 [VIM1], At1g66050 [VIM2], and At5g39550 [VIM3]) are expressed at an appreciable level in strain Bor-4, Col and Ler based on our RT–PCR analysis (data not shown) and cDNA resources available through public databases (e.g., http://www.arabidopsis.org, http://mpss.udel.edu/at). Neither the vim2 nor the vim3 T-DNA insertion allele examined in Col caused a centromere repeat hypomethylation phenotype (Supplementary Fig. 5).
Centromeric small interfering RNA (siRNA) populations are not significantly changed in Col vim1 plants
Several characterized Arabidopsis DNA hypomethylation mutations primarily affect small RNA accumulation, which reduces RNA-directed DNA methylation (Chan et al. 2005). Consequently, we compared the abundance of siRNAs corresponding to the 180-bp centromere repeats in Col and Col vim1-2 plants using RNA gel blot analysis (Supplementary Fig. 6). We did not find a significant change in the abundance of centromeric small RNA populations associated with the VIM1 genotype, suggesting that the vim1-2 mutation reduces centromere DNA methylation without affecting siRNA accumulation.
VIM1 is a methylcytosine-binding protein
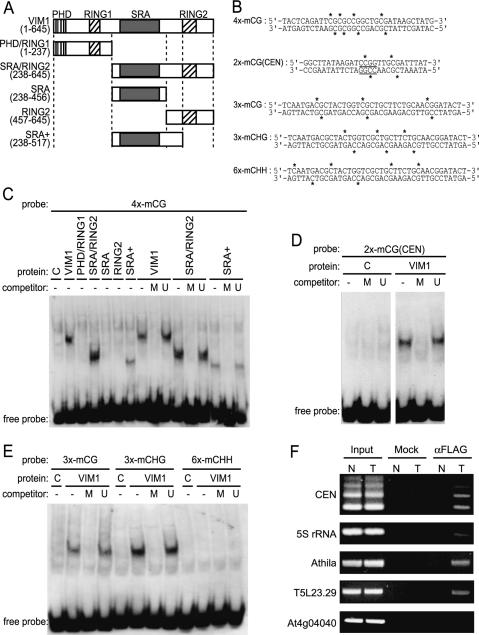
The similarity shared among the SRA domains of VIM1 and three mammalian proteins, human ICBP90 and NIRF, and murine Np95 (Fig. 3D,E; Fujimori et al. 1998; Mori et al. 2002) prompted us to investigate whether VIM1 is a methylcytosine-binding protein because the SRA domain in these mammalian proteins was shown to bind methyl-CpG-modified double-stranded DNA (Unoki et al. 2004). Full-length VIM1 protein and various VIM1 subdomains (Fig. 4) expressed in vitro were tested for their ability to bind methylated double-stranded oligonucleotides by electrophoretic mobility shift assay (EMSA). We demonstrated that full-length VIM1 bound to a double-stranded oligonucleotide (4x-mCG) containing four symmetrical 5mCpG sites in a methylation-dependent manner (Fig. 4A–C). VIM1 bound the same oligonucleotide sequence containing only a single 5mCpG site, but did so more weakly (data not shown). Because vim1 mutations lead to a reduction in cytosine methylation in the 180-bp centromere repeats, we tested and confirmed VIM1’s ability to interact specifically with an oligonucleotide corresponding to the region of the 180-bp repeat sequence containing the 5′-CCGG-3′ site assayed (Fig. 4B,D). Next, we delimited the minimal region of VIM1 that interacted in a methylation-dependent manner with the 4x-mCG probe to 279 amino acids comprising the entire SRA domain and the interval between the SRA and second RING domain (Fig. 4A,C, lanes marked SRA+). While the SRA domain is required for methylcytosine-binding activity, this domain is not sufficient (Fig. 4A,C, lane marked SRA).
Figure 4.
VIM1 is a methylcytosine-binding protein. (A) Schematic of the VIM1 protein and derivatives tested for methylcytosine-binding activity; amino acid coordinates are shown in the parentheses. Domain designations include PHD domain, SRA domain, and RING domains. (B) Oligonucleotide probes used in the methylcytosine-binding assays shown in C–E. Asterisks indicate the positions of 5mC residues. The underlined 5′-CCGG-3′ site corresponds to the HpaII assayed in Figure 1. (C) VIM1 protein derivatives were expressed in wheat germ transcription/translation extracts (C, vector-only control) and incubated with the digoxigenin-endlabeled 4x-mCG probe in the presence of 62-fold excess unlabeled competitor oligonucleotides. (U) Unmethylated; (M) methylated; (−) no competitor. EMSA of incubation products is shown; the position of the unbound free probe is indicated at the bottom of the image. The same abbreviations and symbols are used in C–E. (D) EMSA demonstrating the ability of full-length VIM1 to bind a methylated oligonucleotide probe based on the 180-bp centromere repeat sequence. (E) EMSA of interaction of full-length VIM1 with identical probes methylated in different cytosine contexts. (F) ChIP was performed on nuclei prepared from Col (N, nontransgenic) and transgenic (T) plants expressing Flag-VIM1. After immunoprecipitation, samples were used as templates for amplification of different genomic sequences, which are indicated at the left of the images of the resulting ethidium-stained gels. (Input) 10% of input; (Mock) precipitation in the absence of antibody; (αFlag) precipitation with anti-Flag antibody.
Plant genomes contain cytosine methylation in three different sequence contexts: two symmetrical sites, CpG and CpHpG; and the asymmetric site, CpHpH (Chan et al. 2005). We assayed the specificity of VIM1’s methylcytosine-binding activity using a set of double-stranded oligonucleotides of identical primary sequence that contained cytosine methylation in only one of the three possible sequence contexts (Fig. 4B,E). As expected, VIM1 bound the oligonucleotide modified at 5mCpG sites. VIM1 was unable to bind the 5mCpHpH-containing oligonucleotide, but interacted in a methylation-dependent manner with the 5mCpHpG-containing oligonucleotide. Therefore, VIM1 recognizes cytosine methylation in two different sequence contexts that occur in plant genomes, including centromere repeat arrays.
After demonstrating that VIM1 has methylcytosine-binding activity in vitro, we next determined whether VIM1 associates with methylated DNA in vivo. We employed a chromatin immunoprecipitation (ChIP) assay on nuclei prepared from wild-type Col and transgenic Arabidopsis plants expressing Flag-VIM1. Cross-linked chromatin complexes were precipitated with or without anti-Flag antibody, and the precipitated DNA samples were used as templates to amplify centromeric 180-bp repeats. As seen in Figure 4F, the 180-bp centromere repeat arrays were associated with the VIM1 protein. The unmethylated locus At4g04040 was not associated with VIM1 (Gendrel et al. 2002; Lippman et al. 2004). We also investigated three other methylated genomic loci, including the tandemly repeated 5S rRNA genes (5S rRNA), repetitive Athila retrotransposable elements (Athila), and a Cinful-like retrotransposon (T5L23.29) (Gendrel et al. 2002). The anti-Flag antibody precipitated all methylated genomic sequences tested. Our results indicate that VIM1 associates with methylated DNA both in vitro and in vivo. The specificity of vim1 DNA hypomethylation effects may stem from a heightened sensitivity of centromeric chromatin to loss of VIM1 function.
