Abstract
CD4+ T cells, specific for transforming latent infection with the Epstein Barr virus (EBV), consistently recognize the nuclear antigen 1 of EBV (EBNA1). EBNA1-specific effector CD4+ T cells are primarily T-helper 1 (TH1) polarized. Here we show that most healthy EBV carriers have such IFN-secreting EBNA1-specific CD4+ T cells at a frequency of 0.03% of circulating CD4+ T cells. In addition, healthy carriers have a large pool of CD4+ T cells that proliferated in response to EBNA1 and consisted of distinct memory-cell subsets. Despite continuous antigen presence due to persistent EBV infection, half of the proliferating EBNA1-specific CD4+ T cells belonged to the central-memory compartment (TCM). The remaining EBNA1-specific CD4+ T cells displayed an effector-memory phenotype (TEM), of which a minority rapidly secreted IFN upon stimulation with EBNA1. Based on chemokine receptor analysis, all EBNA1-specific TCM CD4+ T cells were TH1 committed. Our results suggest that protective immune control of chronic infections, like EBV, includes a substantial reservoir of TCM CD4+ TH1 precursors, which continuously fuels TH1-polarized effector cells.
Introduction
The human γ-herpesvirus Epstein Barr virus (EBV) infects more than 90% of the adult population. Latent infection by EBV is associated with human cancers of epithelial and B-cell origin, including 95% of nasopharyngeal carcinoma1 and 90% of endemic Burkitt lymphoma, and it is associated with 40% to 60% of Hodgkin lymphoma cases.2 Although all latently infected B cells have the potential for malignant transformation, most carriers remain free of tumors due to an effective immune control of the virus. This becomes apparent in conditions of immune suppression. Increased frequencies of EBV-associated malignancies can be observed during immunosuppressive therapy3 in patients with hereditary immunodeficiencies4,5 or with immunocompromising coinfections like HIV-induced AIDS.6 T cells are an essential part of EBV-specific immune control. This is evident from the successful treatment of posttransplantation lymphoproliferative disease (PTLD) with the adoptive transfer of EBV-specific T cells.7,8
This study focuses on the CD4+ T-cell response of healthy virus carriers to the latent EBV antigen Epstein-Barr nuclear antigen 1 (EBNA1), which is expressed in all EBV-associated malignancies. Maintenance of EBNA1 expression is due to its crucial function in viral persistence within proliferating cells. EBV DNA only rarely integrates into the host-cell genome9 and is carried as circular DNA episome by most latently EBV-infected cells.10,11 The EBNA1 protein initiates replication of viral episomes prior to mitosis and anchors viral DNA to mitotic chromosomes during cell division.12 By this mechanism, EBV replicates in proliferating cells without viral particle formation during latent EBV infection.
EBNA1 should be a critical target of protective immunity because it is expressed in all EBV-positive proliferating cells and is crucial for viral persistence. However, earlier studies have emphasized escape of EBNA1 from CD8+ T-cell detection due to an amino terminal gly-ala repeat domain.13–15 Since it is consistently recognized by CD4+ T cells in healthy EBV carriers,16,17 it was appreciated that EBNA1 is a promising T-cell antigen to target EBV-transformed B cells.18 EBNA1-specific effector CD4+ T cells are primarily T-helper 1 (TH1) polarized,19 can target EBV-transformed B cells,16,19–21 and prevent EBV-mediated B-cell transformation in vitro.22 EBNA1-specific CD4+ T cells can also mediate rejection of EBNA1-positive Burkitt lymphoma transplanted into mice.23 Recently, EBNA1-specific CD8+ T cells, in association with a few HLA class I haplotypes, have also been reported to recognize EBV-transformed B cells24–26 and to prevent their outgrowth in vitro.25 Therefore, both EBNA1-specific CD4+ and CD8+ T cells are able to recognize EBV-transformed cells and deserve to be studied in more detail.
In this study, we characterized EBNA1-specific CD4+ T cells in healthy EBV carriers by flow cytometry–based intracellular cytokine staining and proliferation assays, using an overlapping peptide library of EBNA1. We determined both the frequency and the phenotype of EBNA1-specific IFNγ-expressing and proliferating CD4+ T cells in peripheral blood of 20 healthy EBV carriers. Our data show that protective EBV-specific immune control contains a small population of readily IFNγ-secreting effector-memory T (TEM) cells, which are supported by a larger reservoir of central-memory (TCM) TH1 precursor cells that proliferate in response to EBNA1. The balance of TCM and TEM CD4+ T cells in the immune response against the latent viral antigen EBNA1 suggests that protective immune control of persistent viral infections requires a reservoir of central-memory T cells that fuels efficient T-cell responses.
Materials and methods
Culture medium
Culture medium RPMI 1640 (GIBCO, Grand Island, NY) was supplemented with 5% pooled human serum (PHS; Mediatech, Herndon, VA) and 20 μg/mL gentamicin (BioWhittaker, Walkersville, MD).
Peptide preparation
Peptides were synthesized in collaboration with the Proteomics Resource Center, Rockefeller University. All peptides were created using a Protein Technologies SYMPHONY multiple-peptide synthesizer (Rainin Instruments, Tucson, AZ) on Wang resin (P-alkoxy-benzyl alcohol resin; BACHEM Bioscience, King of Prussia, PA, and Midwest Bio-Tech, Fishers, IN) using F-moc (9-fluorenylmethyloxycarbonyl) nitrogen terminal–protected amino acids (aa's; Anaspec, San Jose, CA). Couplings were conducted using HBTU (2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate) and HOBT (1-hydroxybenzotriazole) in NMP (N-methylpyrrolidinone) as the primary solvent. Simultaneous resin cleavage and side-chain deprotection were achieved by treatment with concentrated, sequencing-grade trifluoroacetic acid with triisopropylsilane, water, and also ethanedithiol (if indicated by Cys or Met in sequence) added as ion scavengers in a ratio of 95:2:2:1 (all chemical reagents purchased from Fisher, Hampton, NH; Fluka, Steinheim, Switzerland; and Anaspec). Peptides were then released in 8 M acetic acid, filtered from resin, rotary evaporated, and redissolved in high-performance liquid chromatography (HPLC)–grade water for lyophilization. All crude lyophilized products were subsequently analyzed by reverse-phase HPLC (Waters Chromatography, Milford, MA) using a Merck Chromolith Performance C18 column (West Point, PA). Individual peptide integrity was determined and verified by matrix-assisted laser desorption ionization-mass spectrometry using a Perkin-Elmer/Applied Biosystems Voyager (PE/ABI, Foster City, CA) spectrometer system. Overlapping peptides of 12- to 22-aa length (average of 15-aa length) with 11-aa overlap were designed for the EBNA1400-641 sequence of the B95.8 EBV strain using the peptide-generator tool of the HIV sequence database at the Los Alamos National Laboratory.27 For CD8+ T-cell epitopes from EBV and CMV, we synthesized nonamer peptides that are part of the CEF control peptide pool of the NIH AIDS Research & Reference Reagent Program.28,29
Collection and preparation of peripheral blood
The Rockefeller University Internal Review Board approved this study. After volunteers provided informed consent, whole blood was collected via venipuncture under sterile conditions while following universal precautions. Blood was collected into heparinized vacutainers (BD Vacutainer, Franklin Lakes, NJ) or heparinized syringes. An aliquot was immediately used for the whole-blood intracellular cytokine assay. The remaining volume, when available, was used for peripheral blood mononuclear cell (PBMC) isolation. PBMCs were separated by density gradient centrifugation on Ficoll-Hypaque (Amersham Pharmacia Biotech, Piscataway, NJ). PBMCs were depleted of platelets by several washing steps in PBS and then resuspended in RPMI. PBMCs from blood-bank leukocyte concentrates were also separated in the same manner.
