Abstract
J-proteins and Hsp70 chaperones function together in diverse cellular processes. We identified a cytosolic J-protein, Jjj1, of Saccharomyces cerevisiae that is associated with 60S ribosomal particles. Unlike Zuo1, a 60S subunit-associated J-protein that is a component of the chaperone machinery that binds nascent polypeptide chains upon their exit from the ribosome, Jjj1 plays a role in ribosome biogenesis. Cells lacking Jjj1 have phenotypes very similar to those lacking Rei1, a ribosome biogenesis factor associated with pre-60S ribosomal particles in the cytosol. Jjj1 stimulated the ATPase activity of the general cytosolic Hsp70 Ssa, but not Ssb, Zuo1's ribosome-associated Hsp70 partner. Overexpression of Jjj1, which is normally ≈40-fold less abundant than Zuo1, can partially rescue the phenotypes of cells lacking Zuo1 as well as cells lacking Ssb. Together, these results are consistent with the idea that Jjj1 normally functions with Ssa in a late, cytosolic step of the biogenesis of 60S ribosomal subunits. In addition, because of its ability to bind 60S subunits, we hypothesize that Jjj1, when overexpressed, is able to partially substitute for the Zuo1:Ssb chaperone machinery by recruiting Ssa to the ribosome, facilitating its interaction with nascent polypeptide chains.
Keywords: Hsp70, Rei1, ribosome biogenesis, Hsp40
J-proteins, also known as Hsp40s, are obligate cochaperones of Hsp70-type molecular chaperones. Together, these chaperones participate in a wide variety of cellular functions, including protein folding, translocation of proteins across organellar membranes, and disassembly of protein complexes (1). An Hsp70's interaction with client proteins is modulated by its nucleotide-binding state, with ATP fostering rapid interaction with client proteins. This interaction is then stabilized upon ATP hydrolysis (2). J-proteins play a critical role by stimulating the ATPase activity of their Hsp70 partner through interaction of their J-domains, thereby promoting association of the Hsp70 with the substrate molecule.
The number of J-proteins in a particular cellular compartment typically exceeds that of Hsp70s (1). The cytosol of the yeast Saccharomyces cerevisiae contains 13 J-proteins (3) but only two classes of Hsp70s: the Ssas (encoded by SSA1–4) and Ssbs (encoded by SSB1–2) (4). The Ssa class has a variety of cellular functions, including translocation of proteins into the endoplasmic reticulum and mitochondria, uncoating of clathrin-coated vesicles, and general protein folding (4). A number of different J-proteins have been shown to work with Ssa in these processes. On the other hand, the more specialized Ssbs, which are predominantly ribosome-associated, can be cross-linked to very short nascent polypeptides (5) and are thought to function in the protection of newly synthesized polypeptides as they exit the ribosome (6, 7). Unlike the Ssas, which are essential, cells lacking the Ssbs grow slowly, particularly at low temperatures, and are hypersensitive to cations (8).
Although all J-proteins contain the canonical J-domain, some have additional sequences that allow them to carry out specialized functions. In some cases these sequences may serve to tether the J-protein to a particular cellular location or to recruit additional factors that are important for their function. For example, Zuo1 is tethered to the ribosome, where it functions as the Ssbs' J-protein partner (9). Strains lacking the Ssbs, Zuo1, or Ssz1, a regulatory protein that forms a heterodimer with Zuo1, have the same phenotypes, consistent with the three proteins acting together as a chaperone machine (5, 6).
During our analysis of the J-proteins of S. cerevisiae, we found that an uncharacterized J-protein of the yeast cytosol, Jjj1, contains a region, in addition to the J-domain, that is similar to a region of Zuo1. Therefore, we sought to determine Jjj1's function. We found that Jjj1, which is ≈40-fold less abundant than Zuo1, is also ribosome-associated. However, Jjj1 has a role independent of Zuo1 in the biogenesis of 60S ribosomal subunits. Despite this unique role of Jjj1, overexpression of Jjj1 can partially rescue the growth defects of cells lacking Zuo1, indicating a partial overlap in function between these two J-proteins.
Results
Jjj1 and Zuo1 Comigrate with 60S Ribosome Subunits.
A comparison of the amino acid sequence of the ribosome-associated J-protein Zuo1 with other J-proteins of S. cerevisiae revealed an 82-aa region between amino acids 205 and 287 that shares 50% similarity with amino acids 179–261 of Jjj1. Other than the J-domains, the remainder of Zuo1 and Jjj1 are strikingly different. Unlike most J-proteins, Zuo1 has an internal J-domain. Immediately C-terminal to the Zuo1-like region is a segment rich in positively charged amino acids. On the other hand Jjj1, like most J-proteins, has an N-terminal J-domain. In addition, Jjj1 has two putative C2H2-type zinc fingers that flank a region rich in charged amino acids, although, in the case of Jjj1, this region has an overall negative charge.
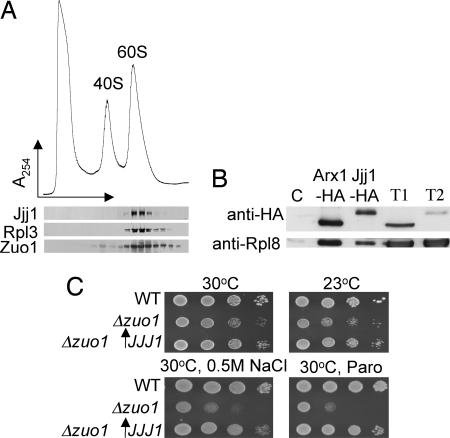
The 82-aa region that is conserved between Jjj1 and Zuo1 is not present in other J-proteins of S. cerevisiae. This unique similarity motivated us to ask whether Jjj1, like Zuo1, is a ribosome-associated J-protein. To test this idea, whole-cell lysate was prepared in magnesium-free buffer from WT cells and centrifuged through a magnesium-free sucrose gradient to separate the 40S and 60S ribosomal subunits (10). The fractions were subjected to immunoblot analysis. Jjj1 and Zuo1 comigrated almost exclusively with 60S subunits, consistent with 60S association (Fig. 1A). Furthermore, Rpl8, a protein of the 60S subunit, could be coimmunoprecipitated with Jjj1, confirming that Jjj1 associates with ribosomes (Fig. 1B)
Fig. 1.