VIM1 is concentrated at chromocenters
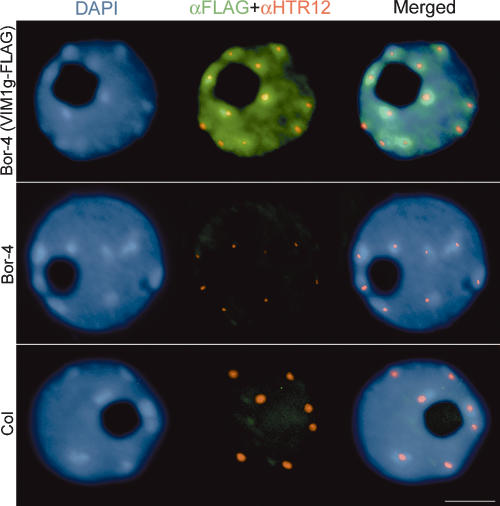
We next studied the subnuclear localization of VIM1 relative to interphase chromatin organization. The VIM1 genomic coding sequence, expressed under control of its native promoter, was engineered to express full-length VIM1 fused to the Flag epitope at the C terminus (VIM1g-Flag) in transgenic plants. Fixed nuclei from Col, Bor-4, and transgenic Bor-4 expressing VIM1g-Flag were immunolabeled with anti-Flag and anti-HTR12 antibodies. Introduction of the VIM1g-Flag construct complemented three centromere-related phenotypes in Bor-4: decondensation of 180-bp centromere repeats (data not shown), altered HTR12 localization (Fig. 5, middle column; Supplementary Table 2), and centromere DNA hypomethylation (Fig. 3C). These results demonstrate that the epitope-tagged VIM1 is functional. Epitope-tagged VIM1 protein was localized to the nucleus, where it showed a broad distribution (excluding the nucleolus) but enrichment in the heterochromatic chromocenters in the majority of the cells examined (Fig. 5, top row; Supplementary Table 3). VIM1 localization is consistent with our ChIP results (Fig. 4F) and the known distribution of methylated DNA sequences, which tend to be concentrated in the chromocenters.
Figure 5.
Subnuclear localization of VIM1. Immunolocalization of the epitope-tagged VIM1 (αFlag) and HTR12 (αHTR12) was performed on nuclei derived from nontransgenic Bor-4 and Col plants, as well as transgenic Bor-4 plants expressing VIM1-Flag under control of the native VIM1 promoter (VIM1g-Flag). DAPI was used as a DNA counterstain. Bar, 5 μm.
VIM1–histone interaction
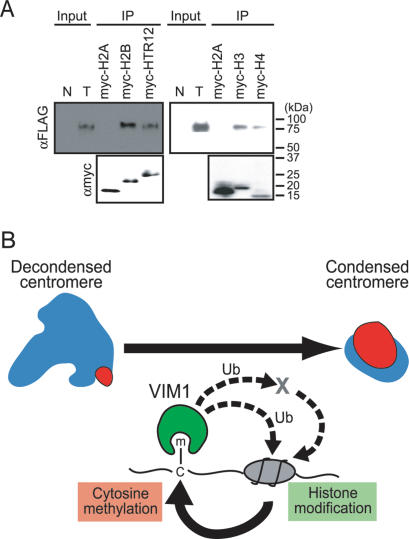
The centromere phenotypes of vim1 mutants, including the apparent changes in HTR12 localization, coupled with the observation that the SRA domain of murine Np95 interacts differentially with core histones (Citterio et al. 2004), led us to test whether VIM1 physically interacts with HTR12. An epitope-tagged HTR12 (N-terminal c-myc tag) was expressed in Escherichia coli and partially purified using an anti-myc conjugated resin. In parallel, we expressed and partially purified recombinant versions of other Arabidopsis histones, including myc-H2A (HTA2), myc-H2B (HTB2), myc-H3 (HTR3), and myc-H4 (HFO4). Purified recombinant histones were incubated with extracts from transgenic plants overexpressing Flag-VIM1. Proteins associated with the immobilized recombinant histones were subject to Western blot analysis to monitor the recovery of the Flag-VIM1 protein. As shown in Figure 6A, Flag-VIM1 interacted with three of the core histones (with the exception of H2A [HTA2]), as well as the centromere-specific histone H3, HTR12.
Figure 6.
VIM1 at the DNA methylation–chromatin interface. (A) VIM1 interacts with histones. (Top panel) Immobilized myc-tagged histones were incubated with protein extracts from transgenic plants expressing Flag-VIM1, and the associated proteins were collected and detected by Western blot analysis using an anti-Flag antibody to detect Flag-VIM1. Input lanes correspond to total protein from either nontransgenic (N) or transgenic (T) plants (1/240×). The bottom panel shows detection of myc-tagged histones in the samples using an anti-myc antibody. (B) A model for the role of VIM1 in the maintenance of centromeric heterochromatin. The centromere decondensation phenotype is illustrated on the top left. (Blue) 180-bp centromere repeat; (red) HTR12. The VIM1 biochemical activities illustrated at the bottom of the figure are necessary for centromere condensation (top right). Demonstrated VIM1 activities include direct binding of methylcytosine (m–C) and histone interaction (direct or indirect). VIM1 is proposed to act as a ubiquitin (Ub) ligase to modify centromere chromatin structure through direct modification of nucleosomal histones or through modification of one or more chromatin effector proteins (X). An alteration in centromeric chromatin structure leads to DNA hypomethylation and decondensation of the centromere repeats.
Discussion
We describe the identification and isolation of the Arabidopsis VIM1 gene encoding a methylcytosine-binding protein required for maintenance of centromere DNA methylation and proper interphase centromere organization. The VIM1 gene was originally identified by a spontaneous loss-of-function mutation in a wild accession that did not exhibit a striking morphological phenotype. Plants are remarkably tolerant of dramatic alterations in DNA methylation relative to mammals. However, several Arabidopsis mutants with altered cytosine methylation suffer developmental abnormalities and reduced fertility under optimal laboratory conditions, suggesting that these mutants would compete poorly in natural settings. The persistence of the vim1-1 mutation in natural populations indicates that the alteration in centromere organization and DNA methylation caused by loss of VIM1 can be tolerated in the wild, and highlights the viability of mining natural variation for novel alleles that affect epigenetic regulation.