EBNA1 and VCA ELISA assays
EBNA1- and viral capsid antigen (VCA)–specific antibodies were quantified from human plasma by EBNA1 and VCA enzyme-linked immunosorbent assays (ELISAs) according to the manufacturer's instructions (Sigma/Diamedix; Miami, FL). Plasma samples from volunteers were diluted at 1:20 and 1:10 and run in triplicates.
Whole-blood intracellular cytokine assay
Aliquots of 500 μL fresh whole blood were placed into 15-mL snap-top round-bottom polypropylene tubes (Falcon, BD Labware, Bedford, MA) for a 6-hour incubation at 37°C in 5% CO2. Each reaction was carried out in the presence of 1 μg/mL costimulatory monoclonal antibodies to CD28 and CD49d (BD Biosciences, Immunocytometry Systems, San Jose, CA). Each tube also included 10 μg/mL Brefeldin A (Sigma, St Louis, MO). CD4+ T-cell–positive controls included 750 ng (1.5 μg/mL) Staphylococcal enterotoxin B (SEB) or influenza A strain Aichi/68 (Charles River Laboratories, North Franklin, CT) at a concentration of 106 hemaglutinin units (HAU/mL). In addition, nonameric CD8+ T-cell epitopes of HCMV and EBV (noted as CMV and EBV in figures; see Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article)29 were used as peptide controls. EBNA1 peptide mixtures of 10 overlapping peptides per reaction were added to a final concentration of 35 μM, or 3.5 μM per peptide per reaction. After examining peptide concentrations between 1 μM and 5 μM, 3.5 μM was determined to provide the most consistent responses in EBV-seropositive volunteers, with the lowest background in EBV-seronegative donors. After the 6-hour incubation, 2 μL of 0.5 M EDTA was added to each aliquot, and the samples were gently mixed and left at room temperature for 5 minutes. Finally, each sample received 4.5 mL of fluorescence-activated cell sorter (FACS) lysing buffer (BD Imunocytometry Systems), and each sample was vigorously vortexed for 5 seconds and left at room temperature for 10 minutes prior to storage at −80°C overnight.
Samples were thawed at room temperature and transferred to FACS tubes. After centrifugation, cells were resuspended in 500 μL of permeabilization solution (0.1% BSA, 0.1% saponin, in PBS) and remained at room temperature for 10 minutes. Permeabilization solution was decanted after a second centrifugation, and the cells were stained with mouse antihuman directly fluorochrome-labeled antibodies for 10 minutes at room temperature. Assays were divided into duplicates or triplicates and stained with the indicated combinations of IL-2–FITC, CD27-FITC, CD28-FITC, CD45RO-FITC, CD45RA-FITC, CD62L-FITC, CCR7-FITC, CD69-FITC, IFNγ-PE, CD3-PerCp, CD4-APC, or CD8-APC (all antibodies from BD Pharmingen, San Diego, CA). Cells were washed with permeabilization solution and resuspended in 200 μL FACS buffer solution (0.1% sodium azide in PBS). The samples were analyzed on a FACScalibur flow cytometer (BD Biosciences, San Jose, CA). Over one million events or 3 × 105 lymphocytes could be analyzed from each sample. After gating on lymphocytes based on size, CD4+CD3+ T cells were analyzed for expression of IFNγ or IL-2. Expression of more than 10 IFNγ-positive T cells above twice the frequency observed to background were defined as a positive response. Approximately 2 × 105 CD4+ T cells were evaluated for IFNγ expression per reaction. Gates to identify IFNγ- or IL-2–positive cells were determined by comparison to mouse isotype controls of IgG1-PE and IgG2b-FITC for IFNγ-PE and IL-2–FITC, respectively.
Proliferation assay by CFSE dilution
Peripheral blood mononuclear cells (PBMCs) were isolated from blood samples via density centrifugation. PBMCs were washed in PBS and incubated at 37°C in 0.3 μM CFSE (carboxyfluorescein succinimidyl ester; Molecular Probes, Eugene, OR) in PBS at a concentration of 107 cells per mL for 10 minutes. Where indicated, CD3+CD4+CD62L+ T cells were CFSE labeled after purification via flow-cytometric cell sorting on a FACSVantageSE sorter. Cells were washed in PBS and resuspended in 5% PHS with 1 μg/mL costimulatory monoclonal antibodies to CD28 and CD49d at a concentration of 1.7 × 106 cells per mL. PBMCs were distributed at 1 mL or 1.7 × 106 cells per well into 48-well plates. Known CD8+ T-cell epitopes of HCMV and EBV (noted as CMV and EBV in figures; Table S1) were used as controls. In addition, noncognate EBNA1 peptide subpools served as negative controls. Additional positive controls included Staphylococcus enterotoxin B (SEB; Sigma) at a final concentration of 125 ng/mL and infection of PBMCs with influenza A strain Aichi/68 (Charles River Laboratories). Influenza A infection was achieved by resuspending PBMCs in RPMI at 107 cells per mL with 50 000 HAU/mL influenza A for 1 hour at 37°C. After infection, cells were washed in RPMI and resuspended in 5% PHS for the 6-day proliferation. EBNA1 peptides were brought to a final concentration of 3.5 μM of each peptide per well. At the conclusion of a 6-day incubation at 37°C and 5% CO2, cells were harvested and washed once in PBS and stained with the indicated combinations of directly fluorochrome-labeled antibodies against CD3-PE, CD4-PerCp, CD8-PerCP, CCR7-PE, CD62L-PE, CD27-PE, CD28-PE, CD45RA-PE, CD45RO-PE, CXCR3-APC, CCR4-APC, or CXCR5-APC (all BD Pharmingen) for 10 minutes at room temperature. The cells were washed once with PBS and resuspended in 200 μL FACS buffer (0.01% sodium azide in PBS) prior to FACS analysis. The samples were analyzed on the FACScalibur flow cytometer gating on lymphocytes based on size. CD3+CD4+ or CD4+CD62L+ T cells were gated for analysis of proliferation of CD4+ T cells relative to phenotypic markers. Proliferating cells were grouped by the number of divisions completed, and the number of precursors was calculated; gating and calculations for precursor frequencies were performed with FlowJo (Tree Star, Ashland, OR) software. To determine the percentage of precursor EBNA1-specific CD4+ T cells, the sum of precursors (total number calculated from the percentage of surviving cells) was divided by the total number of originally plated cells.
Dendritic-cell generation and expansion of EBNA1-specific T cells in vitro
Dendritic cells were generated from PBMCs by culturing monocyte-enriched fractions in 6-well plates in 3 mL RPMI containing 1% human plasma, 1000 IU/mL GM-CSF (Immunex, Seattle, WA), and 500 U/mL IL-4 (Peprotech, Rocky Hill, NJ). CD14− cells were collected and frozen on day 0 for later use. Cultures remained at 37°C and cytokines were added to cultures of CD14+ monocytes on days 0, 2, and 4. On day 5, immature DCs were collected in original medium and transferred to new 6-well plates at a concentration of 106 cells/well. Half of the culture medium was replaced with fresh RPMI plus 1% human plasma and the DCs were matured with 1000 U/mL IL-6, 10 ng/mL IL-1β, 10 ng/mL TNF-α (R&D, Minneapolis, MN), and 1 μg/mL prostaglandin E2 (Sigma). Mature DCs were collected on day 7 and pulsed with overlapping peptides of EBNA1 for 1 hour at 37°C in RPMI at a final peptide concentration of 1 μM for each peptide. Autologous CD14− PBMCs (collected on day 0) were cocultured with matured DCs at a ratio of 30:1 (1 × 106 PBMCs and 3 × 104 DCs, respectively) in 2 mL of 5% PHS per well in a 24-well plate. The cocultures were incubated for 9 days at 37°C. On day 3, rIL-2 (Chiron, Emeryville, CA) was added to the cultures at a final concentration of 50 U/mL. On day 9, cells were collected and analyzed via ELISPOT for IFNγ secretion.