Overexpression of Jjj1, which comigrates with 60S subunits, partially rescues Δzuo1. (A) Lysate from a WT strain was prepared in magnesium-free buffer and separated on a 15–30% sucrose gradient. The OD254 was monitored (Upper). Fractions were collected and analyzed for the presence of Jjj1, Zuo1, and Rpl3 by immunoblotting (Lower). (B) Extracts from cells containing plasmid borne Jjj1-HA or, as a control, genomically integrated Arx1-HA, were incubated with anti-HA antibody and protein A beads. Precipitated proteins were eluted from protein A beads in sample buffer and separated by SDS/PAGE. Total protein extract from Arx-HA cells (T1) and Jjj1-HA cells (T2) as well as immunoprecipitation from an untagged strain (C) were included as controls. Immunoblotting was performed by using antibodies specific for the HA or Rpl8 epitopes. (C) Tenfold serial dilutions of cells were spotted on minimal media plates with indicated additions and incubated at indicated temperatures; pRS415GPD JJJ1 (↑JJJ1); paromomycin (Paro).
Overexpression of Jjj1 Partially Rescues Growth Defects of Δzuo1.
The apparent ribosome association of both Zuo1 and Jjj1 raised the question as to whether Zuo1 and Jjj1 perform overlapping functions. To test this idea, we first placed the coding region of Jjj1 under the control of the constitutive GPD promoter, which led to an ≈60-fold overexpression of Jjj1 (data not shown), or an estimated 130,000 molecules per cell (11). Δzuo1 cells grow slowly, particularly at 30°C and below, and are hypersensitive to a wide variety of cations, including Na+, Li+, and the cationic translation-inhibiting drug, paromomycin, compared with WT cells (8). Overexpression of Jjj1 partially rescued the slow-growth and cation-sensitivity phenotypes caused by the absence of Zuo1, suggesting that Jjj1 can, in part, substitute for Zuo1 (Fig. 1C).
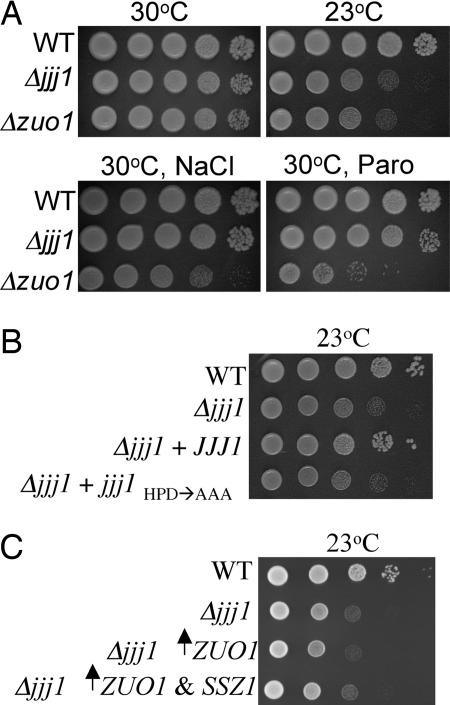
To determine whether the reverse, that is, whether Zuo1 could function for Jjj1, was also true, we first created a Δjjj1 strain and tested for phenotypic effects of the absence of Jjj1. Deletion of JJJ1 also resulted in cold sensitivity (Fig. 2A). This function of Jjj1 depends on its cochaperone activity, because a mutant protein in which the highly conserved histidine–proline–aspartic acid (HPD) motif of the J-domain was replaced with three alanines was unable to restore normal growth, even though it was expressed at levels similar to WT protein (Fig. 2B, data not shown). The Δjjj1 strain did not show any hypersensitivity to NaCl or paromomycin (Fig. 2A). We then tested whether overexpression of Zuo1 affected the cold sensitivity of Δjjj1 cells. However, neither increased expression of Zuo1 alone nor Zuo1 and Ssz1 together obviously improved the growth of Δjjj1 cells, suggesting that Jjj1 carries out a cellular function distinct from that of Zuo1 (Fig. 2C).
Fig. 2.
Jjj1 is a J-protein with a function distinct from Zuo1. Approximately equal numbers of cells of the indicated genotypes were diluted 10-fold, spotted on plates, and incubated under conditions indicated on rich media (A), minimal media; pRS316 JJJ1(+JJJ1) or pRS316 jjj1HPD→AAA (+jjj1HPD→AAA) (B), and minimal media; pRS416TEF ZUO1 (↑ZUO1); pRS415GPD SSZ1 (↑SSZ1) (C).
Δjjj1 Cells Have a Defect in 60S Ribosomal Subunit Biogenesis.