Loss of VIM1 function causes CpG and CpHpG hypomethylation at the centromere, but has no significant effect on DNA methylation of the FWA gene or pericentromeric sequences (5S rRNA genes and Athila retrotransposable elements). The apparent specificity of vim1 effects contrasts with the rather broad spectrum of genomic targets affected by mutations in other Arabidopsis genes involved in cytosine methylation (Rangwala and Richards 2004; Chan et al. 2005). Another interesting feature of the vim1-induced hypomethylation is that the centromeric repeats are completely remethylated in F1 hybrids created by outcrossing with wild-type plants. This behavior contrasts with the inheritance of the hypomethylated state of the centromere induced by ddm1 or met1 in crosses with wild-type plants. The efficient re-establishment of DNA methylation on the centromere repeats may reflect the presence in vim1 mutants of centromeric siRNA populations, which may target these repeats for de novo methylation (Lippman et al. 2003). The altered interphase organization of the centromere observed in vim1 mutants is also unique. Both ddm1 and met1 mutations cause a decondensation of pericentromeric sequences, which become dispersed from chromocenters while the 180-bp centromere repeats remain condensed within the chromocenters (Soppe et al. 2002). In contrast, vim1 mutants display a decondensation of the 180-bp centromere repeats, suggesting that VIM1 plays a role in centromere organization that is distinct from its function in DNA methylation.
VIM1 is a methylcytosine-binding protein that interacts with 5mCpG and 5mCpHpG
Methylcytosine-binding proteins bind 5mCpG dinucleotides regardless of sequence context. These proteins recruit a variety of HDAC complexes and chromatin remodeling factors, leading to chromatin compaction, and, consequently, to transcriptional repression (Klose and Bird 2006). In mammals, MeCP1 and MeCP2 (methyl-CpG-binding proteins 1 and 2) were the first protein or complexes identified with high affinity for methylated DNA (Meehan et al. 1989). Other mammalian methyl-CpG-binding proteins have been identified subsequently, on the basis of the ability to bind methylated sequences in vitro and/or the possession of a conserved MBD motif, including MBD2, MBD4, and Kaiso (which lacks an MBD) (Hendrich and Bird 1998; Prokhortchouk et al. 2001). The biological importance of MBD proteins is demonstrated by the wide range of severe phenotypes that result when genes encoding these proteins are mutated (Amir et al. 1999; Hendrich et al. 2001). In contrast to the considerable data on the role of mammalian methylcytosine-binding proteins, knowledge of plant methylcytosine-binding proteins is very limited. Arabidopsis contains 13 genes encoding MBD motifs. Three Arabidopsis MBD proteins (MBD5, MBD6, and MBD7) have been shown to bind symmetrically methylated CpG sites in vitro and two reports indicate that Arabidopsis MBD5 can also bind 5mCpHpH (Ito et al. 2003; Scebba et al. 2003; Zemach and Grafi 2003). The role of these MBD proteins in plant epigenetic regulation has not yet been characterized.
VIM1 has several interesting features as a methylcytosine-binding protein. First, VIM1 lacks an MBD domain and binds methylcytosine through a region encompassing the SRA domain. We note that the SRA domain in murine Np95 and related mammalian proteins is sufficient for 5mCpG binding (Unoki et al. 2004), but the region in VIM1 required for methylcytosine-binding includes the SRA domain and an additional segment (up to 61 amino acids) adjacent to this domain. Second, VIM1 is able to bind oligonucleotides that contain a single 5mCpG site. Third, VIM1 interacts with both 5mCpG and 5mCpHpG. This is the first report of a methylcytosine-binding protein able to bind 5mCpHpG, suggesting that VIM1 may play an important role in interpreting one of the non-CpG methylation systems found in plants.
Differences between VIM1 and mammalian proteins sharing structural similarity
VIM1 is a methylcytosine-binding protein that has a PHD domain, two RING finger domains, and an SRA domain. Proteins showing structural and amino acid similarity to VIM1 are found in animals as well as plants. The mammalian proteins most closely related to VIM1, human ICBP90 and NIRF, along with murine Np95, appear to function as transcriptional regulators tied into both cell cycle control and DNA repair (Hopfner et al. 2000; Bonapace et al. 2002; Muto et al. 2002; Arima et al. 2004). Despite their similarities, VIM1 shows several distinct differences from these mammalian proteins. First, the mammalian proteins have only one RING domain and an extra ubiquitin-like domain, and amino acid similarity between VIM1 and the mammalian proteins is restricted to the SRA domain. Second, VIM1 is associated with a range of heavily methylated genomic sequences, suggesting that VIM1 could play a role in maintaining heterochromatin throughout the genome. In contrast, ICBP90 regulates the transcription of specific genes, such as topoisomerase-IIα and Retinoblastoma protein 1, as well as participates in the maintenance of silencing of tumor suppressor genes when they become ectopically methylated (Hopfner et al. 2000; Unoki et al. 2004; Jeanblanc et al. 2005). Third, VIM1 is enriched at chromocenters, but Np95 and ICBP90 are broadly localized in the nucleus during interphase; Np95 is globally associated with chromatin (Miura et al. 2001), whereas ICBP90 is distributed throughout the entire nucleoplasm (Arima et al. 2004; Jeanblanc et al. 2005). Fourth, loss of Np95 or ICBP90 function has not yet been reported to affect DNA methylation of their target sequences.
Model for VIM1 action
Our working model is that VIM1 acts as a bridge between cytosine methylation and histone modification in a mutually reinforcing heterochromatin signaling loop that is required for proper centromere organization and centromere DNA methylation (Fig. 6B). VIM1 interacts with core histones and is able to bind 5mC-modified DNA, placing it physically at the interface between DNA methylation and chromatin packaging. The possibility that VIM1 is involved in histone modification is suggested by the demonstration that the SRA domain of ICBP90, Np95, and NIRF can recruit mammalian HDAC1 (Unoki et al. 2004). In addition, Np95, via its RING domain, possesses histone monoubiquitin ligase activity (Citterio et al. 2004). Although histone ubiquitylation is often associated with transcriptional activation, there are examples where this modification correlates with transcriptionally silent heterochromatin (Emre and Berger 2004; Wang et al. 2004). We note the recent discovery that a cullin 4-dependent E3 ubiquitin ligase, complexed with Rik1 and the HMT Clr4, plays a crucial role in formation of centromeric heterochromatin in fission yeast (Horn et al. 2005; Jia et al. 2005; Li et al. 2005; Thon et al. 2005). The in vivo substrates of the Rik1-associated E3 ubiquitin ligase complex are not known, but histone H2B copurified with the Rik1 complex, and this complex exhibited H2B polyubiquitylation activity in vitro (Horn et al. 2005).
Based on these considerations, we propose that VIM1 binds to methylated centromeric DNA and alters histone modification in the centromeric chromatin by recruiting HDACs and/or by ubiquitylation of core histones. The demonstrated interaction with VIM1 and histones H2B, H3, H4, and HTR12 makes these core histones good candidates for substrates of RING-domain mediated ubiquitylation. Alternatively, VIM1 might ubiquitylate a chromatin effector (or effectors), leading to an alteration of centromeric heterochromatin. Alterations in centromeric chromatin are postulated to lead to a reduction in centromeric DNA methylation due to the inefficient recruitment of DNMTs, such as CMT3 (recruited to heterochromatic histone methylation marks) (Lindroth et al. 2004) and DRM2 (which contains ubiquitin-binding domains) (Cao et al. 2000). Our results suggest that this new class of methylcytosine-binding proteins may play a different role in epigenetic regulation compared with MBD-class methylcytosine-binding proteins, which have not been demonstrated to participate in maintenance of genomic cytosine methylation patterns or centromeric heterochromatin.