Enzyme-linked immunospot assay for IFNγ-secreting cells
Enzyme-linked immunospot (ELISPOT) assays were performed as described previously.16 Multiscreen plates (Millipore, Bedford, MA) were coated for 1 hour at 37°C with 10 μg/mL IFNγ-specific antibody Mab 1-D1K (Mabtech, Mariemont, OH). Plates were blocked with 5% PHS in RPMI for 1 hour at 37°C. Cells from the coculture were stimulated at 2 × 105 per well with all overlapping EBNA1 peptides or subpools I-V, dividing the EBNA1 peptides in 4 subpools of 10 and 1 subpool of 11 peptides (1 μM final concentration for each peptide). After 18 hours, the plates were incubated with the biotinylated secondary IFNγ-specific antibody Mab 7-B6-1 (Mabtech) for 2 hours at 37°C, followed by avidin-bound horseradish peroxidase (Vectastain ABC kit; Vector Laboratories, Burlingame, CA) for 1 hour at room temperature. Between incubations, plates were washed 3 times with PBS with 0.05% Tween20. Spots were developed for 5 minutes in stable 3,3-diaminobenzidine (DAB; Research Genetics, Huntsville, AL), washed with sterile water, and air dried before counting with an AID EliSpot reader (AID Autoimmun Diagnostika GmbH, Strassberg, Germany).
Statistical analysis
Volunteer CD4+ T-cell responses were evaluated for statistical significance using the paired Student t test. Percentage of IFNγ-positive CD4+ T cells in response to EBNA1 was compared with responses to no stimuli and the CD8+ T-cell epitopes for CMV. Likewise, EBNA1-specific proliferation was analyzed for statistical significance by comparing the percentage of CFSE-dilute CD4+ T cells in response to EBNA1 relative to the percentage of CFSE-dilute CD4+ T cells in response to no stimuli and the CD8+ T-cell epitopes for CMV.
Results
Overlapping peptide mixtures of EBNA1 can induce cytokine secretion and proliferation of an EBNA1-specific CD4+ T-cell clone
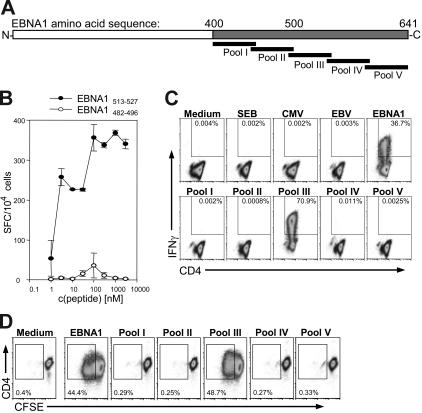
To evaluate the immune response to EBNA1, we constructed overlapping peptide mixtures representing the EBNA1 C-terminal domain (aa's 400-641), which contains all but one described CD4+ T-cell epitope.17,20,30 In order to generate internal controls in an out-bred human volunteer population, the peptide pools were divided into 5 distinct pools with the expectation that one subpool would elicit CD4+ T-cell stimulation in one but not another individual (Figure 1A). To test the efficacy of the peptide pools to elicit IFNγ secretion and proliferation of EBNA1-specific CD4+ T cells, we used the HLA-DR1–restricted EBNA1513-527-specific A4.E116 CD4+ T-cell clone.20 The secretion of IFNγ in response to the cognate EBNA1 peptide (EBNA1513-527) was detectable at peptide concentrations as low as 1 to 10 nM; however, the cells showed no appreciable response to the negative control peptide (EBNA1482-496) at any of the tested concentrations (Figure 1B). According to its specificity, A4.E116 secreted IFNγ in response to the total library of 51 EBNA1 peptides and one of the 10 member subpools, subpool III (aa's 499-548), both containing the cognate peptide. Thirty-seven percent to 71% of the clonal cells produced IFNγ to the cognate peptide pools (Figure 1C). There was no IFNγ expression in response to media, SEB, CMV peptides, EBV peptides, or the other EBNA1 peptide subpools. Similarly, the A4.E116 clone proliferated in response to the peptide pools containing the cognate peptide epitope without any appreciable proliferation to all other stimuli (Figure 1D). Almost half of the clonal cells in the final culture, 44.4% to 48.7%, had diluted CFSE after stimulation for 6 days with peptide pools containing the cognate EBNA1 epitope. These data indicate that complex mixtures of EBNA1 peptides can reliably stimulate IFNγ production and proliferation of EBNA1-specific CD4+ T cells.
Figure 1.
The EBNA1-specific CD4+ T-cell clone A4. E116 responds to EBNA1 peptide pools containing the EBNA1 peptide sequence 513-527. (A) Scheme of EBNA1 amino acid sequence represented by overlapping peptide pools (EBNA1, all EBNA1 peptides; pool I, aa's 400-461; pool II, aa's 452-508; pool III, aa's 499-548; pool IV, aa's 539-593; and pool V, aa's 584-641). (B) ELISPOT assay indicating spots per 104 cells in response to dilutions of peptide 513-527, and a noncognate peptide sequence as a negative control. Error bars indicate standard deviations of duplicates. (C) Intracellular cytokine staining assay for IFNγ in response to EBNA1 peptide pools, known CD8+ T-cell epitopes from CMV and EBV (Table S1), medium alone, and Staphylococcus enterotoxin B (SEB); note significantly positive response to the pool of all EBNA1 peptides and pool III, which both contain peptide 515-529. (D) CFSE proliferation assay in response to EBNA1 peptide pools. The data are representative of 3 independent experiments.
Healthy EBV carriers have CD4+ T-cell responses to overlapping EBNA1 peptide library
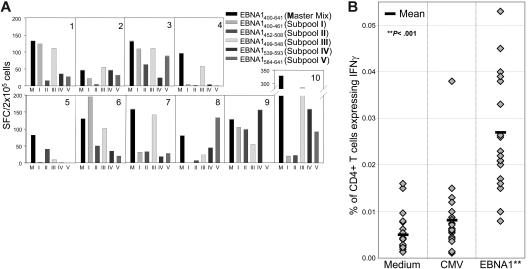
Initially, we tested the EBNA1 peptide recognition from 20 individual blood-bank leukocyte concentrates. PBMCs were isolated from leukocyte concentrates and were expanded for 1 week with dendritic cells plus all 51 EBNA1 peptides; then IFNγ secretion in response to the total EBNA1 peptide pools and the 5 EBNA1 subpools was analyzed by ELISPOT assays. EBNA1-specific CD4+ T cells could be expanded from all 20 leukocyte concentrates. Each leukocyte concentrate recognized different subpools (Figure 2A; Table S2). Since no single EBNA1 subpool elicited a response from each volunteer, we could infer that there were no contaminants of mitogens present in any single subpool. These data indicate that among healthy EBV carriers, no immunodominance of one peptide or particular subpool could be documented within the C-terminal domain of EBNA1. This analysis indicated that our peptide pool was able to expand EBNA1-specific CD4+ T cells from all healthy EBV carriers.
Figure 2.
