Abstract
The roles of DNA methyltransferase-2 (DNMT2) enzymes are controversial; whether DNMT2 functions primarily as a nuclear DNA methyltransferase or as a cytoplasmic tRNA methyltransferase, and whether DNMT2 activity impacts development, as dnmt2 mutant mice or Drosophila lack phenotypes. Here we show that morpholino knockdown of Dnmt2 protein in zebrafish embryos confers differentiation defects in particular organs, including the retina, liver, and brain. Importantly, proper organ differentiation required Dnmt2 activity in the cytoplasm, not in the nucleus. Furthermore, zebrafish Dnmt2 methylates an RNA species of ∼80 bases, consistent with tRNA methylation. Thus, Dnmt2 promotes zebrafish development, likely through cytoplasmic RNA methylation.
Keywords: dnmt2, zebrafish, methylation, liver, retina, tRNA
DNA methylation provides a mechanism for the stable and heritable silencing of transcription (Goll and Bestor 2005); methylated cytosines in CpG-rich regions are bound by transcriptional repressors of the methyl-binding domain (MBD) families, which interact with chromatin-silencing machinery (Klose and Bird 2006). Methylation of cytosines is catalyzed by DNA methyltransferases (DNMTs), which can be classified into three families: DNMT1/2/3 (Goll and Bestor 2005). Generally, vertebrates bear only one Dnmt1 gene and one Dnmt2 gene, but contain several Dnmt3-family genes. Changes in DNA methylation accompany embryonic development (Morgan et al. 2005), and mice lacking Dnmt1 or Dnmt3b die during early embryogenesis, whereas those lacking Dnmt3a die 4 wk after birth (Li et al. 1992; Okano et al. 1999). In zebrafish, Dnmt1 promotes tissues-specific terminal differentiation of multiple organs (Rai et al. 2006).
In contrast to the DNMT1 and DNMT3 families, the functional importance of the DNMT2 family in vertebrates has remained elusive; mice lacking Dnmt2 did not display any defects, and no reduction in global DNA methylation levels were observed in Dnmt2−/− ES cells (Okano et al. 1998). Drosophila bear extremely low levels of DNA methylation; however, mutation of Dnmt2 led to methylation loss (Kunert et al. 2003). Also, Dnmt2 overexpression in flies moderately increased DNA methylation, but the increased methylation observed was not restricted to a CpG context, as cytosine methylation of CpA and CpT was observed (Kunert et al. 2003). These results would appear to support a role for Dnmt2 in DNA methylation; however, omission of Dnmt2 in flies did not confer an observable phenotype (Kunert et al. 2003). Certain lower eukaryotes like Schizosaccharomyces pombe and Dictostylieum bear a Dnmt2 homolog but lack Dnmt1 or Dnmt3 homologs (Goll and Bestor 2005; Jeltsch et al. 2006). S. pombe lacks detectable DNA methylation, suggesting that its Dnmt2 ortholog has an alternative function, although no phenotype resulted from Dnmt2 (PMT1) omission (Wilkinson et al. 1995). In Dictostylieum, <0.2% of CpG regions are methylated, and this methylation is largely restricted to repetitive regions and retrotransposons (Katoh et al. 2006). Here, loss of the distant Dnmt2 homolog dnmA reduced methylation levels twofold and conferred a late developmental defect, raising the possibility that repeat methylation has a developmental consequence (Katoh et al. 2006). DNMT2 displays very weak DNA methyltransferase activity in vitro, in spite of a high degree of identity with DNMT1 and DNMT3B in its catalytic domain, enzymes with relatively robust activity in vitro (Hermann et al. 2003; Goll and Bestor 2005). Thus, certain studies provide a measure of support for DNMT2 as a DNA methyltransferase, while other studies question whether DNA is the primary target.
In support of an alternative target, DNMT2 was recently shown to reside mainly in the cytoplasm in human cells (Goll et al. 2006). Importantly, DNMT2 was shown to methylate cytosine 38 in the anti-codon loop of tRNAAsp in mice, Arabidopsis and Drosophila (Goll et al. 2006). However, as no function is known for tRNAAsp methylation at that position, and as there is no phenotype conferred in higher eukaryotes from loss of Dnmt2 (Goll et al. 2006), the biological purpose of the enzymes of the DNMT2 class in vertebrates remains unknown. These studies raise at least two important questions: First, what biological process in higher eukaryotes relies on Dnmt2? Second, does DNMT2 protein function in the nucleus, consistent with DNA methylation, or rather in the cytoplasm, through an RNA target such as tRNAs? Here we show that the catalytic activity of Dnmt2 in the cytoplasm, but not the nucleus, is important for proper terminal differentiation of particular organs in the zebrafish, establishing a role for this protein in vertebrate development.