Materials and methods
Plant material
Seeds of natural Arabidopsis strains and T-DNA insertion mutants (Alonso et al. 2003) were obtained from the Arabidopsis Biological Resources Center. The natural strains examined were part of the 96-member collection assembled by Nordborg et al. (2005). We also examined the original collection of Bor accessions made by J. Relichová (Masaryk University, Brno, Czech Republic) to confirm the presence to the vim1-1 allele in natural isolates. Plants were grown in a controlled environmental chamber at 22°C under long day conditions (16 h light per day).
DNA blot hybridization
Genomic DNA was digested with HpaII, MspI, or HhaI according to the manufacturer’s (New England Biolabs) instructions. Radiolabeled probes were generated by random priming, and blots were prepared and hybridized using standard methods. The following probes were generated from purified cloned inserts: 180-bp repeat (CEN) clone, pARR20-1 (Vongs et al. 1993), and 5S rRNA gene clone pCT4.1 (Campell et al. 1992). The Athila LTR and FWA probes were derived by PCR from genomic DNA (Soppe et al. 2000; Lindroth et al. 2001).
Construction of plant expression vectors and generation of transgenic plants
The 5.5-kb Col genomic DNA fragment containing 2.0 kb of the promoter region and the whole predicted ORF of VIM1 was PCR-amplified using two primers: 5′-CACCACGACCTTCG GAAGATGCAAGAA-3′ and 5′-CCTGATGGTCGCAGAAA CTGTTGC-3′. The fragment was cloned into pENTR-D TOPO (Invitrogen) and the resulting VIM1 insert was recombined into pEarlyGate302 (Earley et al. 2006). This construct was transformed into Agrobacterium tumefaciens (LBA4404) and was introduced into Bor-4 plants by in planta transformation (Bechtold and Pelletier 1998). A full-length VIM1 cDNA clone in pENTR-D TOPO was recombined into pEarlyGate202 (Earley et al. 2006) to add an N-terminal Flag tag for the ChIP and histone-binding assays. The resulting constructs were introduced into Col plants by standard infiltration protocols.
EMSA
Full-length VIM1 cDNA PCR product (amino acids 1–645) or deletion mutants—designated PHD/RING1 (amino acids 1–237), SRA/RING2 (amino acids 238–645), SRA (amino acids 238–456), RING2 (amino acids 457–645), and SRA+ (amino acids 238–517)—were cloned into the pDEST17 vector (Invitrogen). We synthesized VIM1 or derivatives in vitro using a T7 RNA polymerase-coupled wheat germ extract system (Promega TNTR). For binding assays, double-stranded probe DNA was end-labeled using the DIG gel shift kit (Roche) according to the manufacturer’s instructions. Labeled DNA (62 fmol) were incubated with in vitro translated VIM1 or its derivatives in the presence or absence of competitors (3.85 pmol). Binding reactions were carried out in 25 mM HEPES (pH 7.6), 50 mM KCl, 0.1 mM EDTA (pH 8.0), 12.5 mM MgCl2, 1 mM DTT, and 5% (w/v) glycerol; 1.5 μg poly-dIdC was added as a nonspecific competitor. After incubation for 30 min at room temperature, the reaction mixture was subjected to electrophoresis.
ChIP
ChIP was performed as described in Grendel et al. (2002) on nuclei prepared from wild-type and Arabidopsis plants expressing Flag-VIM1 using anti-Flag (Sigma). Precipitated DNAs were subjected to PCR using the following primers (see Supplementary Table 2): CEN-F + CEN-R; 5S rRNA-F + 5S rRNA-R; Athila-F + Athila-R; T5L23.29-F + T5L23.29-R; At4g04040-F + At4g04040-R. Amplification of CEN, 5S rRNA, Athila, or At4g04040 was performed for 26, 22, 30, or 32 cycles, respectively.
Immunolocalization/DNA-FISH
Root meristems from 14-d-old plants were excised and nuclei were extracted as described previously (Lawrence et al. 2004). After post-fixation in 4% formaldehyde/PBS (phosphate-buffered saline), washes in PBS, and blocking at 37°C, slides were exposed overnight to primary antisera mouse anti-Flag (1:100, Sigma) and rabbit anti-HTR12 (Talbert et al. 2002) in PBS and 0.5% blocking reagent (Roche). After washes in PBS, slides were incubated at 37°C with anti-mouse-FITC diluted 1:100 (Sigma) or goat anti-rabbit-TRITC (1:300, Sigma). For combined protein/DNA-FISH localization experiments, slides were first subjected to immunolocalization, and then post-fixed in 4% formaldehyde/PBS followed by in situ hybridization. FISH using the CEN repeat probe (pARR20-1) labeled with biotin-dUTP and DIG-dUTP was performed as described (Pontes et al. 2004). CEN repeats were detected using either goat anti-biotin conjugated with avidin (1:200, Vector Laboratories) followed by streptavidin-Alexa 543 (Molecular Probes) or anti-digoxeninin (1:200, Roche) and rabbit anti-mouse Alexa 488 (Molecular Probes). DNA was counterstained with DAPI (1 μg/mL) in Vectashield (Vector Laboratories). Nuclei were examined using a Nikon Eclipse E800i epifluorescence microscope, with images collected using a Photometrics Coolsnap ES Mono digital camera. The images were pseudocolored, merged, and processed using Adobe Photoshop (Adobe Systems).
VIM1–histone interaction
Epitope-tagged (myc) recombinant histones were expressed in E. coli BL21(AI) (Invitrogen) and cells were lysed by sonication in a buffer containing 500 mM NaCl and 20 mM Tris-HCl (pH 7.5). For purification, anti-c-Myc agarose conjugate (Sigma) was added to cleared lysates and incubated for 2 h at 4°C. Beads were washed in buffer containing 150 mM NaCl, 20 mM Tris-HCl (pH 7.5), 5 mM MgCl2, and 0.5% NP-40. Total proteins were extracted from transgenic plants overexpressing Flag-VIM1 by grinding tissue in an extraction buffer containing 150 mM NaCl, 20 mM Tris-HCl (pH 7.5), and 5 mM MgCl2. Bead-bound myc-tagged histones were incubated with extracts from transgenic plants overexpressing Flag-VIM1 overnight at 4°C. Beads were washed as described above. Proteins were eluted by adding SDS-PAGE sample buffer and were analyzed by SDS-PAGE electrophoresis followed by Western blotting using anti-myc or anti-Flag antibodies (Sigma).