EBNA1-specific CD4+ T-cell responses among 40 healthy volunteers. (A) IFNγ secretion of PBMCs in response to all EBNA1 peptides (Master mix) and subpools I-V by ELISPOT assays after stimulation for 1 week with autologous dendritic cells pulsed with all EBNA1 peptides. Ten of 20 representative PBMC stimulations from leukocyte concentrates are shown. (B) Each data point represents the frequency of IFNγ-expressing CD4+ T cells from each volunteer to one of 3 stimuli. The stimuli are indicated along the x-axis: medium (no stimulus), CMV (HCMV-derived CD8+ T-cell epitopes; Table S1), and EBNA1 (the master mix of all 51 overlapping peptides of EBNA1; Table S1). The frequency of IFNγ-positive CD4+ T cells of each individual is indicated by a diamond. The average frequency of IFNγ-positive CD4+ T cells for each of the 3 stimuli is indicated by the black bars.
The quantification of EBNA1-specific CD4+ T cells was assessed ex vivo from healthy EBV carriers. Healthy EBV carriers were identified by the detection of IgG antibodies to VCA or EBNA1 by commercial ELISA assays (data not shown). In an analysis of 21 consecutive volunteers, only one was seronegative for both antibodies. EBNA1-specific CD4+ T cells were identified via intracellular cytokine staining for IFNγ; we adapted previously used ELISPOT criteria to identify positive T-cell responses. Positive cells were identified relative to isotype controls; and a positive-responding individual was defined as having frequencies of at least 2-fold over background, consisting of at least 10 IFNγ-positive cells.31,32
While previous studies had focused on the quantification and characterization of predominantly lytic viral antigen–specific CD4+ T-cell responses,33–35 we adapted ex vivo whole-blood intracellular cytokine staining36 and CFSE proliferation assay to analyze CD3+CD4+ T-cell responses against the latent viral antigen EBNA1 in a cohort of 20 healthy EBV carriers. Intracellular cytokine staining for IFNγ identified 18 (90%) of 20 volunteers with detectable CD4+ T cells that produced IFNγ in response to EBNA1. Among these EBNA1 responders, the frequency of CD4+ T cells expressing IFNγ in response to EBNA1 varied from 0.008% to as high as 0.053% CD4+ T cells, with a mean of 0.027% (Figure 2B; Table S2).
In contrast, medium alone and major histocompatibility complex (MHC) class I–restricted CMV peptides elicited only 0.005% (0.007% to 0.016%) and 0.008% (0.0013% cells to 0.038%) IFNγ-producing CD4+ T cells, respectively (P < .001; EBNA1 versus controls). In 15 of 20 healthy volunteers, surface levels of the CD69 activation marker were evaluated, and we found that only activated CD4+ T cells secreted IFNγ in response to the EBNA1 peptides (data not shown). Only a minority of EBNA1-specific CD4+ T cells produced IL-2, predominantly in conjunction with IFNγ (Figure S1; data not shown). These data indicate that 0.03% of peripheral blood CD3+CD4+ T cells recognize the latent EBV antigen EBNA1 by IFNγ secretion.
To show that our healthy cohort of EBV carriers had representative EBV-specific T-cell immunity, we analyzed previously described CD8+ T-cell responses against dominant MHC class I ligands of EBV and CMV as positive controls.29 Positive responses to MHC class I–restricted peptides from CMV ranged from 0.013% to 4.1% of CD8+ T cells expressing IFNγ and were found in 8 (40%) of 20 volunteers. Positive responses to MHC class I–restricted peptides from EBV ranged from 0.028% to 1.21% CD8+ T cells expressing IFNγ and were found in 14 (70%) of 20 volunteers (Figure S2). These results are consistent with frequencies of EBV- and CMV-specific CD8+ T cells previously published37 and suggest that EBV-specific IFNγ-secreting CD8+ T cells are maintained at 10-fold higher frequency than EBV-specific CD4+ T cells.
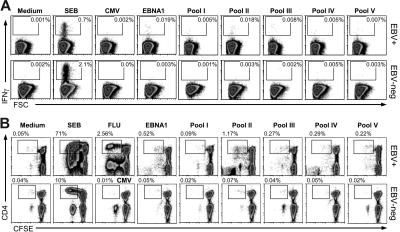
Using the CFSE proliferation assay, we evaluated EBNA1-specific CD4+ T-cell responses in 10 healthy EBV carriers, of whom we obtained larger blood volumes to extend the analysis beyond the ex vivo whole-blood assay (Figure 3B; Table S2). Eight of 10 of these volunteers had significant CD4+ T-cell proliferation in response to EBNA1 peptide pools. Close examination of 6-day CFSE proliferation assays revealed that EBNA1-specific CD4+ T cells underwent up to 5 divisions in response to stimulation with EBNA1 peptides (Figure 3B). Additional proliferation assays of 3-, 5-, and 7-day duration were examined (data not shown); however, we found that 6 days was optimal for maintaining cell survival while displaying detectable proliferation.
Figure 3.
CD4+ T cells respond consistently to EBNA1 with IFNγ secretion and proliferation. EBNA1-specific CD4+ T cells were analyzed by intracellular IFNγ staining and CFSE proliferation assay. CD4+ T-cell responses in response to medium (no stimulus), Staphylococcal enterotoxin B (SEB), HCMV-derived CD8+ T-cell epitopes (CMV), all EBNA1 peptides (EBNA1), and subpools of EBNA1 peptides are shown. (A) Whole-blood assay after gating on lymphocytes based on size and on CD4+ T cells. The frequency of IFNγ-positive CD4+ T cells is indicated. Forward-scatter (FSC; x-axis) and intracellular cytokine staining for INFγ (y-axis) are depicted. The top row represents detected IFNγ responses from one of 20 EBV-positive carriers and the bottom row from one EBV-negative volunteer upon stimulation with the indicated stimuli. Gates for IFNγ-positive cells are based on isotype controls. (B) CFSE dilution assays characterize CD4+ T-cell proliferation in response to the indicated antigens and influenza virus infection (FLU). The frequencies of CD4+ T cells with diluted CFSE are indicated. The top row displays CD4+ T-cell responses from one representative of 20 healthy volunteers. The proliferation responses of an EBV-negative volunteer are depicted in the bottom row.
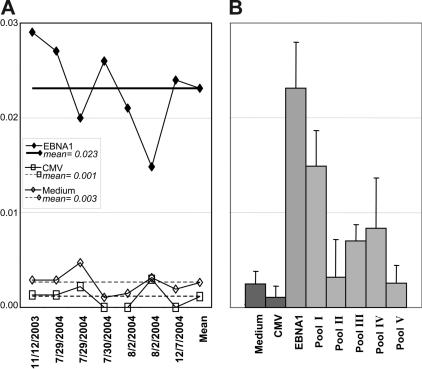
The EBNA1-specific CD4+ T-cell responses were stable over time and detection could be reproduced with our assays. Three volunteers were evaluated at 7 or more different time points over 1 year. The frequencies of EBNA1-specific CD4+ T cells were similar at each time point (Figure 4A). We suggest that the minor variations in EBNA1-specific CD4+ T-cell frequency that we observed reflect fluctuations in the EBV-specific immune control, since we did not observe similar changes in the control stimulations. In addition, the healthy EBV carriers detected EBNA1 at every time point with the same subpool recognition hierarchy (Figure 4B). These data indicate that during well-controlled persistent EBV infection, the frequency and specificity of EBNA1-specific CD4+ T-cell responses remain fairly constant.
Figure 4.