Results and Discussion
To examine the role of dnmt2 in zebrafish development, we first characterized the expression pattern of dnmt2 using whole-mount in situ hybridization during early embryonic development. (We note that, by convention, zebrafish genes are depicted in lowercase.) dnmt2 RNA was present at the eight-cell stage, prior to zygotic transcription, suggesting that a pool of dnmt2 mRNA is contributed from the mother (Supplementary Fig. 1A). dnmt2 RNA was expressed ubiquitously from the bud stage onward. At 24 h post-fertilization (hpf), dnmt2 RNA was present in the eyes and the brain, and faint but specific staining was seen in the liver at 72 hpf (Supplementary Fig. 1A). Quantitative RT–PCR revealed that dnmt2 RNA levels are highest following fertilization, diminish at 6 hpf stage until 30 hpf, and are then partially restored during 48–120 hpf (Supplementary Fig. 1B). Dnmt2 protein abundance was very low during the early period just following fertilization (Supplementary Fig. 1C), suggesting either an inhibition of translation or high turnover of dnmt2 RNA during this period. Alternatively, a dnmt2 psuedogene may be present that is detected by in situ analysis. Regardless, Dnmt2 protein was absent before 36 hpf, and then accumulated in accordance with the increased mRNA abundance during the 36–96 hpf time period (Supplementary Fig. 1C).
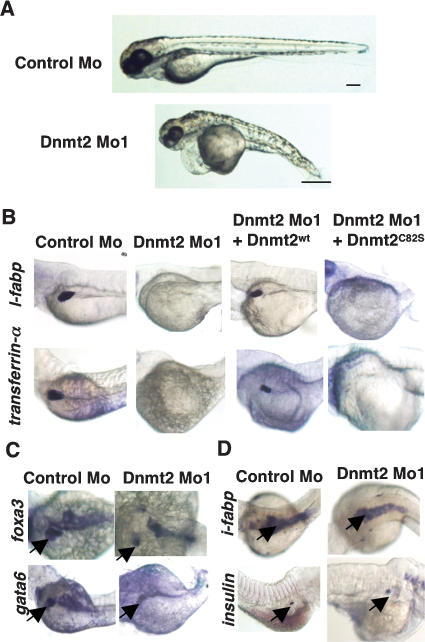
To determine whether the loss of Dnmt2 protein impacts development, we greatly reduced Dnmt2 protein levels by employing two different antisense morpholino oligonucleotides (morpholinos), in separate experiments. One morpholino targeted the AUG codon and flanking RNA to block translation initiation, whereas the other morpholino targeted the exon1–intron1 boundary region of the RNA, to block splicing. Each morpholino was injected separately into embryos at the one-cell stage, which elicited large reductions in protein levels (Supplementary Fig. 2A). Consistent with their impact on the same RNA target, both morpholinos elicited the same spectrum of phenotypes (described below). For brevity, only results with the morpholino that blocks translation are presented in the main text, with phenotypes elicited by the alternative morpholino in Supplementary Figure 2. Dnmt2 morphants displayed abnormalities in overall body structure at all time points following the bud stage (Fig. 1A; data not shown). At 80 hpf, Dnmt2 morphant embryos were approximately half the size of embryos injected with a control morpholino (hereafter termed control morphants) (Fig. 1A).
Figure 1.
Specific developmental defects of Dnmt2 morphants. (A) Gross morphology of Dnmt2 morphants at 80 hpf, displaying pericardial edema, a primitive yolk, a thin trunk, and deformed eyes, brain, somites, and tail. Bar, 0.5 mm. (B–D) Dnmt2 morphants display tissue-specific differentiation defects. Whole-mount in situ of embryos at 80 hpf. Rows display expression of mRNA encoding l-fabp or transferrin-α (A), gata6 or foxa3 (B), and i-fabp or insulin (C). Columns display control morphants, Dnmt2 morphants, or Dnmt2 morphants coinjected with mRNA encoding Dnmt2WT or Dnmt2C82S. Dnmt2 morphants lose the expression of terminal differentiation marker of liver l-fabp and transferrin-α, whereas they retain expression of markers of liver precursors (gata6 and foxa3) and markers of terminal differentiation of intestine (i-fabp) and endocrine pancreas (insulin). Arrows denote the cells expressing the respective marker.
The reduced size observed could reflect a general developmental delay and/or a more specific impairment of particular tissues. To address this question, we examined the developmental progress of particular organs in Dnmt2 morphants. Here, we tested early markers of development to examine organ specification, as well as markers expressed later in development, to examine organ differentiation. Representative embryos are displayed in the figures, with statistics for all embryos examined in Supplementary Table 1. Our examinations began with the liver where dnmt2 is expressed at low levels (Supplmentary Fig. 1A, black arrow). Two markers of late differentiation, liver fatty acid-binding protein (l-fabp) (Denovan-Wright et al. 2000) and transferrin-α (Mudumana et al. 2004), are normally expressed in embryos beginning at 48 hpf. Remarkably, these markers were either completely absent or highly reduced in Dnmt2 morphants when compared with the age-matched control morphants, even at 80 hpf (Fig. 1B). However, two early markers of liver precursor cells, foxa3 and gata6 (Field et al. 2003), were present at 80 hpf, though the size (or number) of foxa3-positive cells was moderately reduced (Fig. 1C). This established that Dnmt2 morphants, like control morphants, specify a set of cells at this location as liver precursors (Fig. 1C), but fail to conduct late differentiation.
