Abstract
Activation-induced cytidine deaminase (AID), which is essential to both class switch recombination and somatic hypermutation of the Ig gene, is expressed in many types of human B cell lymphoma/leukemia. AID is a potent mutator because it is involved in DNA breakage not only of Ig but also of other genes, including proto-oncogenes. Recent studies suggest that AID is required for chromosomal translocation involving cmyc and Ig loci. However, it is unclear whether AID plays other roles in tumorigenesis. We examined the effect of AID deficiency on the generation of surface Ig-positive B cell lymphomas in Emu-cmyc transgenic mice. Almost all lymphomas that developed in AID-deficient transgenic mice were pre-B cell lymphomas, whereas control transgenic mice had predominantly B cell lymphomas, indicating that AID is required for development of B but not pre-B cell lymphomas from cmyc overexpressing tumor progenitors. Thus, AID may play multiple roles in B cell lymphomagenesis.
Keywords: somatic hypermutation, Pim1, secondary hit, clonal expansion
Somatic hypermutation (SHM) originally was considered to take place specifically in the IgV genes (1). Subsequently, several genes other than those encoding Igs were shown to be targets of SHM in activated B cells and human B cell lymphomas. Such target genes include MYC, IG alpha, PAX5, BCL6, and PIM1 (2–4). Activation-induced cytidine deaminase (AID) has been shown to be essential for SHM, gene conversion, and class switch recombination (CSR), three types of genetic alteration induced by antigen stimulation of B lymphocytes (5–9).
Original studies on human lymphomas indicated that AID is expressed in B cell lymphomas of germinal center (GC) origin, such as diffuse large B cell lymphoma, Burkitt's lymphoma, and follicular lymphoma, where AID is expressed physiologically (3, 10, 11). However, more recent analyses have extended AID expression to other types of human B cell lymphoma, including chronic lymphocytic leukemia, Hodgkin's lymphoma, mantle cell lymphoma, mucosa-associated lymphoid tissue lymphoma, mediastinal B cell lymphoma, hairy cell leukemia, and acute lymphocytic leukemia (12–16). The results of these studies suggest that AID may be involved in the pathogenesis of human B cell malignancy, including not only GC-derived B cell lymphoma but also almost all other types of human B cell lymphoma.
Studies on AID transgenic (Tg) animals have revealed that all individual mice develop T cell lymphomas in which genes for non-Igs such as T cell receptor (tcr), cmyc, pim1, cd4, and cd5 accumulate massive mutations in the region 3′ proximal to each promoter (17, 18). The AID Tg mice also develop B cell lymphoma, albeit much less frequently (I-m.O. and T.H., unpublished data). Studies also report AID to be essential for the mouse chromosomal translocation T(12;15), which corresponds to the human chromosomal translocation t(8;14), the hallmark of endemic Burkitt's lymphoma, in IL-6 Tg mice (19, 20). However, these animals did not develop plasmacytoma; instead, they developed polyclonal adenopathy (19). By contrast, Unniraman et al. (21) reported that AID is not required for chromosomal translocation but is important for outgrowth of clones with the translocation in pristine-treated mice. However, Unniraman et al. did not explicitly assess tumor formation either. Therefore, a direct mechanistic link between AID and B cell lymphomagenesis has yet to be established.
It is believed that B cell lymphomagenesis requires not only overexpression of cmyc but also other genetic “hits” (22). To examine the role of AID in B cell tumorigenesis, we studied the influence of AID deficiency on the generation of pre-B or B cell lymphomas in Emu-cmyc Tg mice. We found that AID deficiency induced a marked phenotypic change from predominantly B to predominantly pre-B cell lymphomas in Emu-cmyc Tg mice. We examined whether AID may facilitate the accumulation of the secondary genetic hits that are required for malignant transformation of cmyc-overexpressing B cell tumor progenitors in Emu-cmyc Tg animals. We found that Pim1, one of the targets of aberrant SHM in T lymphomas of AID Tg mice (18), was mutated in B cell lymphoma but not in pre-B cell lymphoma. These results suggest that AID appears to facilitate the development of B cell lymphomas in cmyc Tg mice by introducing second hits.
Results and Discussion
Phenotypic Changes in Emu-cmyc Tg Tumors on AID+/+, AID+/−, and AID−/− Backgrounds.