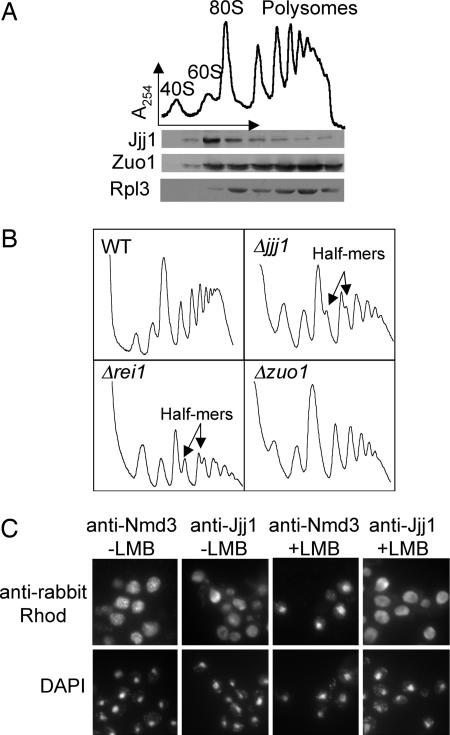
The possibility that Jjj1 performs a function different from Zuo1 led us to look more carefully at the issue of ribosome association. Total cellular extract from WT cells was separated on a sucrose gradient, and fractions were analyzed for the presence of Jjj1, Zuo1, and Rpl3 of the large ribosomal subunit. As expected from previous results (12), Zuo1 comigrated with ribosomes and was found distributed throughout the gradient in fractions containing free 60S subunits, 80S monosomes and polysomes, similar to Rpl3 (Fig. 3A). However, Jjj1 comigrated primarily with 60S and, perhaps, 80S particles, with some trailing into the polysomal fractions (Fig. 3A). This difference in fractionation also pointed to a difference in function between Jjj1 and Zuo1.
Fig. 3.
Cells lacking Jjj1, a strictly cytosolic protein, accumulate half-mer ribosomes at 23°C. (A) Ten OD254 units of lysate from WT cells grown at 30°C were centrifuged through a 5–50% sucrose gradient and the OD254 monitored (Upper). Fractions collected were analyzed by immunoblotting for the presence of Jjj1, Zuo1, and Rpl3 (Lower). (B) Ten OD254 units of lysate from the indicated strains grown at 23°C were centrifuged through 7–47% sucrose gradients, and the OD254 was monitored. (C) Log-phase cells of the LMB-sensitive strain AJY1539 were incubated in LMB (0.1 μg/ml) for 30 min and then fixed with formaldehyde and subjected to indirect immunofluorescence microscopy by using antibodies specific for Jjj1 or Nmd3. Localization was then monitored by using anti-rabbit rhodamine-conjugated antibodies. DAPI was used to visualize nuclei.
Comigration of Jjj1 with 60S subunits and the slow growth at low temperatures of cells lacking Jjj1 is reminiscent of Rei1, a recently identified cytosolic factor required for biogenesis of 60S subunits (13–15). Deletion of REI1 causes a decrease in 60S levels and the appearance of “half-mer” ribosomes (14, 15). Half-mers are interpreted as 48S preinitiation complexes stalled on mRNA because of a lack of mature large ribosomal subunits, which can result from either decreased levels of mature 60S subunits or from a 40S–60S subunit joining defect (16). Based on these similarities, we decided to analyze cellular extracts from a Δjjj1 strain and, as a control, a Δrei1 strain, grown at 23°C, by sucrose gradient centrifugation. Compared with a WT strain, the Δjjj1 and Δrei1 mutants showed a significant accumulation of half-mer ribosomes (Fig. 3B). In contrast, analysis of extracts from a Δzuo1 strain grown at 23°C did not, consistent with the inability of increased expression of Zuo1 to rescue the cold sensitivity of Δjjj1.
Strains having defects in 60S subunit biogenesis often have reduced steady-state levels of mature 60S. To assess the levels of 60S subunits in the Δjjj1 strain, total cellular extracts from WT, Δjjj1, and Δrei1 cells grown at 23°C were prepared and analyzed in magnesium-free buffer to separate 40S and 60S subunits. The area under each peak was measured, and the 60S/40S ratio was determined for each strain. The 60S/40S ratio for the WT strain was 1.92 ± 0.06, similar to the previously reported ratio of 2 (17). For Δrei1, the 60S/40S ratio was 1.36 ± 0.09 and for Δjjj1, 1.44 ± 0.05. Thus, the Δjjj1 strain had an ≈26% lower 60S/40S ratio than did WT cells. The Δrei1 strain, which has been reported to have fewer 60S subunits (14, 15), had a similar reduction in 60S subunits (30%). The 60S/40S ratio for the Δzuo1 strain was indistinguishable from that of WT, 2.03 ± 0.08.
Absence of the 60S Biogenesis Factor Arx1 Suppresses the Δjjj1 Phenotype.
Although factors involved in the biogenesis of the large subunit often cosediment with free 60S subunits, some act in the nucleus and are exported with the pre-60S particle to the cytosol. These shuttling factors are then reimported into the nucleus after their release from the 60S particle. Jjj1 has been reported to be cytosolic (18). To determine whether Jjj1 is potentially a shuttling pre-60S factor, we used a leptomycin B (LMB)-sensitive strain harboring the T539C mutation in the 60S exporter Crm1. If Jjj1 shuttles, we would expect it to be trapped in the nucleus when ribosome export is blocked by addition of LMB. However, in the presence of the Crm1 inhibitor LMB, Jjj1 remained cystosolic (Fig. 3C), whereas the 60S export adapter Nmd3 relocalized to the nucleus, as reported (19, 20). We conclude that Jjj1 functions solely in the cytosol.