Importantly, two organs clearly express markers associated with late differentiation. In the intestine, expression of intestinal fatty acid-binding protein (i-fabp) was relatively normal, as was insulin expression in the endocrine pancreas (Fig. 1D; Biemar et al. 2001; Mudumana et al. 2004). With wild-type zebrafish, i-fabp expression in the intestine initiates ∼72 hpf and insulin expression in the endocrine pancreas after 36 hpf. This suggests that although Dnmt2 morphants are smaller than wild-type or control morphants at 80 hpf, they do not simply show a general developmental delay at the 24- to 36-hpf stage. Taken together, our results suggest that certain organs require Dnmt2 for proper late differentiation.
Two additional sets of experiments established that the developmental defects observed were a consequence of reduction in Dnmt2. First, an identical impact on organ differentiation was observed with an alternative morpholino designed to block mRNA splicing (Supplementary Fig. 2B). Second, we prepared by in vitro transcription an RNA encoding full-length V5-tagged Dnmt2 that was refractory to the affect of the Dnmt2 morpholino, as it bore an alternative sequence around the translation start site. Coinjection of this RNA with the Dnmt2 morpholino rescued l-fabp and transferrin-α expression at high frequency (Fig. 1B; Supplementary Table 1). Furthermore, Dnmt2 catalytic activity was required, as an RNA encoding a V5-tagged mutant Dnmt2 derivative that bears an amino acid substitution at the predicted catalytic residue (C82S, based on human DNMT2) did not rescue liver differentiation (Fig. 1B), although it was expressed at the same level as active Dnmt2 (Supplementary Fig. 2D). Interestingly, an RNA encoding active human DNMT2, but not a catalytically inactive derivative (C79S), rescued the liver differentiation defects conferred by injection of the zebrafish Dnmt2 morpholino, suggesting a conservation of Dnmt2 function in zebrafish and mammals (Supplementary Fig. 3A).
As dnmt2 is expressed in the retina at all embryonic stages, we also tested for eye defects in Dnmt2 morphant embryos. Like other organs, the Dnmt2 morphant eye is slightly smaller than the eye of control morphants. Transverse retinal sections of Dnmt2 morphants at 80 hpf revealed multiple defects in the retina and in the lens (Fig. 2A). First, a duplication of part of the ventral retina is observed (Fig. 2A, yellow arrow). Second, the retinal pigmented epithelium is present in the dorsal retina (Fig. 2A, red arrow), but absent from the ventral retina. Third, whereas the neuroepithelial area is typically composed of multiple cell types in wild-type zebrafish and control morphants at 80 hpf, this region in age-matched Dnmt2 morphants appears to be composed of a single cell type. The overall morphology of this eye section was reminiscent of retinal progenitor cells of the eyes of wild-type fish approaching 24 hpf, suggesting a developmental arrest in the retina.
Figure 2.
Dnmt2 morphants display a developmental delay in the eye. (A) Cross-sections of wild-type and Dnmt2 morphant eyes at 80 hpf stained with toluidine blue. Note the presence of retinal pigmented epithelium only in the dorsal side (red arrow) and duplication of a portion of ventral retina (yellow arrow). (B) Whole-mount in situ for pcna, vsx2, neurod, crx, zash1a, and gso1 in embryos injected with control morpholino or Dnmt2 morpholino alone, or coinjected with V5-Dnmt2WT. Expression of all markers except zash1a (which was examined at 48 hpf) was analyzed at 80 hpf. Note the continued expression of markers of progenitors (pcna and vsx2), and the loss of expression of markers of specified precursors (neurod, crx, and zash1a).
To stage these cells, we examined several markers of eye development. First, these cells expressed proliferating cell nuclear antigen (pcna), a marker of proliferation present in eye progenitor cells (Fig. 2B). Expression of two other early markers, vsx2 (Passini et al. 1997) and pax6.2, were higher in Dnmt2 morphants and present in all cells. In contrast, control morphants displayed weak staining restricted to a subset of cells (Fig. 2B; Supplementary Fig. 4). Retinal progenitors differentiate into neuroepithelial precursors, which then generate specialized precursors of cells such as ganglion, amacrine, bipolar, horizontal, and Muller glia cells, as well as the common cones and rods (Link et al. 2000; Hatakeyama and Kageyama 2004). These precursors then terminally differentiate into their respective final mature form. To test whether the retinal cells of Dnmt2 morphants are defective in differentiation, we tested for markers of the specialized precursor cells such as atoh7 (the zebrafish homolog MATH5), neurod (Inoue et al. 2002), crx (Furukawa et al. 1997), and zash1a (MASH1 homolog) (Tomita et al. 1996; Hatakeyama and Kageyama 2004). Interestingly, each of these markers was either highly reduced or completely absent in Dnmt2 morphant eyes (Fig. 2B; Supplementary Fig. 4). Dnmt2 morphants also lacked markers of terminal differentiation for various cell types, including Muller Glia (glutamine synthase, gso1) and cones (opsins) (Supplementary Fig. 4). Taken together, these data strongly suggest that Dnmt2 is required for differentiation of retinal progenitors into the precursors of specialized retinal neuroepithelial cell types.