Acknowledgments
We thank T. Dittmer, H.-f. Kuo, S. Rangwala, T.R. Smith, and H. Yi for comments on the manuscript. We are grateful to J. Relichová for providing Bor natural accessions, and J. Haag (histone constructs) and S. Henikoff (anti-HTR12 antibody) for providing reagents. This work was supported by grants from the National Science Foundation to E.J.R (MCB-0321990) and the Monsanto Company. DNA-FISH and immunolocalization was performed by O.P., who was supported by fellowship SFRH/BPD/17508/2004 from the Fundação para a Ciência e Tecnologia (Portugal) and by NIH grants R01GM60380 and R01GM077590 to C.S.P. We thank the Arabidopsis Biological Resource Center at The Ohio State University for providing Arabidopsis strains and SALK T-DNA insertion mutants.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1512007
References
- Alonso J.M., Stepanova A.N., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Stepanova A.N., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Zimmerman J., Barajas P., Cheuk R., Barajas P., Cheuk R., Cheuk R., et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Amir R.E., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y., Wan M., Tran C.Q., Francke U., Zoghbi H.Y., Tran C.Q., Francke U., Zoghbi H.Y., Francke U., Zoghbi H.Y., Zoghbi H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Arima Y., Hirota T., Bronner C., Mousli M., Fujiwara T., Niwa S., Ishikawa H., Saya H., Hirota T., Bronner C., Mousli M., Fujiwara T., Niwa S., Ishikawa H., Saya H., Bronner C., Mousli M., Fujiwara T., Niwa S., Ishikawa H., Saya H., Mousli M., Fujiwara T., Niwa S., Ishikawa H., Saya H., Fujiwara T., Niwa S., Ishikawa H., Saya H., Niwa S., Ishikawa H., Saya H., Ishikawa H., Saya H., Saya H. Down-regulation of nuclear protein ICBP90 by p53/p21Cip1/WAF1-dependent DNA-damage checkpoint signals contributes to cell cycle arrest at G1/S transition. Genes Cells. 2004;9:131–142. doi: 10.1111/j.1356-9597.2004.00710.x. [DOI] [PubMed] [Google Scholar]
- Bechtold N., Pelletier G., Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- Bonapace I.M., Latella L., Papait R., Nicassio F., Sacco A., Muto M., Crescenzi M., Di Fiore P.P., Latella L., Papait R., Nicassio F., Sacco A., Muto M., Crescenzi M., Di Fiore P.P., Papait R., Nicassio F., Sacco A., Muto M., Crescenzi M., Di Fiore P.P., Nicassio F., Sacco A., Muto M., Crescenzi M., Di Fiore P.P., Sacco A., Muto M., Crescenzi M., Di Fiore P.P., Muto M., Crescenzi M., Di Fiore P.P., Crescenzi M., Di Fiore P.P., Di Fiore P.P. Np95 is regulated by E1A during mitotic reactivation of terminally differentiated cells and is essential for S phase entry. J. Cell Biol. 2002;157:909–914. doi: 10.1083/jcb.200201025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campell B.R., Song Y., Posch T.E., Cullis C.A., Town C.D., Song Y., Posch T.E., Cullis C.A., Town C.D., Posch T.E., Cullis C.A., Town C.D., Cullis C.A., Town C.D., Town C.D. Sequence and organization of 5S ribosomal RNA-encoding genes of Arabidopsis thaliana. Gene. 1992;112:225–228. doi: 10.1016/0378-1119(92)90380-8. [DOI] [PubMed] [Google Scholar]
- Cao X., Springer N.M., Muszynski M.G., Phillips R.L., Kaeppler S., Jacobsen S.E., Springer N.M., Muszynski M.G., Phillips R.L., Kaeppler S., Jacobsen S.E., Muszynski M.G., Phillips R.L., Kaeppler S., Jacobsen S.E., Phillips R.L., Kaeppler S., Jacobsen S.E., Kaeppler S., Jacobsen S.E., Jacobsen S.E. Conserved plant genes with similarity to mammalian de novo DNA methyltransferases. Proc. Natl. Acad. Sci. 2000;97:4979–4984. doi: 10.1073/pnas.97.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.W., Henderson I.R., Jacobsen S.E., Henderson I.R., Jacobsen S.E., Jacobsen S.E. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 2005;6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- Citterio E., Papait R., Nicassio F., Vecchi M., Gomiero P., Mantovani R., Di Fiore P.P., Bonapace I.M., Papait R., Nicassio F., Vecchi M., Gomiero P., Mantovani R., Di Fiore P.P., Bonapace I.M., Nicassio F., Vecchi M., Gomiero P., Mantovani R., Di Fiore P.P., Bonapace I.M., Vecchi M., Gomiero P., Mantovani R., Di Fiore P.P., Bonapace I.M., Gomiero P., Mantovani R., Di Fiore P.P., Bonapace I.M., Mantovani R., Di Fiore P.P., Bonapace I.M., Di Fiore P.P., Bonapace I.M., Bonapace I.M. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol. Cell. Biol. 2004;24:2526–2535. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig J.M. Heterochromatin—Many flavours, common themes. Bioessays. 2005;27:17–28. doi: 10.1002/bies.20145. [DOI] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S., Opper K., Juehne T., Song K., Pikaard C.S., Juehne T., Song K., Pikaard C.S., Song K., Pikaard C.S., Pikaard C.S. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Emre N.C., Berger S.L., Berger S.L. Histone H2B ubiquitylation and deubiquitylation in genomic regulation. Cold Spring Harbor Symp. Quant. Biol. 2004;69:289–299. doi: 10.1101/sqb.2004.69.289. [DOI] [PubMed] [Google Scholar]
- Fujimori A., Matsuda Y., Takemoto Y., Hashimoto Y., Kubo E., Araki R., Fukumura R., Mita K., Tatsumi K., Muto M., Matsuda Y., Takemoto Y., Hashimoto Y., Kubo E., Araki R., Fukumura R., Mita K., Tatsumi K., Muto M., Takemoto Y., Hashimoto Y., Kubo E., Araki R., Fukumura R., Mita K., Tatsumi K., Muto M., Hashimoto Y., Kubo E., Araki R., Fukumura R., Mita K., Tatsumi K., Muto M., Kubo E., Araki R., Fukumura R., Mita K., Tatsumi K., Muto M., Araki R., Fukumura R., Mita K., Tatsumi K., Muto M., Fukumura R., Mita K., Tatsumi K., Muto M., Mita K., Tatsumi K., Muto M., Tatsumi K., Muto M., Muto M. Cloning and mapping of Np95 gene which encodes a novel nuclear protein associated with cell proliferation. Mamm. Genome. 1998;9:1032–1035. doi: 10.1007/s003359900920. [DOI] [PubMed] [Google Scholar]