Frequencies of EBNA1-specific CD4+ T-cell responses are stable over time. (A) The frequency of EBNA1-specific IFNγ-expressing CD4+ T cells (◆) was assessed at the indicated time points in comparison to the frequencies of IFNγ-expressing CD4+ T cells in response to medium (◇) and CMV-derived CD8+ T-cell epitopes (□). (B) The average EBNA1 subpool (EBNA1400-461; Table S1) recognition was determined from the assays conducted at the 7 different time points, indicated in panel A. Standard deviations are depicted. The data are representative for 3 healthy EBV carriers that were analyzed at 7 or more time points.
EBNA1-specific CD4+ T-cell proliferation is derived from a large pool of CD4+ T cells
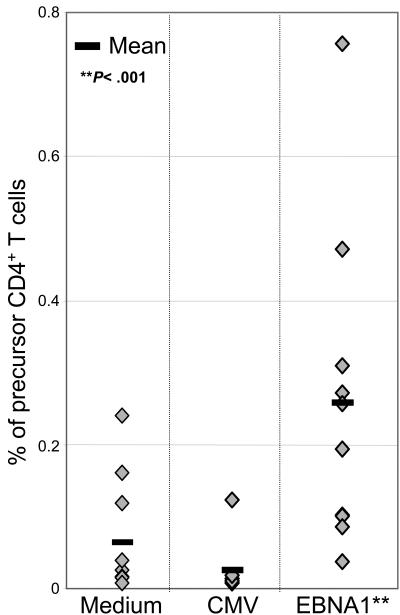
While comparing IFNγ-secreting with proliferating EBNA1-specific CD4+ T cells, we noticed that the hierarchy of EBNA1 subpool recognition sometimes differed between IFNγ secretion and proliferation of CD4+ T cells (recognition of subpool IV in the top rows in Figure 3A-B). Using FlowJo software, we were able to gate on populations of cells from each cellular division and determine the progenitor frequency of EBNA1-specific CD4+ T cells. Progenitors of proliferating EBNA1-specific CD4+ T cells were much more abundant than EBNA1-specific IFNγ-expressing CD4+ T cells (eg, subpool II recognition by the healthy EBV carrier in Figure 3B). Precursor frequencies ranged from 0.038% to 0.76% of total CD4+ T cells, with a mean of 0.26% (Figure 5; Table S3). In contrast, only 0.064% (0.01%-0.24%) and 0.032% (0.008%-0.12%) CD4+ T cells proliferated in response to medium- or CMV-derived HLA class I–restricted peptides, respectively (P < .001; EBNA1 versus controls). In all our assays, the percentage of precursors was calculated from the percentage of precursors multiplied with the total number of surviving cells and divided by the total number of original cells input at the start of the cultures. These data indicate that in the peripheral blood of healthy EBV carriers, there are 2 distinct populations of EBNA1-specific CD4+ T cells: a small, immediate TH1-polarized response as defined by rapid ex vivo IFNγ production; and a larger population of proliferating EBNA1-specific CD4+ T cells.
Figure 5.
Frequency of EBNA1-specific proliferating CD4+ T-cell precursors. Each data point represents the frequency of CD4+ T-cell precursor cells from the original culture from each volunteer relative to one of 3 stimuli. The stimuli are indicated along the x-axis: medium (nothing added), CMV (HCMV-derived CD8+ T-cell epitopes; Table S1), and EBNA1 (the master mix of all 51 overlapping peptides of EBNA1; Table S1). The frequency of precursors from each individual is indicated by a diamond. The average frequency of precursors from each of the 3 stimuli is indicated by the black bars.
EBNA1-specific CD4+ T cells consist of effector- and central-memory compartments
In order to define these populations further, we ascertained surface phenotypes of both populations. We evaluated CD27, CD28, CD45RA, and CD45RO expression, which had been proposed to distinguish differentiation phenotypes of CD4+ and CD8+ T cells.33,38 In addition, we assessed the expression of CCR7 and CD62L on EBNA1-specific T cells, which would indicate T-cell homing capacity to secondary lymphoid tissues as well as central (TCM)–versus effector-memory (TEM) T-cell distribution.39
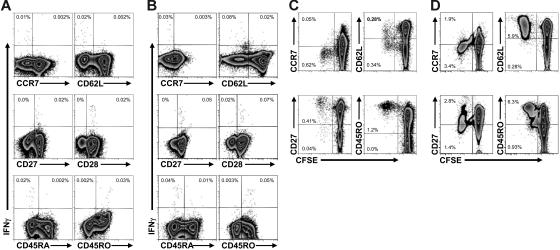
By definition, IFNγ-secreting EBNA1-specific CD4+ T cells are TH1-polarized effector T cells. These cells expressed CD45RO and lacked CD45RA; in addition, the expression of both CD27 and CD28 argue that these cells are memory T cells early into their differentiation.33,38 The absence of both CCR7 and CD62L indicate that the TH1-polarized EBNA1-specific CD4+ T cells are TEM cells (Figure 6A). The phenotype of these EBNA1-specific CD4+ TEM cells was similar to influenza A virus–specific CD4+ T cells (Figure 6B).
Figure 6.
Phenotype of EBNA1- and influenza-specific IFNγ-secreting and proliferating CD4+ T cells. EBNA1- and influenza-specific CD4+ T-cell responses of one representative volunteer are shown. (A) After gating on CD3+CD4+ T cells, EBNA1-specific cells were identified by intracellular IFNγ staining (y-axis), and the expression of the indicated surface markers was analyzed (x-axis). The frequencies of EBNA1-specific CD3+CD4+ cells are shown in the appropriate quadrants. Gates were determined following an analysis with isotype controls (data not shown). One of 3 experiments is shown. (B) As described for panel A but characterizing IFNγ-positive influenza-specific CD4+ T cells. One of 2 experiments is shown. (C) After gating on CD3+CD4+ T cells, EBNA1-specific proliferation was identified by CFSE dilution (x-axis), and the expression of the indicated surface markers was analyzed (y-axis). The frequencies of CFSE dilute or proliferating EBNA1-specific CD4+ cells are shown in the appropriate quadrants. One of 3 experiments is shown. (D) As described for panel C but following CD4+ T-cell proliferation after influenza stimulation. One of 2 experiments is shown.
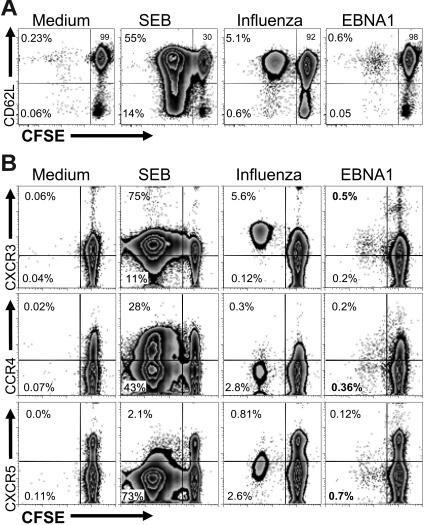
The population of CD4+ T cells that proliferated in response to EBNA1 had the same CD45RO+CD45RA−CD27+CD28+ phenotype as the IFNγ-secreting CD4+ T cells; however, proliferating cells contained both CD62L+ and CD62L− T-cell subsets (Figure 6C). Although proliferating influenza A–specific CD4+ T cells were also CD45RO+CD45RA−CD27+CD28+, these proliferating influenza A–specific CD4+ T cells were nearly all CD62L+ (Figure 6D). Unlike EBV, the influenza A virus does not persist in vivo as a chronic infection and, therefore, influenza A–specific CD4+ T cells seem to persist nearly entirely as central-memory T cells after virus clearance. To confirm our observations, we used flow-cytometric cell sorting to isolate CD3+CD4+CD62L+ cells from PBMCs prior to the CFSE dilution assay. We found that nearly all proliferating influenza- and EBNA1-specific CD4+ T cells maintain their CD62L surface expression through the duration of the 6-day assay (Figure 7A). Since CD62L expression on CD45RO+ T cells has been associated with central memory,39 these findings suggest that the persistent EBV infection leads to an EBNA1-specific CD4+ T-cell population that contains a substantial proportion of central-memory T cells.