Finally, we examined the development of the brain in Dnmt2 morphants. At 24 hpf, Dnmt2 morphants lack expression of the early proneural marker neurogenin-1 (ngn-1) (Korzh et al. 1998) in the hypothalamus and diencephalon, whereas other regions of the brain still expressed ngn-1 (Supplementary Fig. 5). These results suggest a role for Dnmt2 in neurogenesis in specific regions of the brain.
One clear important question is whether Dnmt2 morphant phenotypes result from defects in DNA methylation in the nucleus, or RNA methylation in the cytoplasm. In keeping with studies in mice, we did not observe a reduction in global DNA methylation levels in Dnmt2 morphants at 72 hpf (Supplementary Fig. 6). Therefore, if Dnmt2 is indeed a DNA methyltransferase in vivo, then its function is clearly limited to particular loci. However, the Dnmt2 morphant phenotypes revealed above enabled us to directly test whether zebrafish Dnmt2 functions primarily in the cytoplasm or the nucleus. We reasoned that if defects in DNA methylation underlie Dnmt2 morphant phenotypes, then a Dnmt2 derivative that is restricted to the nucleus should rescue the developmental defects conferred by the Dnmt2 morpholino, whereas a Dnmt2 derivative that resides only in the cytoplasm will not. Conversely, if Dnmt2 morphant phenotypes were due to the loss of RNA methylation activity in the cytoplasm, then a Dnmt2 derivative that resides only in the cytoplasm should rescue. To sequester Dnmt2 in nucleus, we added the nuclear localization signal (NLS) from SV40 T antigen to the N terminus of Dnmt2, termed NLS-Dnmt2. To retain Dnmt2 in the cytoplasm, we added the nuclear export sequence (NES) of mitogen-activated protein kinase kinase (MAPKK) to the N terminus of Dnmt2, termed NES-Dnmt2. To examine derivative expression and compartmentalization, both derivatives bore either a GFP or a V5 tag at the C terminus. Partitioning of these derivates could not be examined in developing fish embryos due to high levels of background autofluorescence and relatively low abundance resulting from construct dilution during embryogenesis. Therefore, we examined their partitioning by performing confocal microscopy in a zebrafish fibroblast cell line (ZF4), as well as a human RKO cell line. Partitioning was highly efficient; the NLS-tagged Dnmt2 derivative appeared entirely nuclear, whereas the NES-tagged derivative appeared entirely cytoplasmic (Fig. 3A).
Figure 3.
Cytoplasmic localization of Dnmt2 rescues Dnmt2 morphant phenotypes. (A) Confocal images of GFP and TOPRO-3 fluorescence in ZF4 and RKO cells transfected with empty vector, or vectors containing tagged Dnmt2WT, NLS-Dnmt2, mNLS-Dnmt2, or NES-Dnmt2. (B) Whole-mount in situ of embryos at 80 hpf, stained for expression of l-fabp and vsx2. Columns depict embryos from control morphants, Dnmt2 morphants, or Dnmt2 morphants coinjected with NLS-Dnmt2, mNLS-Dnmt2, or NES-Dnmt2.
To assess the activity of these Dnmt2 derivatives in zebrafish, DNA constructs encoding each were separately coinjected along with the Dnmt2 morpholino, and injected fish were monitored for the ability of the construct to rescue the expression of l-fabp in the liver and vsx2 in the eye. Remarkably, the NLS-Dnmt2 derivative failed to rescue expression, whereas NES-Dnmt2 derivative rescued their expression at high efficiency (Fig. 3B). Similar results were obtained with two other markers, transferrin-α and crx, in the liver and eye, respectively (Supplementary Table 1; data not shown). Importantly, both derivatives were expressed at similar levels (Supplementary Fig. 7). As a further control, we replaced a single lysine with alanine in the NLS of the NLS-Dnmt2 derivative (mNLS-Dnmt2); a substitution that efficiently compromised NLS function, restoring mNLS-Dnmt2 to the cytoplasm (Fig. 3A). Remarkably, the mNLS derivative was highly effective in rescuing markers of late differentiation in the liver and eye, and is produced at the same level as the other Dnmt2 derivatives (Fig. 3B; Supplementary Fig. 7). Taken together, these data demonstrate that Dnmt2 utilizes its catalytic activity in the cytoplasm to promote late differentiation of particular organs.