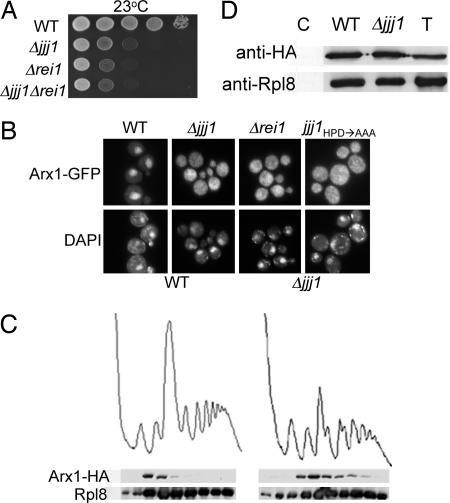
Because Rei1 is also a solely cytosolic factor involved in 60S ribosome biogenesis, and both Δrei1 and Δjjj1 cells have a cold-sensitive phenotype, we compared the growth of the individual mutants with that of the double mutant, Δjjj1 Δrei1. Δrei1 cells grew slightly less well at 23°C compared with Δjjj1 cells. However, Δjjj1 Δrei1 cells grew indistinguishably from cells lacking only Rei1 (Fig. 4A), suggesting to us that Jjj1 and Rei1 might function together, because loss of a second protein in a pathway often does not cause additional harm to the cell.
Fig. 4.
Arx1 fails to recycle in Δjjj1 cells. (A) Approximately equal numbers of cells of the indicated genotypes were diluted 10-fold, spotted onto rich media, and incubated at 23°C. (B) Localization of Arx1-GFP was monitored by fluorescence microscopy in the indicated strains. All cells were grown at 25°C in rich media, except Δjjj1 carrying pRS316 jjj1HPD→AAA (jjj1HPD→AAA), which was cultured in minimal media lacking uracil. (C) Lysates from 25°C-grown cells containing chromosomally integrated ARX1-HA were centrifuged through 7–47% sucrose gradients and the OD254 monitored (Upper). Fractions were analyzed for the presence of Arx1-HA and Rpl8 by immunoblot analysis (Lower). (D) Extracts from WT or Δjjj1 cells containing genomically integrated Arx1-HA were incubated with anti-HA antibody and protein A beads. Precipitated proteins were eluted from protein A beads in sample buffer and separated by SDS/PAGE. Total protein extract (T) as well as immunoprecipitation from an untagged strain (C) were included as controls. Immunoblotting was performed by using antibodies specific for the HA or Rpl8.
Rei1 has been reported to play a role in the cytosol-to-nuclear recycling of pre-60S biogenesis factors, such as Arx1, that are bound to pre-60S particles upon their exit to the cytosol (14, 15). In a WT strain, the majority of the fluorescent signal produced by Arx1-GFP is found in the nucleus, with a faint cytoplasmic signal being consistent with nuclear-cytoplasmic transport of Arx1-bound pre-60S subunits (14, 15). In the absence of Rei1, however, Arx1-GFP is primarily cytosolic (14, 15), and the protein persists on the 60S ribosomal subunit (14), suggesting that it is not recycled properly to the nucleus.
To determine whether Arx1 is recycled properly in a Δjjj1 strain, we monitored the Arx1-GFP signal in this strain. Similar to Δrei1, at 25°C Arx1-GFP is predominantly found in the cytosol in Δjjj1 cells, suggesting that it may remain bound to the 60S subunit (Fig. 4B). Indeed, in the absence of Jjj1, Arx1 still comigrated with 60S ribosomal subunits (Fig. 4C), and the 60S ribosomal protein Rpl8 was coimmunoprecipitated with Arx1 (Fig. 4D), indicative of continued association of Arx1 with 60S subunits in the cytosol. This role of Jjj1 depends on its cochaperone activity, because Arx1-GFP is also primarily cytosolic in the jjj1HPD→AAA mutant (Fig. 4B). The fact that, in the absence of Jjj1, Arx1 migrates further into the gradient may indicate that 60S subunits containing Arx1 are able to participate, at least marginally, in subunit joining and thus translation initiation. Regardless, we conclude that Jjj1 is required for proper recycling of Arx1.
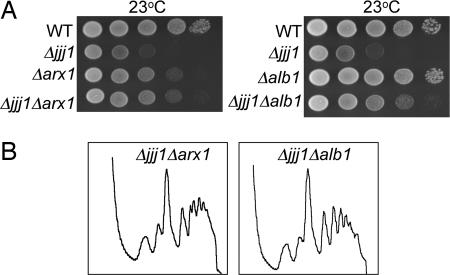
The deletion of ARX1 has also been shown to suppress the growth defect of a Δrei1 strain (14, 15), presumably because the continued presence of Arx1 on the 60S subunit is detrimental to the final stages of 60S maturation (14). Therefore, we compared the phenotypes of Δjjj1, Δarx1, and Δjjj1 Δarx1 strains. Δarx1 cells, although slightly cold sensitive, grew better at 23°C than Δjjj1 cells. Δjjj1 Δarx1 cells grew as well as Δarx1 cells (Fig. 5A). Thus deletion of ARX1 suppresses the cold-sensitive growth defect caused by the absence of either Rei1 or Jjj1. The absence of Arx1 has also been reported to substantially alleviate the half-mer accumulation of a Δrei1 strain (14). We tested the effect of deletion of ARX1 on the polysome profile of a Δjjj1 cells. Distinct half-mer peaks were absent in profiles of Δjjj1 Δarx1 cells. (Figs. 3B and 5B). In addition, the ratio of 60S to 40S subunits in Δjjj1 Δarx1 was 1.76 ± 0.07 compared with 1.92 ± 0.06 in WT and 1.44 ± 0.05 in Δjjj1. We conclude that deletion of ARX1 at least partially relieved the 60S biogenesis defect of a Δjjj1 strain. The fact that the phenotype of Δjjj1 is more severe than deletion of ARX1 suggests that the consequences of persistent association of Arx1 with 60S subunits may be more severe than those for its complete absence.