Figure 7.
Stability of the central-memory phenotype and chemokine receptor expression on proliferating EBNA1-specific CD4+ T cells. (A) CFSE-labeled CD3+CD4+CD62L+ cells, purified by flow-cytometric cell sorting, were stimulated with the indicated antigens: medium (no stimulus), Staphylococcus enterotoxin B (SEB), influenza A infection (FLU), and the cognate EBNA1 peptide pool I. After 6 days, CD3+CD4+ T cells were analyzed for CD62L expression (y-axis) and CFSE dilution (x-axis). The frequencies of CD3+CD4+ T cells in each quadrant are indicated. One representative of 2 experiments is shown. (B) After gating on CD3+CD4+CD62L+ T cells, proliferation in response to 4 stimuli (medium, no stimulus; SEB, Staphylococcus enterotoxin B; influenza, influenza infection; and EBNA1, master mix of all 51 overlapping peptides of EBNA1) was identified by CFSE dilution (x-axis), and the expression of the indicated chemokine receptors (CXCR3, CCR4, and CXCRR5) was analyzed (y-axis). The frequency of CFSE dilute, and therefore proliferating, CD4+ cells is indicated in the appropriate quadrants. Gates and quadrants were determined after analysis with isotype controls (data not shown). One of 3 experiments is shown.
To better evaluate the CD62L+CD4+ T cells, chemokine receptor expression among these cells was determined. It has been shown that CXCR3, CCR4, and CXCR5 can be used to differentiate pre–T-helper polarization status among central-memory CD4+ T cells.40 We found that these EBNA1-specific CD3+CD4+CD62L+-proliferating T cells were predominantly CXCR3+CCR4−CXCR5− (Figure 7B). This suggests that these proliferating cells are central-memory TH1 precursors. These data suggest that the EBNA1-specific CD4+ T-cell population consists of a small subset of TH1-polarized TEM cells that immediately secrete IFNγ and a larger population of TCM TH1 precursor cells that readily proliferate in response to EBNA1.
Discussion
EBNA1 is the only EBV protein expressed in all EBV-associated malignancies, and the characterization of EBNA1-specific T-cell immunity might be crucial to understand how healthy EBV carriers prevent tumorigenesis by EBV. In this study, we evaluated the CD4+ T-cell responses of 40 EBV-positive healthy volunteers to overlapping peptides of the EBNA1 C-terminal domain (aa's 400-641), and we describe the memory CD4+ T-cell compartments specific for the latent viral antigen EBNA1. We detected IFNγ expression in response to EBNA1 among all 20 blood bank–derived leukocyte concentrates after 1 week of expansion in vitro. As detected by intracellular cytokine staining for IFNγ, an additional 18 of 20 volunteers carried EBNA1-specific CD4+ T cells (90%) with average frequencies of 0.027% of circulating CD4+ T cells. Most of the volunteers' responses to EBNA1 consisted of multiple specificities, with 2 to 3 EBNA1 peptide subpools usually being recognized. We also determined that an average of 0.26% of circulating CD4+ T cells proliferated in response to EBNA1, of which around half maintained a CD62L+ central-memory phenotype. Our data indicate that EBNA1-specific CD4+ T cells consist of 2 distinct populations: a small population of readily IFNγ-secreting TH1-polarized TEM cells and a larger population of TCM TH1 precursor cells. We believe that our current strategy of intracellular cytokine staining for IFNγ combined with analysis of proliferation in response to pools of EBNA1 peptides enables an accurate assessment of the immune response to EBNA1. Studies to employ this strategy to monitor EBV-specific T-cell responses in patients with EBV-associated malignancies are currently underway in our laboratory.
Using overlapping peptide pools of a specific protein allows for a better assessment of antigen-specific CD4+ T-cell responses. Previously, we used Escherichia coli–derived recombinant EBNA1 to quantify IFNγ-producing EBNA1458-641-specific CD4+ T cells in peripheral blood and found a frequency of 0.5% of peripheral blood CD4+ T cells.20 Although the recombinant EBNA1 yielded higher frequencies of IFNγ-positive CD4+ T cells than the overlapping peptide mixture used in this study, we find the latter antigen formulation more reliable. The preparation of recombinant EBNA1 has intrinsic variability between preparations, sometimes leading to unspecific responses in EBV-seronegative donors. In addition, subdividing the EBNA1 peptide library into subpools allows us to control for specificity with noncognate EBNA1 subpools. Finally, the use of overlapping peptides provides an easy method to narrow down the specificity of EBNA1-directed T-cell responses, which were reproducible over time in several donors (Figure 4). We propose that the small fluctuations in the frequency of EBNA1-specific CD4+ T cells that we observed in our longitudinal analysis follow variations in the EBV load. Accordingly, the ratio between central- and effector-memory subsets might change due to the level of EBV reactivation, recruiting central-memory CD4+ T cells to the effector-memory pool upon increased latent EBV infection and accumulation of central-memory T cells during phases of low antigenic load. The capacity of EBNA1-specific central-memory CD4+ T cells to be rapidly recruited as effector T cells might contribute to the robust EBV immune control, protecting from spikes in viral replication.
This study reports for the first time ex vivo the memory CD4+ T-cell compartments specific for a latent viral antigen. Unlike CD4+ T-cell responses to lytic antigens, the EBNA1-specific CD4+ T-cell response presents with a lower frequency due to lower antigen expression levels in EBV-infected B cells. In contrast, EBV-specific CD8+ T cells are around 10-fold more frequent than EBV-specific CD4+ T cells during both acute viral infection of infectious mononucleosis and persistent infection. For example, while EBNA3C386-400-specific CD4+ T cells can reach 0.03% of peripheral blood CD4+ T cells,33 EBNA3A-specific CD8+ T-cell responses have been found to reach 0.1% to 2.0%.37 Consistent with these previous reports, we found IFNγ-secreting EBV-specific CD8+ T-cell responses also in the range from 0.03% to 1.21% of CD8+ T cells in our healthy volunteer cohort (Figure S2; data not shown), whereas EBNA1-specific IFNγ-positive CD4+ T cells were 10-fold less frequent. Therefore, EBNA1-specific IFNγ-secreting CD4+ T-cell responses are as infrequent as other latent EBV antigen–specific CD4+ T-cell responses.