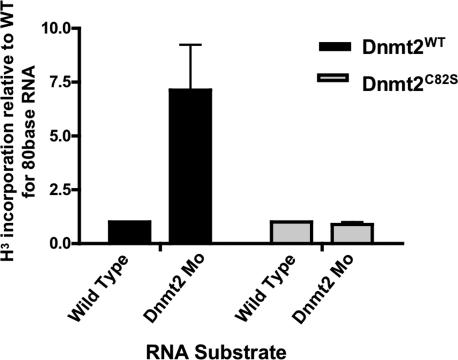
To better define the catalytic role of zebrafish Dnmt2, we determined whether zebrafish Dnmt2 functions as an RNA methyltransferase. In Arabidopsis, methylation of tRNAAsp by Dnmt2 alters tRNAAsp mobility in an acrylamide gel (Goll et al. 2006). We verified this observation in Arabidopsis dnmt2 mutants, but found no alteration in the mobility of zebrafish tRNAAsp in Dnmt2 morphants (Supplementary Fig. 8). We therefore employed a more direct test by examining the ability of zebrafish Dnmt2 to methylate small RNAs isolated from zebrafish, including tRNAs. Active Dnmt2, or the inactive derivative Dnmt2C82S derivative were overexpressed in 293 cells and immunopurified using an antibody against the V5 epitope, present on both derivatives. With both derivatives, immunoisolation was effective and equivalent (Supplementary Fig. 9). For the RNA substrate, we reasoned that RNA purified from a Dnmt2 morphant would lack the Dnmt2-dependent modification, and therefore would serve as a substrate for modification by our isolated Dnmt2 enzyme. Therefore, we purified RNA from either wild-type zebrafish embryos (72 hpf) or Dnmt2 morphants (72 hpf). Both substrates were incubated separately with either immunopurified wild-type Dnmt2 or the inactive Dnmt2C82S derivative, in the presence of tritium-labeled S-adenosylmethionine (H3-AdoMet). RNA species were separated on an acrylamide gel, and all bands present in the 10-base to 100-base range were separately excised and their methylation (tritium incorporation) quantified by scintillation counting (Supplementary Fig. 9). Interestingly, almost all bands lacked detectable methylation, with the notable exception of the band containing an RNA species of ∼80 bases, the size of tRNAAsp in zebrafish (sized in Supplementary Fig. 8). Here, we observed a sevenfold increase in the methylation of this ∼80-base RNA species derived from Dnmt2 morphant embryos compared with this RNA species derived from age-matched wild-type embryos (Fig. 4). This observation demonstrates that zebrafish Dnmt2 methylates an RNA species of ∼80 bases, consistent with recent results showing the methylation of tRNAAsp by Dnmt2 enzymes in other organisms.
Figure 4.
Zebrafish Dnmt2 is an RNA methyltransferase. Total RNA was purified from wild-type and Dnmt2 morphant zebrafish embryos at 72 hpf and subjected to a H3-labeled methyltransfer reaction using immunoprecipitated wild-type or catalytically inactive (C82S) zebrafish Dnmt2. After the reaction, recovered RNA was separated on a denaturing UREA-PAGE (see Supplementary Fig. 9). The band corresponding to the size of tRNAAsp (∼80 bp) (see Supplementary Fig. 8) was excised and H3 incorporation was measured by scintillation counting. The bar graph depicts the fold increase in H3 counts per minute from the band containing tRNAAsp (band 5, ∼80 bases) (Supplementary Fig. 8) from Dnmt2 morphants compared with wild-type zebrafish embryos. The two assays at the left utilized wild-type zebrafish Dnmt2 protein, whereas the two at the right utilized the catalytically inactive (C82S) enzyme. Assays represent three biological replicates; error bars are ±1 SD.
Taken together, our data provide the first evidence that a DNMT2-family enzyme impacts metazoan development, and strongly suggests that the biologically important function of Dnmt2 involves RNA methylation in the cytoplasm. We have shown that knockdown of Dnmt2 in zebrafish embryos confers defects in the terminal differentiation of liver, abnormal neurogenesis in the hypothalamus and diencephalon, and differentiation defects in the retina. These defects could be efficiently rescued by catalytically active zebrafish or human Dnmt2 derivatives, showing conservation of function. These phenotypes in zebrafish were unexpected, as loss of DNMT2 in mice or Drosophila did not confer observable phenotypes. Thus, a clear question emerging from our work is why zebrafish rely on Dnmt2 for proper development, whereas mice and flies do not. One possible explanation is that mice and flies have redundant mechanisms that compensate for the loss of Dnmt2, mechanisms that are absent (or less robust) in zebrafish. Important recent work revealing Dnmt2 as a tRNA methyltransferase challenged earlier conceptions that the primary function of Dnmt2 is to methylate DNA in the nucleus (Goll et al. 2006). Our demonstration that Dnmt2 activity is required in the cytoplasm provides evidence that the functionally relevant substrates are cytoplasmic RNA molecules, with tRNAAsp emerging as a clear candidate. A question for future studies is whether the modification state of tRNAAsp underlies the developmental defects observed, or whether these defects are due to Dnmt2 action on an alternative cytoplasmic RNA substrate. Vertebrates contain a large repertoire of noncoding RNAs of much lower abundance than tRNAs, which limits their detection in standard methyltransferase assays. As noncoding RNAs have emerged as important developmental regulators, they remain attractive alternative substrates for Dnmt2 function. Our work here establishes the zebrafish as a tractable vertebrate model system for the further study of Dnmt2 enzymes, their relevant cytoplasmic RNA substrates, and their impact on development.