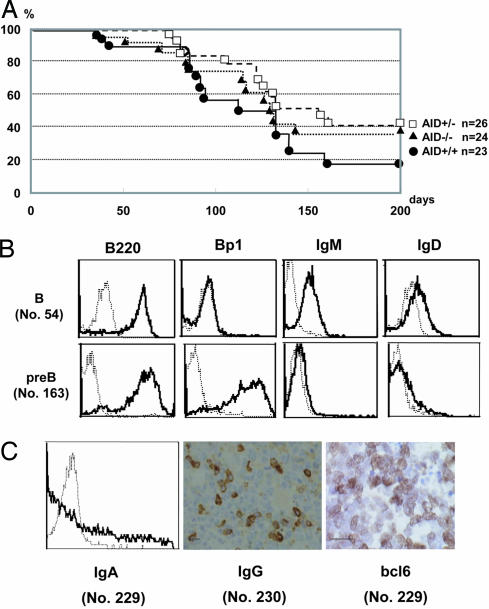
The survival curve of AID−/− Emu-cmyc Tg mice was similar (Fig. 1A) to those of AID+/− and AID+/+ Emu-cmyc Tg mice, except for slightly longer survival in heterozygous and homozygous AID mutants (AID+/+Emu-cmyc Tg mice vs. AID+/− Emu-cmyc Tg mice, P < 0.05; AID+/+ Emu-cmyc Tg mice vs. AID−/− Emu-cmyc Tg mice, P = 0.12). The survival curve of AID+/+ Emu-cmyc Tg mice is similar to that reported in ref. 23. The median survival periods were 112, 157, and 130 days for AID+/+, AID+/−, and AID−/− Emu-cmyc Tg mice, respectively. The tumors that developed in AID−/− Emu-cmyc Tg mice were morphologically indistinguishable from those occurring in AID+/+ and AID+/− Emu-cmyc Tg mice. In all cases, the lymph node architecture was completely destroyed. The predominant tumor cell was large and cleaved, and it had the appearance of an immunoblast: a large vesicular nucleus with prominent central nucleoli and a thick nuclear membrane, as reported in ref. 24. This histological feature revealed that the tumor resembled the human immunoblastic type of large B cell lymphoma or immunoblastic lymphoma in the Revised European–American Classification of Lymphoid Neoplasms (REAL classification).
Fig. 1.
Survival analysis and representatives of lymphomas in Emu-cmyc Tg mice. (A) Kaplan–Meier analysis of overall survival in AID+/+, AID+/−, and AID−/− Emu-cmyc Tg mice. There were no marked differences in myc-induced lymphomagenesis among AID+/+, AID+/−, and AID−/− Emu-cmyc Tg mice. Kaplan–Meier survival curves of AID+/+, AID+/−, and AID−/− Emu-cmyc Tg mice show that the average survival periods were 112, 157, and 130 days, respectively. Vertical lines indicate ages of surviving mice, and “n” indicates the number of mice in each group. (B) Flow cytometric analysis of B220, Bp1, IgM, and IgD expression typical in B cell lymphoma no. 54 (Upper) and pre-B cell lymphoma no. 163 (Lower). Each lymphoma is listed in Table 1. (C) Expression of IgA, IgG, and bcl6 in B cell lymphomas. (Left) Flow cytometric staining of IgA in no. 229 lymphoma. (Center) Immunohistological staining of IgG in no. 230 lymphoma. (Right) Immunohistological staining of bcl6 in no. 229 lymphoma. Table 1 defines lymphoma numbers. (Scale bars: 100 μm.)
The lymphomas that developed in Emu-cmyc Tg mice were categorized into pre-B and B cell lymphomas, depending on surface Ig expression (24, 25). The pre-B and B cell lymphomas were defined in this report by B220+Ig−Bp1+IgD− and B220+Ig+Bp1−, respectively (26–28). The B cell lymphomas expressed IgM, IgG, or IgA. The IgD was expressed on many but not all IgM+ B cell lymphomas (Fig. 1B and Table 1). The majority (6/8) of tumors in Emu-cmyc Tg AID+/+ mice were B cell lymphomas, with the remaining being pre-B cell lymphomas. These results are in good agreement with the previous report that 70% of the tumors in Emu-cmyc Tg mice were B cell lymphomas, with the remaining tumors being pre-B cell lymphomas (25). In contrast, almost all lymphomas (9/10) in AID−/− Emu-cmyc Tg mice were pre-B cell lymphomas.
Table 1.