EBNA1, however, is expressed in both lytic and latent EBV infection,41 yet the phenotype of EBNA1-specific CD4+ T cells is most consistent with the reported phenotype of latent EBV antigen–specific CD8+ T cells.38 Unlike lytic EBV antigen–specific CD8+ T cells, latent EBV antigen–specific CD8+ T cells are frequently CD28+42 and express the memory isoform of CD45, CD45RO.42 EBNA1-specific TH1-polarized CD4+ T cells displayed mainly a CD45RA−CD45RO+CD27+CD28+ phenotype and are therefore CD4+ TEM cells that display the same phenotype as latent EBV antigen–specific CD8+ T cells (Figure 6A). With this phenotype they are clearly distinct from HCMV-specific CD8+ T cells that display a CD45RA−CD45RO+CD27−CD28− phenotype.38
In contrast to the readily IFNγ-secreting EBNA1-specific CD4+ TEM cells, we found proliferating EBNA1-specific CD4+ T cells to persist at a 5- to 10-fold higher frequency of circulating CD4+ T cells. Our data suggest that these higher frequencies of proliferating EBNA1-specific CD4+ T cells reflect a significant proportion of EBNA1-specific CD4+ TCM precursor cells. We detected a substantial proportion of CD62L+ EBNA1-specific proliferating CD4+ T cells, which is consistent with these cells being of central-memory phenotype.39 Although both CD62L and CCR7 have been described to distinguish central- from effector-memory T cells,39 unlike CCR7, we found that CD62L is not down-regulated on EBNA1-specific TCM cells upon proliferation. The uniformity in CD62L expression of influenza-specific T cells (Figure 6D) indicates that after clearance of an acute viral infection, T-cell memory is maintained predominantly as central-memory T cells. Accordingly, we suggest that the proliferating CD62L+ EBNA1-specific T cells (45% for proliferating CD4+ T cells of 3 separate donors) are central-memory T cells instrumental for the immune control of persistent EBV infection, whereas CD62L− EBNA1-specific T cells reflect effector-memory T cells that have been activated from this central-memory compartment due to persistent antigenic challenge. These findings are consistent with the hypothesis that during immune-controlled persistent viral infections effector-memory T cells (CD62L−) coexist with central-memory T cells (CD62L+), whereas after clearance of acute infection preferentially central-memory T cells remain.
Thus, we used CD62L to characterize EBNA1-specific CD4+ TCM cells in more detail. Our studies revealed that the proliferating EBNA1-specific CD62L+CD4+ T cells are, like the TEM cells, predominantly CD45RO+CD45RA−CD28+CD27+ but also CXCR3+CCR4−CXCR5− central-memory TH1-committed CD4+ T cells.40 Given the substantial proportion of this population, we suggest that these TCM TH1 precursors are a vital part of the immune control of EBV among asymptomatic carriers. These TH1 precursors are likely to be the reservoir from which the immediate EBNA1-specific CD4+ TEM cells are derived. This population may regulate the frequency of circulating EBNA1-specific CD4+ TEM cells based on antigen load, an important stopgap in preventing EBV-driven lymphoproliferative disease.
Our studies characterize the phenotype and frequency of EBNA1-specific CD4+ T cells, which are capable of exerting immune control over all latently EBV-infected proliferating cells. We propose that the EBV-specific immune control is mediated by TH1-polarized adaptive immune responses, which, as shown here, consist of EBNA1-specific CD4+ T cells of central- and effector-memory cells. Of those, only the effector-memory T cells display rapid effector functions, whereas the central-memory T cells represent a large population of TH1 precursors that might fuel the TEM cell pool and maintain, at the same time, lasting memory to EBV. Determining any deficit in the EBV-specific immune function among patients with EBV-associated malignancies can now be achieved through comparative studies of EBNA1-specific T cells from patients relative to our findings in healthy volunteers. A lack of EBNA1-specific T-cell immunity or imbalance in the CD4+ T-cell subset composition in these patients would encourage the development of immunotherapies to restore the protective EBNA1-specific T cells and complement current antineoplastic therapies.
Supplementary Material
Acknowledgments
This work was supported in part by the Arnold and Mabel Beckman Foundation, the Alexandrine and Alexander Sinsheimer Foundation, the National Cancer Institute (R01CA108609; C.M.), and a General Clinical Research Center grant (M01-RR00102) from the National Center for Research Resources at the National Institutes of Health (to the Rockefeller University Hospital). K.N.H. is supported by the Clinical Scholar Program of the Rockefeller University Hospital and the Children's Cancer Research Fund (CCRF), a California nonprofit organization. The contents of this publication, however, do not necessarily represent the policies or views of the CCRF.
We thank Ralph Steinman for critically reading the manuscript and the Rockefeller University Resource Centers for Proteomics and Flow Cytometry for expert technical support. We also thank the volunteers who altruistically participated in this study.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: K.N.H., J.U., and B.S. performed research; H.Z. contributed vital new reagents; and K.N.H. and C.M. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christian Münz, Laboratory of Viral Immunobiology, The Rockefeller University, 1230 York Ave, New York, NY 10021; e-mail: munzc@rockefeller.edu.
References
- 1.Chan AT, Teo PM, Johnson PJ. Nasopharyngeal carcinoma. Ann Oncol. 2002;13:1007–1015. doi: 10.1093/annonc/mdf179. [DOI] [PubMed] [Google Scholar]
- 2.Macsween KF, Crawford DH. Epstein-Barr virus-recent advances. Lancet Infect Dis. 2003;3:131–140. doi: 10.1016/s1473-3099(03)00543-7. [DOI] [PubMed] [Google Scholar]
- 3.Nalesnik MA. Clinicopathologic characteristics of post-transplant lymphoproliferative disorders. Recent Results Cancer Res. 2002;159:9–18. doi: 10.1007/978-3-642-56352-2_2. [DOI] [PubMed] [Google Scholar]
- 4.Bar RS, DeLor CJ, Clausen KP, Hurtubise P, Henle W, Hewetson JF. Fatal infectious mononucleosis in a family. N Engl J Med. 1974;290:363–367. doi: 10.1056/NEJM197402142900704. [DOI] [PubMed] [Google Scholar]
- 5.Purtilo DT, Cassel C, Yang JP. Letter: Fatal infectious mononucleosis in familial lymphohistiocytosis. N Engl J Med. 1974;291:736. doi: 10.1056/nejm197410032911415. [DOI] [PubMed] [Google Scholar]
- 6.Boshoff C, Weiss R. AIDS-related malignancies. Nat Rev Immunol. 2002;2:373–382. doi: 10.1038/nrc797. [DOI] [PubMed] [Google Scholar]
- 7.Papadopoulos EB, Ladanyi M, Emanuel D, et al. Infusions of donor leukocytes to treat Epstein-Barr virus-associated lymphoproliferative disorders after allogeneic bone marrow transplantation. N Engl J Med. 1994;330:1185–1191. doi: 10.1056/NEJM199404283301703. [DOI] [PubMed] [Google Scholar]
- 8.Heslop HE, Brenner MK, Rooney CM. Donor T cells to treat EBV-associated lymphoma. N Engl J Med. 1994;331:679–680. doi: 10.1056/NEJM199409083311017. [DOI] [PubMed] [Google Scholar]
- 9.Humme S, Reisbach G, Feederle R, et al. The EBV nuclear antigen 1 (EBNA1) enhances B cell immortalization several thousandfold. Proc Natl Acad Sci U S A. 2003;100:10989–10994. doi: 10.1073/pnas.1832776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams A, Lindahl T. Epstein-Barr virus genomes with properties of circular DNA molecules in carrier cells. Proc Natl Acad Sci U S A. 1975;72:1477–1481. doi: 10.1073/pnas.72.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindahl T, Adams A, Bjursell G, Bornkamm GW, Kaschka-Dierich C, Jehn U. Covalently closed circular duplex DNA of Epstein-Barr virus in a human lymphoid cell line. J Mol Biol. 1976;102:511–530. doi: 10.1016/0022-2836(76)90331-4. [DOI] [PubMed] [Google Scholar]