Materials and methods
Zebrafish morpholino, mRNA, and plasmid injections
Zebrafish stocks and embryo culture were performed as described previously (Nadauld et al. 2004). Morpholinos, mRNA, and plasmids were injected at the one-cell stage. Morpholino oligonucleotides of following sequences were ordered from GeneTools LLC: Dnmt2 Mo1, 5′-AGTC GCTCCGTGTTCTCCATATTTC-3′; Dnmt2 Mo2, 5′-AGAAACAACT CACCCAAAGCCTTCA-3′; Control Mo, 5′-CCTCTTACCTCAGTTA CAATTTATA-3′. Morpholinos were dissolved in 1× Danieau buffer. Zebrafish dnmt2 (GenBank accession no. AB196920) was cloned in pcDNA-DEST40 and pDEST47 vector (Invitrogen) for injections of V5-tagged and GFP-tagged plasmids, respectively. For mRNA injections, zebrafish dnmt2 and human DNMT2 were cloned into pCRII-TOPO vector (Invitrogen). mRNA was transcribed using mMessage Machine kit (Ambion). A nuclear localization signal from SV40 T antigen, PKKKRKV, or a nuclear export signal from MAPKK, LQKKLEELEL, was added to the N terminus to make NLS-Dnmt2 and NES-Dnmt2, respectively. The third lysine in the NLS sequence was mutated to alanine in the mNLS-Dnmt2.
Whole-mount in situ hybridizations and histological analyses
Whole-mount in situ hybridizations were performed as described previously (Nadauld et al. 2004) using digoxigenin-labeled riboprobes for dnmt2, insulin, gata6, foxa3, i-fabp, l-fabp, transferrin-α, atoh7, crx, vsx2, neurod, ngn-1, zash1a, and gso-1. For histological analyses, embryos were fixed in 10% neutral buffered formalin, rinsed in PBS, and embedded in glycol methacrylate (Polysciences). Sections were cut using a Leica microtome, stained in toluidine blue (for eye sections), analyzed with a Zeiss Axiovert100 microscope, and photographed using an Olympus Magnafire color camera.
Cell lines, transfections, and immunohistochemistry
RKO, HEK293, or zebrafish ZF4 cells were cultured as recommended by ATCC. Cells were plated in 12-well plates on coverslips (for localization experiments) or in 10-cm plates (for immunoprecipitation experiments). Transfections were performed using lipofectamine 2000 following manufacturer’s instructions. For localization experiments, cells were fixed 24 h post-transfection and washed in PBS, and TOPRO-3 was added at 1:2000. Cells were then washed and imaged using a FV300 confocal microscope (Olympus).
Western blotting and antibodies
Embryos were collected, homogenized in IPH buffer (50 mM Tris at pH 6.0, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, Protease Inhibitors), and rotated for 10 min at 4°C, and supernatant was collected after centrifugation. The protein was separated by denaturing SDS-PAGE, transferred to a PVDF membrane, and probed with either mouse DNMT2 (Imgenex), V5 (monoclonal [Invitrogen] or rabbit polyclonal [Abcam]), or vinculin (Sigma). Mouse DNMT2 antibody specifically recognizes zebrafish Dnmt2, as only the 38-kDa species is reduced in Dnmt2 morphants derived from either morpholino.
Methyltransferase assay
RNA was purified (Ambion) from wild-type and Dnmt2 morphant embryos. Dnmt2 derivatives were immunoprecipitated from 293 cell extracts and incubated with RNA as described previously (Goll et al. 2006). The RNA was recovered by phenol-chloroform extraction and EtOH precipitation. RNA was separated on a 15% denaturing PAGE gel, and EtBr stained. The bands were excised from the gel, dissolved in gel elution buffer (0.5 M ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA at pH 8.0, 0.1% SDS), and counted on a scintillation counter (Beckman).