- Fuks F., Hurd P.J., Deplus R., Kouzarides T., Hurd P.J., Deplus R., Kouzarides T., Deplus R., Kouzarides T., Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Yordan C., Colot V., Martienssen R.A., Lippman Z., Yordan C., Colot V., Martienssen R.A., Yordan C., Colot V., Martienssen R.A., Colot V., Martienssen R.A., Martienssen R.A. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science. 2002;297:1871–1873. doi: 10.1126/science.1074950. [DOI] [PubMed] [Google Scholar]
- Genereux D.P., Miner B.E., Bergstrom C.T., Laird C.D., Miner B.E., Bergstrom C.T., Laird C.D., Bergstrom C.T., Laird C.D., Laird C.D. A population-epigenetic model to infer site-specific methylation rates from double-stranded DNA methylation patterns. Proc. Natl. Acad. Sci. 2005;102:5802–5807. doi: 10.1073/pnas.0502036102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll M.G., Bestor T.H., Bestor T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Hendrich B., Bird A., Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B., Guy J., Ramsahoye B., Wilson V.A., Bird A., Guy J., Ramsahoye B., Wilson V.A., Bird A., Ramsahoye B., Wilson V.A., Bird A., Wilson V.A., Bird A., Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes & Dev. 2001;15:710–723. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Dalal Y., Dalal Y. Centromeric chromatin: What makes it unique? Curr. Opin. Genet. Dev. 2005;15:177–184. doi: 10.1016/j.gde.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Hopfner R., Mousli M., Jeltsch J.M., Voulgaris A., Lutz Y., Marin C., Bellocq J.P., Oudet P., Bronner C., Mousli M., Jeltsch J.M., Voulgaris A., Lutz Y., Marin C., Bellocq J.P., Oudet P., Bronner C., Jeltsch J.M., Voulgaris A., Lutz Y., Marin C., Bellocq J.P., Oudet P., Bronner C., Voulgaris A., Lutz Y., Marin C., Bellocq J.P., Oudet P., Bronner C., Lutz Y., Marin C., Bellocq J.P., Oudet P., Bronner C., Marin C., Bellocq J.P., Oudet P., Bronner C., Bellocq J.P., Oudet P., Bronner C., Oudet P., Bronner C., Bronner C. ICBP90, a novel human CCAAT binding protein, involved in the regulation of topoisomerase IIα expression. Cancer Res. 2000;60:121–128. [PubMed] [Google Scholar]
- Horn P.J., Bastie J.N., Peterson C.L., Bastie J.N., Peterson C.L., Peterson C.L. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes & Dev. 2005;19:1705–1714. doi: 10.1101/gad.1328005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga K.L., Brower-Toland B., Elgin S.C., Brower-Toland B., Elgin S.C., Elgin S.C. The contradictory definitions of heterochromatin: Transcription and silencing. Chromosoma. 2006;115:110–122. doi: 10.1007/s00412-006-0052-x. [DOI] [PubMed] [Google Scholar]
- Ito M., Koike A., Koizumi N., Sano H., Koike A., Koizumi N., Sano H., Koizumi N., Sano H., Sano H. Methylated DNA-binding proteins from Arabidopsis. Plant Physiol. 2003;133:1747–1754. doi: 10.1104/pp.103.026708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R., Bird A., Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33 (Suppl.):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Jeanblanc M., Mousli M., Hopfner R., Bathami K., Martinet N., Abbady A.Q., Siffert J.C., Mathieu E., Muller C.D., Bronner C., Mousli M., Hopfner R., Bathami K., Martinet N., Abbady A.Q., Siffert J.C., Mathieu E., Muller C.D., Bronner C., Hopfner R., Bathami K., Martinet N., Abbady A.Q., Siffert J.C., Mathieu E., Muller C.D., Bronner C., Bathami K., Martinet N., Abbady A.Q., Siffert J.C., Mathieu E., Muller C.D., Bronner C., Martinet N., Abbady A.Q., Siffert J.C., Mathieu E., Muller C.D., Bronner C., Abbady A.Q., Siffert J.C., Mathieu E., Muller C.D., Bronner C., Siffert J.C., Mathieu E., Muller C.D., Bronner C., Mathieu E., Muller C.D., Bronner C., Muller C.D., Bronner C., Bronner C. The retinoblastoma gene and its product are targeted by ICBP90: A key mechanism in the G1/S transition during the cell cycle. Oncogene. 2005;24:7337–7345. doi: 10.1038/sj.onc.1208878. [DOI] [PubMed] [Google Scholar]
- Jenuwein T., Allis C.D., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jia S., Kobayashi R., Grewal S.I., Kobayashi R., Grewal S.I., Grewal S.I. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat. Cell Biol. 2005;7:1007–1013. doi: 10.1038/ncb1300. [DOI] [PubMed] [Google Scholar]
- Kakutani T., Munakata K., Richards E.J., Hirochika H., Munakata K., Richards E.J., Hirochika H., Richards E.J., Hirochika H., Hirochika H. Meiotically and mitotically stable inheritance of DNA hypomethylation induced by ddm1 mutation of Arabidopsis thaliana. Genetics. 1999;151:831–838. doi: 10.1093/genetics/151.2.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankel M.W., Ramsey D.E., Stokes T.L., Flowers S.K., Haag J.R., Jeddeloh J.A., Riddle N.C., Verbsky M.L., Richards E.J., Ramsey D.E., Stokes T.L., Flowers S.K., Haag J.R., Jeddeloh J.A., Riddle N.C., Verbsky M.L., Richards E.J., Stokes T.L., Flowers S.K., Haag J.R., Jeddeloh J.A., Riddle N.C., Verbsky M.L., Richards E.J., Flowers S.K., Haag J.R., Jeddeloh J.A., Riddle N.C., Verbsky M.L., Richards E.J., Haag J.R., Jeddeloh J.A., Riddle N.C., Verbsky M.L., Richards E.J., Jeddeloh J.A., Riddle N.C., Verbsky M.L., Richards E.J., Riddle N.C., Verbsky M.L., Richards E.J., Verbsky M.L., Richards E.J., Richards E.J. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003;163:1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R.J., Bird A.P., Bird A.P. Genomic DNA methylation: The mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Lawrence R.J., Earley K., Pontes O., Silva M., Chen Z.J., Neves N., Viegas W., Pikaard C.S., Earley K., Pontes O., Silva M., Chen Z.J., Neves N., Viegas W., Pikaard C.S., Pontes O., Silva M., Chen Z.J., Neves N., Viegas W., Pikaard C.S., Silva M., Chen Z.J., Neves N., Viegas W., Pikaard C.S., Chen Z.J., Neves N., Viegas W., Pikaard C.S., Neves N., Viegas W., Pikaard C.S., Viegas W., Pikaard C.S., Pikaard C.S. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell. 2004;13:599–609. doi: 10.1016/s1097-2765(04)00064-4. [DOI] [PubMed] [Google Scholar]