- 12.Kieff E, Rickinson A. Epstein-Barr Virus and its replication. In: Knipe DM, Howley PM, editors. Fields Virology. 3rd ed. Philadelphia, PA: Lippincott-Raven; 2001. pp. 2511–2573. [Google Scholar]
- 13.Levitskaya J, Coram M, Levitsky V, et al. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature. 1995;375:685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- 14.Blake N, Lee S, Redchenko I, et al. Human CD8+ T cell responses to EBV EBNA1: HLA class I presentation of the (Gly-Ala)-containing protein requires exogenous processing. Immunity. 1997;7:791–802. doi: 10.1016/s1074-7613(00)80397-0. [DOI] [PubMed] [Google Scholar]
- 15.Khanna R, Burrows SR, Steigerwald-Mullen PM, Moss DJ, Kurilla MG, Cooper L. Targeting Epstein-Barr virus nuclear antigen 1 (EBNA1) through the class II pathway restores immune recognition by EBNA1-specific cytotoxic T lymphocytes: evidence for HLA-DM-independent processing. Int Immunol. 1997;9:1537–1543. doi: 10.1093/intimm/9.10.1537. [DOI] [PubMed] [Google Scholar]
- 16.Münz C, Bickham KL, Subklewe M, et al. Human CD4+ T lymphocytes consistently respond to the latent Epstein-Barr virus nuclear antigen EBNA1. J Exp Med. 2000;191:1649–1660. doi: 10.1084/jem.191.10.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leen A, Meij P, Redchenko I, et al. Differential immunogenicity of Epstein-Barr virus latent-cycle proteins for human CD4+ T-helper 1 responses. J Virol. 2001;75:8649–8659. doi: 10.1128/JVI.75.18.8649-8659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Münz C. Epstein-Barr virus nuclear antigen 1: from immunologically invisible to a promising T cell target. J Exp Med. 2004;199:1301–1304. doi: 10.1084/jem.20040730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bickham K, Münz C, Tsang ML, et al. EBNA1-specific CD4+ T cells in healthy carriers of Epstein-Barr virus are primarily Th1 in function. J Clin Invest. 2001;107:121–130. doi: 10.1172/JCI10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paludan C, Bickham K, Nikiforow S, et al. EBNA1 specific CD4+ Th1 cells kill Burkitt's lymphoma cells. J Immunol. 2002;169:1593–1603. doi: 10.4049/jimmunol.169.3.1593. [DOI] [PubMed] [Google Scholar]
- 21.Voo KS, Fu T, Heslop HE, Brenner MK, Rooney CM, Wang RF. Identification of HLA-DP3-restricted peptides from EBNA1 recognized by CD4+ T cells. Cancer Res. 2002;62:7195–7199. [PubMed] [Google Scholar]
- 22.Nikiforow S, Bottomly K, Miller G, Münz C. Cytolytic CD4+-T-cell clones reactive to EBNA1 inhibit Epstein-Barr virus-induced B-cell proliferation. J Virol. 2003;77:12088–12104. doi: 10.1128/JVI.77.22.12088-12104.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu T, Voo K, Wang R. Critical role of EBNA1-specific CD4+ T cells in the control of mouse Burkitt lymphoma in vivo. J Clin Invest. 2004;114:542–550. doi: 10.1172/JCI22053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voo KS, Fu T, Wang HY, et al. Evidence for the presentation of major histocompatibility complex class I-restricted Epstein-Barr virus nuclear antigen 1 peptides to CD8+ T lymphocytes. J Exp Med. 2004;199:459–470. doi: 10.1084/jem.20031219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee SP, Brooks JM, Al-Jarrah H, et al. CD8 T cell recognition of endogenously expressed Epstein-Barr virus nuclear antigen 1. J Exp Med. 2004;199:1409–1420. doi: 10.1084/jem.20040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tellam J, Connolly G, Green KJ, et al. Endogenous presentation of CD8+ T cell epitopes from Epstein-Barr virus nuclear antigen 1. J Exp Med. 2004;199:1421–1431. doi: 10.1084/jem.20040191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Los Alamos National Security. PeptGen: HIV Molecular Immunology Database. [Accessed October 29, 2003]; http://www.hiv.lanl.gov/content/hiv-db/PEPTGEN/PeptGenSubmitForm.html. [Google Scholar]
- 28.NIH AIDS Research and Reference Reagent Program. [Accessed October 29, 2003]; www.aidsreagent.org.
- 29.Currier JR, Kuta EG, Turk E, et al. A panel of MHC class I restricted viral peptides for use as a quality control for vaccine trial ELISPOT assays. J Immunol Methods. 2002;260:157–172. doi: 10.1016/s0022-1759(01)00535-x. [DOI] [PubMed] [Google Scholar]
- 30.Long H, Haigh T, Gudgeon N, et al. CD4+ T-cell responses to Epstein-Barr virus (EBV) latent-cycle antigens and the recognition of EBV-transformed lymphoblastoid cell lines. J Virol. 2005;79:4896–4907. doi: 10.1128/JVI.79.8.4896-4907.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dhodapkar MV, Steinman RM, Sapp M, et al. Rapid generation of broad T-cell immunity in humans after a single injection of mature dendritic cells. J Clin Invest. 1999;104:173–180. doi: 10.1172/JCI6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Speiser DE, Pittet MJ, Guillaume P, et al. Ex vivo analysis of human antigen-specific CD8+ T-cell responses: quality assessment of fluorescent HLA-A2 multimer and interferon-gamma ELISPOT assays for patient immune monitoring. J Immunother. 2004;27:298–308. doi: 10.1097/00002371-200407000-00006. [DOI] [PubMed] [Google Scholar]
- 33.Amyes E, Hatton C, Montamat-Sicotte D, et al. Characterization of the CD4+ T cell response to Epstein-Barr virus during primary and persistent infection. J Exp Med. 2003;198:903–911. doi: 10.1084/jem.20022058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amyes E, McMichael A, Callan M. Human CD4+ T cells are predominantly distributed among six phenotypically and functionally distinct subsets. J Immunol. 2005;175:5765–5773. doi: 10.4049/jimmunol.175.9.5765. [DOI] [PubMed] [Google Scholar]
- 35.Maecker HT, Dunn HS, Suni MA, et al. Use of overlapping peptide mixtures as antigens for cytokine flow cytometry. J Immunol Methods. 2001;255:27–40. doi: 10.1016/s0022-1759(01)00416-1. [DOI] [PubMed] [Google Scholar]
- 36.Hanekom W, Hughes J, Mavinkurve M, et al. Novel application of a whole blood intracellular cytokine detection assay to quantitate specific T-cell frequency in field studies. J Immunol Methods. 2004;291:185–195. doi: 10.1016/j.jim.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 37.Tan LC, Gudgeon N, Annels NE, et al. A re-evaluation of the frequency of CD8+ T cells specific for EBV in healthy virus carriers. J Immunol. 1999;162:1827–1835. [PubMed] [Google Scholar]
- 38.Appay V, Dunbar PR, Callan M, et al. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat Med. 2002;8:379–385. doi: 10.1038/nm0402-379. [DOI] [PubMed] [Google Scholar]
- 39.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 40.Rivino L, Messi M, Jarrossay D, Lanzavecchia A, Sallusto F, Geginat J. Chemokine receptor expression identifies pre-T helper (Th)1, pre-Th2, and nonpolarized cells among human CD4+ central memory T cells. J Exp Med. 2004;200:725–735. doi: 10.1084/jem.20040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaefer BC, Strominger JL, Speck SH. A simple reverse transcriptase PCR assay to distinguish EBNA1 gene transcripts associated with type I and II latency from those arising during induction of the viral lytic cycle. J Virol. 1996;70:8204–8208. doi: 10.1128/jvi.70.11.8204-8208.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hislop AD, Gudgeon NH, Callan MF, et al. EBV-specific CD8+ T cell memory: relationships between epitope specificity, cell phenotype, and immediate effector function. J Immunol. 2001;167:2019–2029. doi: 10.4049/jimmunol.167.4.2019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.