Acknowledgments
We thank Richard Dorsky and Ed Levine for helpful suggestions. We are grateful to Mary Goll for advice and Dnmt2 reagents, and Steve Jacobsen for RNA derived from Arabidopsis Dnmt2 mutants. This work was supported by the National Cancer Institute (D.A.J.), the Howard Hughes Medical Institute (B.R.C.), and the Huntsman Cancer Foundation (D.A.J and B.R.C). We thank the DNA peptide resource, DNA sequencing resource, the microarray resource (Brian Dalley), and the centralized zebrafish animal resource (CZAR) at the University of Utah.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1472907
References
- Biemar F., Argenton F., Schmidtke R., Epperlein S., Peers B., Driever W., Argenton F., Schmidtke R., Epperlein S., Peers B., Driever W., Schmidtke R., Epperlein S., Peers B., Driever W., Epperlein S., Peers B., Driever W., Peers B., Driever W., Driever W. Pancreas development in zebrafish: Early dispersed appearance of endocrine hormone expressing cells and their convergence to form the definitive islet. Dev. Biol. 2001;230:189–203. doi: 10.1006/dbio.2000.0103. [DOI] [PubMed] [Google Scholar]
- Denovan-Wright E.M., Pierce M., Sharma M.K., Wright J.M., Pierce M., Sharma M.K., Wright J.M., Sharma M.K., Wright J.M., Wright J.M. cDNA sequence and tissue-specific expression of a basic liver-type fatty acid binding protein in adult zebrafish (Danio rerio) Biochim. Biophys. Acta. 2000;1492:227–232. doi: 10.1016/s0167-4781(00)00102-0. [DOI] [PubMed] [Google Scholar]
- Field H.A., Ober E.A., Roeser T., Stainier D.Y., Ober E.A., Roeser T., Stainier D.Y., Roeser T., Stainier D.Y., Stainier D.Y. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev. Biol. 2003;253:279–290. doi: 10.1016/s0012-1606(02)00017-9. [DOI] [PubMed] [Google Scholar]
- Furukawa T., Morrow E.M., Cepko C.L., Morrow E.M., Cepko C.L., Cepko C.L. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- Goll M.G., Bestor T.H., Bestor T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Goll M.G., Kirpekar F., Maggert K.A., Yoder J.A., Hsieh C.L., Zhang X., Golic K.G., Jacobsen S.E., Bestor T.H., Kirpekar F., Maggert K.A., Yoder J.A., Hsieh C.L., Zhang X., Golic K.G., Jacobsen S.E., Bestor T.H., Maggert K.A., Yoder J.A., Hsieh C.L., Zhang X., Golic K.G., Jacobsen S.E., Bestor T.H., Yoder J.A., Hsieh C.L., Zhang X., Golic K.G., Jacobsen S.E., Bestor T.H., Hsieh C.L., Zhang X., Golic K.G., Jacobsen S.E., Bestor T.H., Zhang X., Golic K.G., Jacobsen S.E., Bestor T.H., Golic K.G., Jacobsen S.E., Bestor T.H., Jacobsen S.E., Bestor T.H., Bestor T.H. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J., Kageyama R., Kageyama R. Retinal cell fate determination and bHLH factors. Semin. Cell Dev. Biol. 2004;15:83–89. doi: 10.1016/j.semcdb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Hermann A., Schmitt S., Jeltsch A., Schmitt S., Jeltsch A., Jeltsch A. The human Dnmt2 has residual DNA-(cytosine-C5) methyltransferase activity. J. Biol. Chem. 2003;278:31717–31721. doi: 10.1074/jbc.M305448200. [DOI] [PubMed] [Google Scholar]
- Inoue T., Hojo M., Bessho Y., Tano Y., Lee J.E., Kageyama R., Hojo M., Bessho Y., Tano Y., Lee J.E., Kageyama R., Bessho Y., Tano Y., Lee J.E., Kageyama R., Tano Y., Lee J.E., Kageyama R., Lee J.E., Kageyama R., Kageyama R. Math3 and NeuroD regulate amacrine cell fate specification in the retina. Development. 2002;129:831–842. doi: 10.1242/dev.129.4.831. [DOI] [PubMed] [Google Scholar]
- Jeltsch A., Nellen W., Lyko F., Nellen W., Lyko F., Lyko F. Two substrates are better than one: Dual specificities for Dnmt2 methyltransferases. Trends Biochem. Sci. 2006;31:306–308. doi: 10.1016/j.tibs.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Katoh M., Curk T., Xu Q., Zupan B., Kuspa A., Shaulsky G., Curk T., Xu Q., Zupan B., Kuspa A., Shaulsky G., Xu Q., Zupan B., Kuspa A., Shaulsky G., Zupan B., Kuspa A., Shaulsky G., Kuspa A., Shaulsky G., Shaulsky G. Developmentally regulated DNA methylation in Dictyostelium discoideum. Eukaryot. Cell. 2006;5:18–25. doi: 10.1128/EC.5.1.18-25.