Fig. 5.
Deletion of ARX1 or ALB1 suppresses defects of Δjjj1 cells. (A) Approximately equal numbers of cells of the indicated genotypes were diluted 10-fold and spotted on rich media. Plates were incubated at 23°C. (B) Ten OD254 units of lysate from the indicated strains grown at 23°C were centrifuged through 5–50% sucrose gradients and the OD254 was monitored.
Arx1 is reported to exist in a stable complex with the small, highly basic protein Alb1, and deletion of ALB1 also suppresses the growth defects of a Δrei1 strain (15). We tested the effects of the absence of Alb1 in our system. Deletion of ALB1 suppressed the half-mer defect (Figs. 3B and 5B), the cold-sensitivity of a Δjjj1 strain (Fig. 5A), and the decrease in 60S subunits, as the 60S/40S ratio of Δjjj1 Δalb1 cells was 1.71 ± 0.04, consistent with the idea that Arx1 and Alb1 function together. On the other hand, we saw no effect of deletion of ARX1 or ALB1 on the growth of a Δzuo1 strain (data not shown).
Jjj1 J-Domain Stimulates the ATPase Activity of Ssa, but Not Ssb.
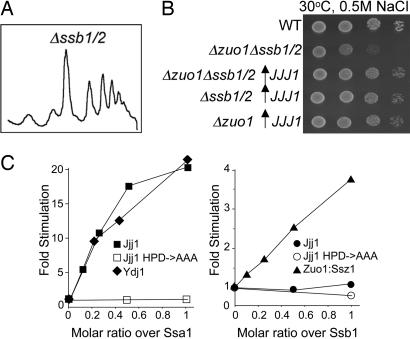
The proper localization of Arx1 depends on the J-domain of Jjj1. Because no J-protein has been found to function alone, but rather always with an Hsp70 partner, we sought to determine the Hsp70 partner of Jjj1. Because Jjj1 and Zuo1 have similarities in their amino acid sequences, and Zuo1 functions with the ribosome-associated Hsp70 Ssb, we first asked whether strains lacking Ssb have a defect in 60S subunit biogenesis, which would be expected if Jjj1 and Ssb functioned together. Total cellular extracts from a Δssb1/2 strain grown at 23°C were analyzed. The polysome profile of this strain was similar to that of a WT strain, with no visible accumulation of half-mer ribosomes (Fig. 6A).
Fig. 6.
Jjj1 stimulates the ATPase activity of Ssa1. (A) Ten OD254 units of lysate from the Δssb1 Δ ssb2 (Δssb1/2) grown at 23°C were centrifuged through a 5–50% sucrose gradient, and the OD254 was monitored. (B) Approximately equal numbers of cells of the indicated genotypes were diluted 10-fold and spotted on minimal media. Plates were incubated at 23°C. (C) Hsp70–ATP-[32P] complexes were incubated in the presence of various concentrations of Jjj1, Jjj1HPD→AAA, Ydj1, or Zuo1–Ssz1 heterodimer. The rates of hydrolysis of ATP were determined, and fold stimulation over the basal rate was plotted.
This lack of evidence for a role of the Ssb-type Hsp70s in ribosome biogenesis suggested to us that Jjj1 did not function with Ssb. We then asked whether overexpression of Jjj1 could partially rescue Δzuo1 even in the absence of Ssb1/2. Not only could expression of Jjj1 from the GPD promoter rescue a Δzuo1 Δssb1/2 strain, but it rescued a Δssb1/2 strain and a Δzuo1 Δssb1/2 Δssz1 strain, which lacks the entire ribosome-associated chaperone complex (Fig. 6B, data not shown).
The results reported above suggested to us that Jjj1 and Ssb may not function as a J-protein:Hsp70 pair. Ssa is the other major Hsp70 of the cytosol. We could not test cells lacking Ssa, as we did those lacking Ssb, because such strains are nonviable. In addition, reduced Ssa function results in pleiotropic effects, including defects in the nucleolus (21), the major site of ribosome biogenesis. However, a critical role of a J-protein is the stimulation of the ATPase activity of its partner Hsp70 and thus, such stimulation serves as an indicator of functional partnerships between J-proteins and Hsp70s. Therefore, we purified Jjj1 and tested whether it was able to stimulate the ATPase activity of Ssa1 or Ssb1, using preformed, radiolabeled ATP-bound Hsp70 complexes. Even at a 0.1:1 ratio with Ssa1, Jjj1 stimulated Ssa1's ATPase activity 5-fold, with 17-fold stimulation occurring at a 0.5:1 ratio (Fig. 6C). This level of stimulation was equivalent to that obtained with Ydj1, a known cytosolic J-protein partner of Ssa1 (9, 22). This stimulation depended on a functional J-domain, as Jjj1HPD→AAA failed to stimulate Ssa1. In contrast, Jjj1 did not significantly stimulate the ATPase activity of Ssb1, although stimulation by the Zuo1–Ssz1 complex was observed, as expected (9). These results suggest that Ssa is the in vivo Hsp70 partner of Jjj1.