- Lehnertz B., Ueda Y., Derijck A.A., Braunschweig U., Perez-Burgos L., Kubicek S., Chen T., Li E., Jenuwein T., Peters A.H., Ueda Y., Derijck A.A., Braunschweig U., Perez-Burgos L., Kubicek S., Chen T., Li E., Jenuwein T., Peters A.H., Derijck A.A., Braunschweig U., Perez-Burgos L., Kubicek S., Chen T., Li E., Jenuwein T., Peters A.H., Braunschweig U., Perez-Burgos L., Kubicek S., Chen T., Li E., Jenuwein T., Peters A.H., Perez-Burgos L., Kubicek S., Chen T., Li E., Jenuwein T., Peters A.H., Kubicek S., Chen T., Li E., Jenuwein T., Peters A.H., Chen T., Li E., Jenuwein T., Peters A.H., Li E., Jenuwein T., Peters A.H., Jenuwein T., Peters A.H., Peters A.H. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- Li F., Goto D.B., Zaratiegui M., Tang X., Martienssen R., Cande W.Z., Goto D.B., Zaratiegui M., Tang X., Martienssen R., Cande W.Z., Zaratiegui M., Tang X., Martienssen R., Cande W.Z., Tang X., Martienssen R., Cande W.Z., Martienssen R., Cande W.Z., Cande W.Z. Two novel proteins, dos1 and dos2, interact with rik1 to regulate heterochromatic RNA interference and histone modification. Curr. Biol. 2005;15:1448–1457. doi: 10.1016/j.cub.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Lindroth A.M., Cao X., Jackson J.P., Zilberman D., McCallum C.M., Henikoff S., Jacobsen S.E., Cao X., Jackson J.P., Zilberman D., McCallum C.M., Henikoff S., Jacobsen S.E., Jackson J.P., Zilberman D., McCallum C.M., Henikoff S., Jacobsen S.E., Zilberman D., McCallum C.M., Henikoff S., Jacobsen S.E., McCallum C.M., Henikoff S., Jacobsen S.E., Henikoff S., Jacobsen S.E., Jacobsen S.E. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292:2077–2080. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- Lindroth A.M., Shultis D., Jasencakova Z., Fuchs J., Johnson L., Schubert D., Patnaik D., Pradhan S., Goodrich J., Schubert I., Shultis D., Jasencakova Z., Fuchs J., Johnson L., Schubert D., Patnaik D., Pradhan S., Goodrich J., Schubert I., Jasencakova Z., Fuchs J., Johnson L., Schubert D., Patnaik D., Pradhan S., Goodrich J., Schubert I., Fuchs J., Johnson L., Schubert D., Patnaik D., Pradhan S., Goodrich J., Schubert I., Johnson L., Schubert D., Patnaik D., Pradhan S., Goodrich J., Schubert I., Schubert D., Patnaik D., Pradhan S., Goodrich J., Schubert I., Patnaik D., Pradhan S., Goodrich J., Schubert I., Pradhan S., Goodrich J., Schubert I., Goodrich J., Schubert I., Schubert I., et al. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 2004;23:4286–4296. doi: 10.1038/sj.emboj.7600430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z., May B., Yordan C., Singer T., Martienssen R., May B., Yordan C., Singer T., Martienssen R., Yordan C., Singer T., Martienssen R., Singer T., Martienssen R., Martienssen R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 2003;1:E67. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z., Gendrel A.V., Black M., Vaughn M.W., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., Gendrel A.V., Black M., Vaughn M.W., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., Black M., Vaughn M.W., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., Vaughn M.W., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., Lavine K., Mittal V., May B., Kasschau K.D., Mittal V., May B., Kasschau K.D., May B., Kasschau K.D., Kasschau K.D., et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- Matzke M.A., Birchler J.A., Birchler J.A. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- Meehan R.R., Lewis J.D., McKay S., Kleiner E.L., Bird A.P., Lewis J.D., McKay S., Kleiner E.L., Bird A.P., McKay S., Kleiner E.L., Bird A.P., Kleiner E.L., Bird A.P., Bird A.P. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- Miura M., Watanabe H., Sasaki T., Tatsumi K., Muto M., Watanabe H., Sasaki T., Tatsumi K., Muto M., Sasaki T., Tatsumi K., Muto M., Tatsumi K., Muto M., Muto M. Dynamic changes in subnuclear NP95 location during the cell cycle and its spatial relationship with DNA replication foci. Exp. Cell Res. 2001;263:202–208. doi: 10.1006/excr.2000.5115. [DOI] [PubMed] [Google Scholar]
- Mori T., Li Y., Hata H., Ono K., Kochi H., Li Y., Hata H., Ono K., Kochi H., Hata H., Ono K., Kochi H., Ono K., Kochi H., Kochi H. NIRF, a novel RING finger protein, is involved in cell-cycle regulation. Biochem. Biophys. Res. Commun. 2002;296:530–536. doi: 10.1016/s0006-291x(02)00890-2. [DOI] [PubMed] [Google Scholar]
- Muto M., Kanari Y., Kubo E., Takabe T., Kurihara T., Fujimori A., Tatsumi K., Kanari Y., Kubo E., Takabe T., Kurihara T., Fujimori A., Tatsumi K., Kubo E., Takabe T., Kurihara T., Fujimori A., Tatsumi K., Takabe T., Kurihara T., Fujimori A., Tatsumi K., Kurihara T., Fujimori A., Tatsumi K., Fujimori A., Tatsumi K., Tatsumi K. Targeted disruption of Np95 gene renders murine embryonic stem cells hypersensitive to DNA damaging agents and DNA replication blocks. J. Biol. Chem. 2002;277:34549–34555. doi: 10.1074/jbc.M205189200. [DOI] [PubMed] [Google Scholar]
- Nightingale K.P., O’Neill L.P., Turner B.M., O’Neill L.P., Turner B.M., Turner B.M. Histone modifications: Signalling receptors and potential elements of a heritable epigenetic code. Curr. Opin. Genet. Dev. 2006;16:125–136. doi: 10.1016/j.gde.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Nordborg M., Hu T.T., Ishino Y., Jhaveri J., Toomajian C., Zheng H., Bakker E., Calabrese P., Gladstone J., Goyal R., Hu T.T., Ishino Y., Jhaveri J., Toomajian C., Zheng H., Bakker E., Calabrese P., Gladstone J., Goyal R., Ishino Y., Jhaveri J., Toomajian C., Zheng H., Bakker E., Calabrese P., Gladstone J., Goyal R., Jhaveri J., Toomajian C., Zheng H., Bakker E., Calabrese P., Gladstone J., Goyal R., Toomajian C., Zheng H., Bakker E., Calabrese P., Gladstone J., Goyal R., Zheng H., Bakker E., Calabrese P., Gladstone J., Goyal R., Bakker E., Calabrese P., Gladstone J., Goyal R., Calabrese P., Gladstone J., Goyal R., Gladstone J., Goyal R., Goyal R., et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelissier T., Tutois S., Deragon J.M., Tourmente S., Genestier S., Picard G., Tutois S., Deragon J.M., Tourmente S., Genestier S., Picard G., Deragon J.M., Tourmente S., Genestier S., Picard G., Tourmente S., Genestier S., Picard G., Genestier S., Picard G., Picard G. Athila, a new retroelement from Arabidopsis thaliana. Plant Mol. Biol. 1995;29:441–452. doi: 10.1007/BF00020976. [DOI] [PubMed] [Google Scholar]