Phenotypical analysis of lymphomas in AID+/+, AID+/−, and AID−/− Emu-cmyc Tg mice
| Lymphoma | B220 | Bp1 | IgM | IgD | IgG/IgA | AID | bcl6 | Pre-B/B |
|---|---|---|---|---|---|---|---|---|
| AID+/+ | ||||||||
| 163 | + | + | − | − | − | − | − | Pre-B |
| 172 | + | +* | − | − | − | − | − | Pre-B |
| 20 | + | −† | + | + | − | + | −† | B |
| 28 | +* | −† | + | + | − | − | −† | B |
| 54 | + | − | + | + | IgG‡ | + | − | B |
| 229 | + | − | − | − | IgA | − | + | B |
| 230 | + | −† | − | − | IgG§ | − | + | B |
| 321 | + | − | + | + | − | − | − | B |
| AID+/− | ||||||||
| 45 | + | − | − | − | − | ND | − | Pro-B¶ |
| 1 | + | +* | − | − | − | − | −† | Pre-B |
| 34 | + | ND | − | − | − | ND | −† | Pre-B |
| 74 | + | + | − | − | − | − | − | Pre-B |
| 86 | + | +* | − | − | − | − | −† | Pre-B |
| 166 | + | + | − | − | − | − | − | Pre-B |
| 185 | + | + | − | − | − | − | − | Pre-B |
| 7 | +* | −† | +‖ | ND | − | − | + | B |
| 60 | + | −† | + | + | − | + | −† | B |
| 112 | + | −† | + | + | − | − | − | B |
| 162 | + | − | + | − | IgG‡ | + | + | B |
| 169 | + | ND | − | − | IgA | − | + | B |
| 216 | + | − | + | − | − | + | + | B |
| 226 | + | − | + | + | − | − | + | B |
| AID−/− | ||||||||
| 5 | + | −† | − | − | − | − | ND | Pre-B |
| 36 | + | +* | − | − | − | ND | −† | Pre-B |
| 130 | + | + | − | − | − | ND | − | Pre-B |
| 161 | + | + | − | − | − | ND | −† | Pre-B |
| 234 | + | + | − | − | − | ND | − | Pre-B |
| 236 | + | + | − | − | − | − | − | Pre-B |
| 237 | + | + | − | − | − | ND | −† | Pre-B |
| 238 | + | + | − | − | − | − | −† | Pre-B |
| 239 | + | + | − | − | − | ND | − | Pre-B |
| 254 | + | − | + | − | − | ND | + | B |
B220, Bp1, and IgM were considered positive when they were expressed in >50% of tumor cells by FACS. IgA, IgD, and bcl6 were considered positive when they were expressed in >20% of tumor cells by FACS (IgA and IgD) or immunohistochemistry (bcl6). AID was positive when mRNA expression of AID was >10% of that found in the total spleen cells by real-time PCR. ND, not determined.
*Strongly positive by RT-PCR.
†Negative by RT-PCR.
‡A few tumor cells (<5%) expressed IgG by immunohistochemistry.
§More than 20% of tumor cells expressed IgG by immunohistochemistry.
¶Defined as pro-B because of the presence of CD43 antigen, which is expressed from hematopoietic stem cells to pre-B cells.
‖Analyzed by immunohistochemistry.
Interestingly, the proportion of B and pre-B (including pro-B) cell lymphomas in AID+/− Emu-cmyc Tg mice was 1:1 (7/14). One lymphoma was classified as pro-B lymphoma because of its expression of the CD43 antigen (29). The relative decrease of B cell lymphomas in AID+/− Emu-cmyc Tg mice compared with AID+/+ Emu-cmyc Tg mice suggests that AID may have weak haploinsufficiency in the development of B cell lymphoma.
Molecular Features of B Cell Lymphomas in Emu-cmyc Tg Mice.
Among the B cell lymphomas analyzed, nos. 54, 229, and 230 in AID+/+ Emu-cmyc Tg mice and nos. 162 and 169 in AID+/− Emu-cmyc Tg mice showed IgG or IgA expression (Table 1), which was assessed by immunohistochemistry or FACS, respectively (Fig. 1C), and confirmed by RT-PCR to detect postswitch products (data not shown). Among these tumors, only nos. 54 and 162 expressed significant levels (>10% of that found in the total spleen cells) of AID. Other AID-negative IgG/IgA-expressing tumors must have once expressed AID because CSR never occurs without AID (7). Some of the IgM-positive B cell lymphomas (nos. 20, 60, and 216) expressed similar amounts of AID, whereas AID expression was not observed in any of the pre-B cell lymphomas. Several human IgM-positive B cell lines are known to express AID, some of which continue to switch or accumulate SHM in vitro (30–32). The majority of tumor cells in nos. 54 and 162 expressed IgM, but their small fractions expressed IgG, suggesting that they may continue to switch.