Discussion
The results reported here strongly support the idea that the cytosolic J-protein Jjj1 plays an important role in a late cytosolic step in the biogenesis of 60S ribosomal subunits. First, Jjj1 comigrates with 60S subunits. Second, Δjjj1 cells are cold sensitive and have a decrease in the amount of 60S subunits compared with WT cells. Third, analysis of Δjjj1 polysome profiles revealed the presence of half-mers. These phenotypes are strikingly similar to those of cells lacking Rei1, a cytosolic factor required in a late step of 60S ribosome biogenesis. It has been postulated that the cytosolic protein Rei1 facilitates removal of two proteins that are also important for 60S subunit biogenesis, Arx1 and Alb1, which exit the nucleus bound to the pre-60S subunit, allowing subsequent steps of subunit maturation (14, 15). In good part, this hypothesis is based on the fact that in the absence of Rei1, Arx1 remains ribosome-associated (14), and both Arx1 and Alb1 fail to recycle to the nucleus (14, 15). Additionally, deletion of either ARX1 or ALB1 suppresses the defects of a Δrei1 mutant (14, 15). Intriguingly, Arx1 also remains ribosome-associated, and fails to recycle, in Δjjj1 cells. The growth defect of Δjjj1 is suppressed by the absence of Arx1 or Alb1 as well.
Given that Δjjj1 and Δrei1 cells have nearly identical phenotypes, how might Jjj1 be functioning in ribosome biogenesis? It is possible that Rei1 and Jjj1 function together, perhaps directly or indirectly causing a conformational change that reduces the affinity of Arx1/Alb1 for the pre-60S ribosome, causing their dissociation. There are precedents for J-protein:Hsp70 machinery being used for the remodeling of protein–nucleic acid and protein–protein complexes. For example, DnaJ and DnaK of Escherichia coli are required for dissociation of λP protein from initiation complex to allow phage λDNA replication (23); in eukaryotic cells, the J-protein auxilin and Hsc70 are required for uncoating of clathrin-coated vesicles (24). The dissociation/recycling of two other pre-60S ribosomal biogenesis factors, Nmd3 and Tif6, from the cytosol are thought to require the GTPases, Lsg1 and Efl1, respectively (25–27). In addition, ATPases of the AAA-ATPase and ABC-family ATPases are required in 60S biogenesis (28, 29). Thus, it is possible that in all of these cases, energy from the hydrolysis of nucleotides may be used for remodeling of pre-60S ribosomal particles to facilitate generation of mature subunits.
The association of two J-proteins, Zuo1 and Jjj1, with the ribosome raises questions of overlap in function as well as whether they share an Hsp70 partner. It is well established that Zuo1 functions in partnership with Ssb (9). On the other hand, our data point to Jjj1 partnering with Ssa. This conclusion is supported by the ability of Jjj1 to stimulate the ATPase activity of Ssa but not Ssb. We have no evidence that Zuo1 plays a role in ribosome biogenesis, because Δzuo1 cells do not accumulate half-mers. In addition, increased expression of Zuo1 does not suppress the defects observed in cells lacking Jjj1. On the other hand, increased expression of Jjj1 partially suppressed both the slow growth and the cation-sensitive phenotype of cells lacking Zuo1. We found that overexpression of Jjj1 was not only able to rescue the Δzuo1 strain, but Δssb1/2, Δssb1/2 Δzuo1, and Δssb1/2 Δzuo1 Δssz1 strains as well, further supporting the idea that Zuo1 and Jjj1 function with different Hsp70 partners.
The data reported here, together with previously published data, suggest to us the following scenario. Normally, Jjj1 preferentially associates with cytosolic pre-60S ribosomal particles and, with Ssa as its Hsp70 partner, facilitates ribosome biogenesis, perhaps aiding in the recycling of factors such as Alb1 and Arx1. Such a function is consistent with its predominant comigration with 60S subunits. Zuo1, which is ≈40 times more abundant than Jjj1 and present in a 1:1 stoichiometry with ribosomes, binds near the ribosome exit site, and, working with its regulatory factor Ssz1 and its Hsp70 partner Ssb, facilitates early stages of protein folding. Although perhaps not a normal occurrence, Jjj1 may also be competent to bind to mature 60S subunits. Indeed, when Jjj1 is overexpressed 20-fold, it all remains ribosome associated (data not shown). Jjj1 may be able to recruit the Hsp70 Ssa to ribosomes, fostering its interaction with nascent polypeptide chains and allowing this chaperone machinery to substitute for Zuo1/Ssb in protein folding. Such an ability of Ssa to perform the function of Ssb is reminiscent of the ability of the human Zuo1 ortholog, Mpp11, to substitute for Zuo1 (30). In this case, Mpp11 partners with the highly conserved Ssa, rather than the fungal-specific Ssb. Mpp11, which stimulates the ATPase activity of Ssa, but not Ssb, can rescue both a Δzuo1 and a Δzuo1 Δssb1/2 mutant (30).
Our unpublished results suggest that Jjj1 and Zuo1 binding to the ribosome may not be mutually exclusive. Even so, the ability of Jjj1 to substitute for Zuo1 may not be surprising, because precise positioning of Hsp70 at the exit site may not be required for its ability to participate in protein folding. We have previously reported that the peptidyl-prolyl isomerase Trigger Factor, a ribosome-associated chaperone on the E. coli ribosome, can functionally substitute for the Zuo1/Ssb/Ssz1 machinery, even though they appear to bind to different sites, because their binding to the ribosome is not mutually exclusive (21). However, Trigger Factor does bind close to the ribosome exit site, through its interaction with Rpl23, the ortholog to Rpl25 of eukaryotes (31). Evidence suggests that Rei1 may associate with pre-60S subunits in the vicinity of the exit tunnel, as Rpl25-GFP, but not Rpl25 tagged with the smaller HA epitope, blocks association of Arx1 with the ribosome (14). Because Jjj1 also is important for proper recycling of Arx1 and can perform some of the functions of Zuo1, which is thought to function with Ssb in nascent chain protection, it is tempting to speculate that it may also be located near the exit of the ribosome tunnel.
Jjj1's function in ribosome biogenesis appears to be specific. However, despite the ability of Jjj1 to partially substitute for Zuo1, the basis of such specificity, be it in its positioning on the ribosome or its direct interaction with other factors as well as the exact mechanism of action of J-protein:Hsp70 machinery in ribosome biogenesis awaits further study.