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R.J., Bird A.P., Bird A.P. Genomic DNA methylation: The mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Korzh V., Sleptsova I., Liao J., He J., Gong Z., Sleptsova I., Liao J., He J., Gong Z., Liao J., He J., Gong Z., He J., Gong Z., Gong Z. Expression of zebrafish bHLH genes ngn1 and nrd defines distinct stages of neural differentiation. Dev. Dyn. 1998;213:92–104. doi: 10.1002/(SICI)1097-0177(199809)213:1<92::AID-AJA9>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Kunert N., Marhold J., Stanke J., Stach D., Lyko F., Marhold J., Stanke J., Stach D., Lyko F., Stanke J., Stach D., Lyko F., Stach D., Lyko F., Lyko F. A Dnmt2-like protein mediates DNA methylation in Drosophila. Development. 2003;130:5083–5090. doi: 10.1242/dev.00716. [DOI] [PubMed] [Google Scholar]
- Li E., Bestor T.H., Jaenisch R., Bestor T.H., Jaenisch R., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Link B.A., Fadool J.M., Malicki J., Dowling J.E., Fadool J.M., Malicki J., Dowling J.E., Malicki J., Dowling J.E., Dowling J.E. The zebrafish young mutation acts non-cell-autonomously to uncouple differentiation from specification for all retinal cells. Development. 2000;127:2177–2188. doi: 10.1242/dev.127.10.2177. [DOI] [PubMed] [Google Scholar]
- Morgan H.D., Santos F., Green K., Dean W., Reik W., Santos F., Green K., Dean W., Reik W., Green K., Dean W., Reik W., Dean W., Reik W., Reik W. Epigenetic reprogramming in mammals. Hum. Mol. Genet. 2005;14:R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- Mudumana S.P., Wan H., Singh M., Korzh V., Gong Z., Wan H., Singh M., Korzh V., Gong Z., Singh M., Korzh V., Gong Z., Korzh V., Gong Z., Gong Z. Expression analyses of zebrafish transferrin, ifabp, and elastaseB mRNAs as differentiation markers for the three major endodermal organs: Liver, intestine, and exocrine pancreas. Dev. Dyn. 2004;230:165–173. doi: 10.1002/dvdy.20032. [DOI] [PubMed] [Google Scholar]
- Nadauld L.D., Sandoval I.T., Chidester S., Yost H.J., Jones D.A., Sandoval I.T., Chidester S., Yost H.J., Jones D.A., Chidester S., Yost H.J., Jones D.A., Yost H.J., Jones D.A., Jones D.A. Adenomatous polyposis coli control of retinoic acid biosynthesis is critical for zebrafish intestinal development and differentiation. J. Biol. Chem. 2004;279:51581–51589. doi: 10.1074/jbc.M408830200. [DOI] [PubMed] [Google Scholar]
- Okano M., Xie S., Li E., Xie S., Li E., Li E. Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res. 1998;26:2536–2540. doi: 10.1093/nar/26.11.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M., Bell D.W., Haber D.A., Li E., Bell D.W., Haber D.A., Li E., Haber D.A., Li E., Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Passini M.A., Levine E.M., Canger A.K., Raymond P.A., Schechter N., Levine E.M., Canger A.K., Raymond P.A., Schechter N., Canger A.K., Raymond P.A., Schechter N., Raymond P.A., Schechter N., Schechter N. Vsx-1 and Vsx-2: Differential expression of two paired-like homeobox genes during zebrafish and goldfish retinogenesis. J. Comp. Neurol. 1997;388:495–505. doi: 10.1002/(sici)1096-9861(19971124)388:3<495::aid-cne11>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Rai K., Nadauld L.D., Chidester S., Manos E.J., James S.R., Karpf A.R., Cairns B.R., Jones D.A., Nadauld L.D., Chidester S., Manos E.J., James S.R., Karpf A.R., Cairns B.R., Jones D.A., Chidester S., Manos E.J., James S.R., Karpf A.R., Cairns B.R., Jones D.A., Manos E.J., James S.R., Karpf A.R., Cairns B.R., Jones D.A., James S.R., Karpf A.R., Cairns B.R., Jones D.A., Karpf A.R., Cairns B.R., Jones D.A., Cairns B.R., Jones D.A., Jones D.A. Zebra fish Dnmt1 and Suv39h1 regulate organ-specific terminal differentiation during development. Mol. Cell. Biol. 2006;26:7077–7085. doi: 10.1128/MCB.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K., Nakanishi S., Guillemot F., Kageyama R., Nakanishi S., Guillemot F., Kageyama R., Guillemot F., Kageyama R., Kageyama R. Mash1 promotes neuronal differentiation in the retina. Genes Cells. 1996;1:765–774. doi: 10.1111/j.1365-2443.1996.tb00016.x. [DOI] [PubMed] [Google Scholar]
- Wilkinson C.R., Bartlett R., Nurse P., Bird A.P., Bartlett R., Nurse P., Bird A.P., Nurse P., Bird A.P., Bird A.P. The fission yeast gene pmt1+ encodes a DNA methyltransferase homologue. Nucleic Acids Res. 1995;23:203–210. doi: 10.1093/nar/23.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]