More than half of the B cell lymphomas (nos. 229, 230, 7, 162, 169, 216, 226, and 254) expressed bcl6 in the nucleus as assessed by histochemical staining (Fig. 1C and Table 1). Because expression of bcl6 and occasional CSR in some B cell lymphomas in Emu-cmyc Tg mice indicates that they are derived from GC/post-GC B cells (33), the V regions in the heavy chain were sequenced to detect SHM. Unexpectedly, the V regions in seven B cell lymphomas from Emu-cmyc Tg mice did not have SHM at all (Table 2). The occurrence of CSR, expression of bcl6, and unmutated V regions provide conflicting results to define the tumor origin in the normal scheme of B cell development (34–36). However, recent studies have revealed that the discrepancy among the incidence of CSR, bcl6 expression, and SHM can happen in pathophysiological conditions. In T cell-independent manners, CSR can occur without SHM at non-GC sites, such as the extrafollicular site (37). The subgroup of human chronic lymphocytic leukemia with unmutated V regions undergoes active CSR (16). Burkitt-like lymphoma cells developing in cmyc knockin mice express bcl6 but do not show SHM in the V regions, which indicates that deregulated expression of cmyc induces expression of bcl6 in cells negative for Ig SHM (38). Most importantly, AID can be expressed even in Ig SHM-negative B lymphomas, regardless of whether they derive from GC or non-GC cells (10, 12–16, 39).
Table 2.
Analysis of SHM in the V region and Pim1 genes in pre-B and B lymphomas in AID+/+ and AID+/− Emu-cmyc transgenic mice
| Lymphoma | Pre-B/B | V region sequence |
Pim1 mutation |
|||
|---|---|---|---|---|---|---|
| VH | JH | Mutation† | Location of mutation | Amino acid substitution | ||
| AID+/+ | ||||||
| 20 | B | V10.3b* | ND | None | None | None |
| 28 | B | Vh7183.3b | JH3 | None | 591T > C | 138S > P |
| 54, 229, and 230 | B | ND | ND | ND | None | None |
| 321 | B | V588* | ND | None | ND | ND |
| 163 and 172 | Pre-B | ND | ND | ND | None | None |
| AID+/− | ||||||
| 7 and 169 | B | ND | ND | ND | None | None |
| 60 | B | V588 | JH2 | None | ND | ND |
| 112 | B | V588 | JH2 | None | 720G > A | None |
| 162 | B | VOX* | ND | None | 720G > A | None |
| 216 | B | V588 | JH2 | None | ND | ND |
| 226 | B | ND | ND | ND | 720G > A | None |
| 1, 45, 74, 86, 166, and 185 | Pre-B | ND | ND | ND | None | None |
The genomic location of the mutations in Pim1 is numbered from the starting site of exon 1 (chromosome 17 at 29, 217, 824). The location of the amino acid substitution in Pim1 protein is numbered from the translational starting site (ENSMUSP00000024811). The genomic region for sequence analysis is that from chromosome 17 from 29, 218, 046 to 29, 218, 893. ND, not determined.
*Analyzed by RT-PCR.
†Homology to rearranged VH segments in the IgBLAST database (www.ncbi.nlm.nih.gov/igblast).
Mutations in Non-Ig Genes in B Cell Lymphoma.
Increasing numbers of genes have been identified as targets of aberrant SHM (2–4, 40). To analyze possible genomic instability induced by AID expression, we examined SHM on several genes. Nine genes [IG kappa, IGH, MB-1 (IG alpha), B29 (IG beta), PAX5, BCL6, MYC, PIM1, and ARHH] are known to have SHM in normal and malignant B cells (2–4, 40). Of these, we focused on Pim1 because it has been reported to act synergistically with cmyc in lymphomagenesis (41) and because we previously found missense mutations in pim1 in the T cell lymphoma of AID Tg mice (18). We determined the nucleotide sequence (848 bp) covering exons 1 through 4 in 10 B cell lymphomas derived from AID+/+ and AID+/− Emu-cmyc Tg mice. We found one missense mutation (S138P) and three silent mutations (720 G to A), whereas pre-B cell lymphomas from eight mice showed no mutations (Table 2). These mutations are not polymorphisms because we identified the mutations by comparing lymphoma sequences with the genomic sequence of the tails of the identical mouse. The locations of the mutations were the same as those found in T cell lymphomas of AID Tg mice (18). Although it has not been determined whether these mutations in pim1 conveyed a survival advantage, there is no doubt that the Pim1 gene was one of the SHM targets in B lymphomas. It therefore is possible that these clones might have accumulated mutations in other positions in and surrounding the Pim1 gene as well as other oncogenes.