Materials and Methods
Yeast Strains, Plasmids, and Genetic Techniques.
JJJ1 was obtained by PCR-amplification of genomic DNA from positions 1–2280 by using Pfu Turbo polymerase (Stratagene, La Jolla, CA) and cloned into the XbaI and BamHI sites in pRS415GPD (32) to generate pRS415GPD JJJ1. WT and mutants of JJJ1, generated by using QuikChange (Stratagene), were cloned into pRS316 (33). To create Jjj1-HA, 3 tandem copies encoding the hemagglutinin tag (HA) were inserted before the stop codon in pRS316 JJJ1. For Zuo1 overexpression, DNA from positions 1–1802 was amplified and cloned into pRS415TEF by using ClaI and BamHI, to generate pRS415TEF ZUO1. To overexpress SSZ1, codons 1–2617 were PCR amplified and cloned into the BamHI and SalI sites of pRS416GPD to generate pRS416GPD SSZ1.
To obtain a null allele of JJJ1, positions −215 to 2115 of the jjj1::KanMX4 cassette were PCR amplified from a homozygous diploid yeast genomic knockout collection (Open Biosystems, Huntsville, AL) (34) and transformed into DS10 (GAL2 his3-11,15 leu2-3112 lys1 lys2 Δtrp1 ura3-52), a derivative of S288C. The KanMX4 cassette was replaced with TRP1 through a marker swap plasmid (35). Δrei1, Δarx1, and Δalb1 were obtained by PCR amplification of the KanMX4-marked genes from the knockout collection and homologous recombination in DS10. To create multiple deletion strains, such as Δssb1::HIS3 Δssb2::TRP1 Δzuo1::URA3 and Δssb1::HIS3 Δssb2::TRP1 Δzuo1::URA3 Δssz1::LYS2, appropriate haploid strains were mated and sporulated (36). Strains having ARX1 tagged with three tandem copies of the HA epitope or GFP inserted in the chromosome at the ARX1 locus were made by homologous recombination in WT BY4741 and Δjjj1 DS10 (37).
Minimal and rich glucose-based media were as described (8). Plates without additions, or with addition of paromomycin (100 μg/ml), were incubated for 2 days at 30°C or 3 days at 23°C, unless otherwise indicated. Plates having 0.5 M NaCl were incubated for 3 days at 30°C. In cases where strains were to be plated on minimal selective media, cells were first transformed with the appropriate empty vector for comparison with strains having test plasmids. All chemicals were obtained from Sigma (St. Louis, MO) unless otherwise stated.
Analysis of Cell Extracts.
Separation of 40S and 60S subunits under low-magnesium conditions was performed as described (10), with slight modifications. One liter of the appropriate yeast culture was grown in yeast peptone dextrose (YPD) medium at 23°C to an optical density of 0.5–1 at 600 nm. Cells were harvested by centrifugation at 4°C and washed with 14 ml of ice-cold breaking buffer B [50 mM Tris·HCl (pH 7.4)/50 mM NaCl/1 mM dithiolthreitol]. Cells were centrifuged at 4,000 × g for 5 min and resuspended in 3.5 ml of breaking buffer B plus 140 units of rNAsin (Promega, Madison, WI). Lysis of cells and clearing of lysate was performed as described (12). Ten OD254 units of cleared lysate were layered on 10-ml 15–30% sucrose gradients prepared in breaking buffer B. Gradients were centrifuged in an SW40 Ti rotor at 4°C at 40,000 rpm for 4.5 h. Fractions of 0.6 ml were collected, and proteins were precipitated with 86% acetone overnight at −20°C before immunoblot analysis. For analysis of 60S/40S ratios, profile images were scanned at identical resolutions, and the areas under each peak were quantified by using NIH Image v1.63. Lysate preparation and sucrose gradient centrifugation for polysome analysis was carried out essentially as described (12, 26).
Jjj1 Purification.
WT and mutant JJJ1 were PCR amplified such that DNA encoding a His tag was inserted at the C terminus. DNA was digested with NdeI and BamHI and then ligated into the pET3a vector (Novagen, San Diego, CA). The plasmids were transformed into BL21 pLys cells lacking DnaK and DnaJ for protein expression. Proteins were induced at 30°C with the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at an OD600 of 0.6 and incubated for an additional 4 h. The cell lysates were prepared by French press, and purification was performed following the His-tag protein purification protocols from Novagen.
Other Methods.
Other procedures were carried out as described: isolation of Hsp70–ATP complexes and ATPase assays (9), microscopy (14), indirect immunofluorescence experiments (19), and LMB experiments (26). Immunoblot detection was carried out by using the ECL system (Amersham Pharmacia Biotech, Piscataway, NJ) according to the manufacturer's suggestion. The anti-Jjj1 antibody was raised in rabbits against a fusion of amino acid 304–590 of Jjj1 to GST. Anti-Rpl3 was a gift from Jon Warner (Albert Einstein College of Medicine, Bronx, NY). Anti-HA was purchased from Covance (Denver, PA). Anti-Rpl8, anti-Nmd3, and anti-Zuo1 were produced as reported (12, 19, 38).