- Pontes O., Neves N., Silva M., Lewis M.S., Madlung A., Comai L., Viegas W., Pikaard C.S., Neves N., Silva M., Lewis M.S., Madlung A., Comai L., Viegas W., Pikaard C.S., Silva M., Lewis M.S., Madlung A., Comai L., Viegas W., Pikaard C.S., Lewis M.S., Madlung A., Comai L., Viegas W., Pikaard C.S., Madlung A., Comai L., Viegas W., Pikaard C.S., Comai L., Viegas W., Pikaard C.S., Viegas W., Pikaard C.S., Pikaard C.S. Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc. Natl. Acad. Sci. 2004;101:18240–18245. doi: 10.1073/pnas.0407258102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhortchouk A., Hendrich B., Jorgensen H., Ruzov A., Wilm M., Georgiev G., Bird A., Prokhortchouk E., Hendrich B., Jorgensen H., Ruzov A., Wilm M., Georgiev G., Bird A., Prokhortchouk E., Jorgensen H., Ruzov A., Wilm M., Georgiev G., Bird A., Prokhortchouk E., Ruzov A., Wilm M., Georgiev G., Bird A., Prokhortchouk E., Wilm M., Georgiev G., Bird A., Prokhortchouk E., Georgiev G., Bird A., Prokhortchouk E., Bird A., Prokhortchouk E., Prokhortchouk E. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes & Dev. 2001;15:1613–1618. doi: 10.1101/gad.198501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangwala S.H., Richards E.J., Richards E.J. The value-added genome: Building and maintaining genomic cytosine methylation landscapes. Curr. Opin. Genet. Dev. 2004;14:686–691. doi: 10.1016/j.gde.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Richards E.J. Inherited epigenetic variation—Revisiting soft inheritance. Nat. Rev. Genet. 2006;7:395–401. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- Saze H., Scheid O.M., Paszkowski J., Scheid O.M., Paszkowski J., Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat. Genet. 2003;34:65–69. doi: 10.1038/ng1138. [DOI] [PubMed] [Google Scholar]
- Scebba F., Bernacchia G., De Bastiani M., Evangelista M., Cantoni R.M., Cella R., Locci M.T., Pitto L., Bernacchia G., De Bastiani M., Evangelista M., Cantoni R.M., Cella R., Locci M.T., Pitto L., De Bastiani M., Evangelista M., Cantoni R.M., Cella R., Locci M.T., Pitto L., Evangelista M., Cantoni R.M., Cella R., Locci M.T., Pitto L., Cantoni R.M., Cella R., Locci M.T., Pitto L., Cella R., Locci M.T., Pitto L., Locci M.T., Pitto L., Pitto L. Arabidopsis MBD proteins show different binding specificities and nuclear localization. Plant Mol. Biol. 2003;53:715–731. doi: 10.1023/B:PLAN.0000019118.56822.a9. [DOI] [PubMed] [Google Scholar]
- Soppe W.J., Jacobsen S.E., Alonso-Blanco C., Jackson J.P., Kakutani T., Koornneef M., Peeters A.J., Jacobsen S.E., Alonso-Blanco C., Jackson J.P., Kakutani T., Koornneef M., Peeters A.J., Alonso-Blanco C., Jackson J.P., Kakutani T., Koornneef M., Peeters A.J., Jackson J.P., Kakutani T., Koornneef M., Peeters A.J., Kakutani T., Koornneef M., Peeters A.J., Koornneef M., Peeters A.J., Peeters A.J. The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol. Cell. 2000;6:791–802. doi: 10.1016/s1097-2765(05)00090-0. [DOI] [PubMed] [Google Scholar]
- Soppe W.J., Jasencakova Z., Houben A., Kakutani T., Meister A., Huang M.S., Jacobsen S.E., Schubert I., Fransz P.F., Jasencakova Z., Houben A., Kakutani T., Meister A., Huang M.S., Jacobsen S.E., Schubert I., Fransz P.F., Houben A., Kakutani T., Meister A., Huang M.S., Jacobsen S.E., Schubert I., Fransz P.F., Kakutani T., Meister A., Huang M.S., Jacobsen S.E., Schubert I., Fransz P.F., Meister A., Huang M.S., Jacobsen S.E., Schubert I., Fransz P.F., Huang M.S., Jacobsen S.E., Schubert I., Fransz P.F., Jacobsen S.E., Schubert I., Fransz P.F., Schubert I., Fransz P.F., Fransz P.F. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 2002;21:6549–6559. doi: 10.1093/emboj/cdf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P.B., Masuelli R., Tyagi A.P., Comai L., Henikoff S., Masuelli R., Tyagi A.P., Comai L., Henikoff S., Tyagi A.P., Comai L., Henikoff S., Comai L., Henikoff S., Henikoff S. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell. 2002;14:1053–1066. doi: 10.1105/tpc.010425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H., Selker E.U., Selker E.U. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- Thon G., Hansen K.R., Altes S.P., Sidhu D., Singh G., Verhein-Hansen J., Bonaduce M.J., Klar A.J., Hansen K.R., Altes S.P., Sidhu D., Singh G., Verhein-Hansen J., Bonaduce M.J., Klar A.J., Altes S.P., Sidhu D., Singh G., Verhein-Hansen J., Bonaduce M.J., Klar A.J., Sidhu D., Singh G., Verhein-Hansen J., Bonaduce M.J., Klar A.J., Singh G., Verhein-Hansen J., Bonaduce M.J., Klar A.J., Verhein-Hansen J., Bonaduce M.J., Klar A.J., Bonaduce M.J., Klar A.J., Klar A.J. The Clr7 and Clr8 directionality factors and the Pcu4 cullin mediate heterochromatin formation in the fission yeast Schizosaccharomyces pombe. Genetics. 2005;171:1583–1595. doi: 10.1534/genetics.105.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unoki M., Nishidate T., Nakamura Y., Nishidate T., Nakamura Y., Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23:7601–7610. doi: 10.1038/sj.onc.1208053. [DOI] [PubMed] [Google Scholar]
- Vongs A., Kakutani T., Martienssen R.A., Richards E.J., Kakutani T., Martienssen R.A., Richards E.J., Martienssen R.A., Richards E.J., Richards E.J. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R.S., Zhang Y., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R.S., Zhang Y., Erdjument-Bromage H., Vidal M., Tempst P., Jones R.S., Zhang Y., Vidal M., Tempst P., Jones R.S., Zhang Y., Tempst P., Jones R.S., Zhang Y., Jones R.S., Zhang Y., Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Zemach A., Grafi G., Grafi G. Characterization of Arabidopsis thaliana methyl-CpG-binding domain (MBD) proteins. Plant J. 2003;34:565–572. doi: 10.1046/j.1365-313x.2003.01756.x. [DOI] [PubMed] [Google Scholar]