Acknowledgments
We thank J. Warner for antibodies specific for L3. This work was supported by National Institutes of Health Grants GM31107 (to E.A.C.) and GM53655 (to A.W.J.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Craig EA, Huang P, Aron R, Andrew A. Rev Physiol Biochem Pharmacol. 2006;156:1–21. doi: 10.1007/s10254-005-0001-0. [DOI] [PubMed] [Google Scholar]
- 2.Bukau B, Weissman J, Horwich A. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 3.Walsh P, Bursac D, Law YC, Cyr D, Lithgow T. EMBO Rep. 2004;5:567–571. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craig EA, Huang P. In: Protein Folding Handbook. Buchner J, Kiefhaber T, editors. Vol 4. Weinheim, Germany: Wiley; 2005. pp. 490–515. [Google Scholar]
- 5.Hundley H, Eisenman H, Walter W, Evans T, Hotokezaka Y, Wiedmann M, Craig E. Proc Natl Acad Sci USA. 2002;99:4203–4208. doi: 10.1073/pnas.062048399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gautschi M, Mun A, Ross S, Rospert S. Proc Natl Acad Sci USA. 2002;99:4209–4214. doi: 10.1073/pnas.062048599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craig EA, Eisenman HC, Hundley HA. Curr Opin Microbiol. 2003;6:157–162. doi: 10.1016/s1369-5274(03)00030-4. [DOI] [PubMed] [Google Scholar]
- 8.Kim S, Craig E. Eukaryotic Cell. 2005;4:82–89. doi: 10.1128/EC.4.1.82-89.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang P, Gautschi M, Walter W, Rospert S, Craig EA. Nat Struct Mol Biol. 2005;12:497–504. doi: 10.1038/nsmb942. [DOI] [PubMed] [Google Scholar]
- 10.Foiani M, Cigan AM, Paddon CJ, Harashima S, Hinnebusch AG. Mol Cell Biol. 1991;11:3203–3216. doi: 10.1128/mcb.11.6.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 12.Yan W, Schilke B, Pfund C, Walter W, Kim S, Craig EA. EMBO J. 1998;17:4809–4817. doi: 10.1093/emboj/17.16.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwase M, Toh-e A. Cell Struct Funct. 2004;29:1–15. doi: 10.1247/csf.29.1. [DOI] [PubMed] [Google Scholar]
- 14.Hung NJ, Johnson AW. Mol Cell Biol. 2006;26:3718–3727. doi: 10.1128/MCB.26.10.3718-3727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lebreton A, Saveanu C, Decourty L, Rain JC, Jacquier A, Fromont-Racine M. J Cell Biol. 2006;173:349–360. doi: 10.1083/jcb.200510080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helser TL, Baan RA, Dahlberg AE. Mol Cell Biol. 1981;1:51–57. doi: 10.1128/mcb.1.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Si K, Maitra U. Mol Cell Biol. 1999;19:1416–1426. doi: 10.1128/mcb.19.2.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 19.Ho JH, Kallstrom G, Johnson AW. J Cell Biol. 2000;151:1057–1066. doi: 10.1083/jcb.151.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gadal O, Strauss D, Kessl J, Trumpower B, Tollervey D, Hurt E. Mol Cell Biol. 2001;21:3405–3415. doi: 10.1128/MCB.21.10.3405-3415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Liang S, Tartakoff AM. EMBO J. 1996;15:6750–6757. [PMC free article] [PubMed] [Google Scholar]
- 22.Cyr DM, Douglas MG. J Biol Chem. 1994;269:9798–9804. [PubMed] [Google Scholar]
- 23.Zylicz M, Ang D, Liberek K, Georgopoulos C. EMBO J. 1989;8:1601–1608. doi: 10.1002/j.1460-2075.1989.tb03544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemmon SK. Curr Biol. 2001;11:R49–R52. doi: 10.1016/s0960-9822(01)00010-0. [DOI] [PubMed] [Google Scholar]
- 25.Senger B, Lafontaine DL, Graindorge JS, Gadal O, Camasses A, Sanni A, Garnier JM, Breitenbach M, Hurt E, Fasiolo F. Mol Cell. 2001;8:1363–1373. doi: 10.1016/s1097-2765(01)00403-8. [DOI] [PubMed] [Google Scholar]
- 26.Kallstrom G, Hedges J, Johnson A. Mol Cell Biol. 2003;23:4344–4355. doi: 10.1128/MCB.23.12.4344-4355.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hedges J, West M, Johnson AW. EMBO J. 2005;24:567–579. doi: 10.1038/sj.emboj.7600547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galani K, Nissan TA, Petfalski E, Tollervey D, Hurt E. J Biol Chem. 2004;279:55411–55418. doi: 10.1074/jbc.M406876200. [DOI] [PubMed] [Google Scholar]
- 29.Dong J, Lai R, Jennings JL, Link AJ, Hinnebusch AG. Mol Cell Biol. 2005;25:9859–9873. doi: 10.1128/MCB.25.22.9859-9873.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hundley HA, Walter W, Bairstow S, Craig EA. Science. 2005;308:1032–1034. doi: 10.1126/science.1109247. [DOI] [PubMed] [Google Scholar]
- 31.Kramer G, Rauch T, Rist W, Vorderwulbecke S, Patzelt H, Schulze-Specking A, Ban N, Deuerling E, Bukau B. Nature. 2002;419:171–174. doi: 10.1038/nature01047. [DOI] [PubMed] [Google Scholar]
- 32.Mumberg D, Muller R, Funk M. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 33.Sikorski RS, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 35.Voth WP, Jiang YW, Stillman DJ. Yeast. 2003;20:985–993. doi: 10.1002/yea.1018. [DOI] [PubMed] [Google Scholar]
- 36.Eisenman H, Craig E. Mol Microbiol. 2004;53:335–344. doi: 10.1111/j.1365-2958.2004.04134.x. [DOI] [PubMed] [Google Scholar]
- 37.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 38.West M, Hedges JB, Chen A, Johnson AW. Mol Cell Biol. 2005;25:3802–3813. doi: 10.1128/MCB.25.9.3802-3813.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]