Some B cell lymphomas in Emu-cmyc Tg mice had the unmutated V region and mutated Pim1 genes (nos. 28, 112, and 162). Under physiological conditions, IgV genes are the preferred targets of the SHM machinery over other oncogenes (3). However, this preference may not be applicable in lymphomas. A subgroup of chronic lymphocytic leukemia has been reported to have mutated bcl6 but unmutated IgV genes (42). Aberrant mutations in non-Ig genes could be enhanced by overexpression of cmyc. Supporting information (SI) Table 3 shows the results of spectral karyotyping analyses of the lymphomas that developed in AID+/+, AID+/−, and AID−/− Emu-cmyc Tg mice. These analyses indicated that none of the lymphomas from the three genetic backgrounds had apparent chromosomal translocations.
Model for the Involvement of AID in B Cell Lymphomagenesis.
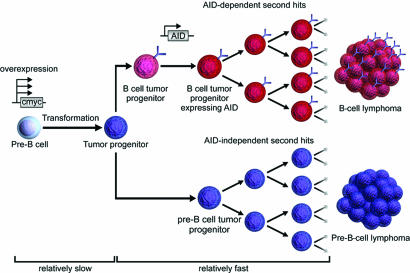
Chromosomal translocations involving cmyc have been suggested to be the triggering but not sufficient event in the development of lymphoma (22, 24, 43). It is likely that B lineage cells in Emu-cmyc Tg mice require several additional genetic alterations for lymphomagenesis. The absence of drastic differences in the survival period among AID+/+, AID+/−, and AID−/− Emu-cmyc Tg mice indicates that AID does not augment the overall efficiency of tumorigenesis. In other words, the rate-limiting step of tumorigenesis resides before AID expression. Because Tg cmyc is expressed at or before the pre-B stage, tumor progenitors are likely to arise slowly at the pre-B cell stage. Pre-B cell lymphomas do not depend on AID, which is in agreement with the fact that AID expression has not been shown in pre-B cells, although a very low level of AID was reported to be expressed in immature B cells (10, 44). Pre-B cell tumor progenitors can differentiate into the B cell stage, as reported in ref. 24. Because AID is required for formation of B cell lymphoma, AID may give additional genetic alterations in B cell tumor progenitors and facilitate outgrowth of B cell lymphomas. Otherwise, B cell tumor progenitors may die out without expansion (Fig. 2). By contrast, tumor progenitors at the pre-B cell stage may gain a final genetic hit spontaneously to grow out selectively and fix tumorigenesis. This phenomenon occurs because pre-B cells proliferate four to eight times faster than B cells do, and this phenomenon is enhanced further by aberrant cmyc expression (45). Because AID is critically involved in chromosomal translocation, as reported recently by Ramiro et al. (20), AID may have dual functions in B cell lymphoma development: initiation by chromosomal translocation and clonal expansion by mutagenesis. Because of its multiple functions in B cell lymphoma development, AID could become a useful therapeutic target.
Fig. 2.
Hypothetical scheme for AID involvement in development of B cell lymphoma in Emu-cmyc Tg mice. Development of B cell or pre-B cell tumor in Emu-cmyc Tg mice derives from pre-B cell progenitors (light blue circles), which already are transformed by overexpression of cmyc. Some tumor progenitor cells (blue circles) differentiate into B cells expressing IgM (light red circles). Some of these IgM-positive B cell tumor progenitors are activated to express AID, which may introduce second hits in the genome (dark red circles), resulting in the generation of B cell lymphoma. Most of the B cell tumor progenitors in AID−/− Emu-cmyc Tg mice fail to grow out without second hits. On the other hand, pre-B tumor progenitor (dark blue circles) can grow in AID−/− Emu-cmyc Tg mice probably because of secondary hits associated with rapid proliferations.
Methods
Mice.
AID-deficient mice (C57BL/6) were interbred with Emu-cmyc Tg mice (24) (inbred C57BL/6 strain; kindly provided by Alan Harris, The Walter and Eliza Hall Institute of Medical Research, Melbourne PO Royal Hospital, Victoria, Australia). The offspring were intercrossed to obtain AID+/+, AID+/−, and AID−/− Emu-myc Tg littermates. Kaplan–Meier curves represent the percentage of overall survival. The log-rank test was used to determine the statistical significance of differences in survival between the different genotypes of Emu-cmyc Tg mice. All mouse protocols were approved by the Institute of Laboratory Animals, Faculty of Medicine, Kyoto University.
FACS.
Single-cell suspensions from the spleens and lymph nodes of mice were stained with the following antibodies: FITC-conjugated anti-IgM (clone II/41), allophycocyanin-conjugated anti-B220 (clone RA3–6B2), phycoerythrin (PE)-conjugated anti-Bp1 (clone FG35.4), PE-conjugated anti-IgD (clone 11–26c), PE-conjugated anti-IgA (clone mA-6E1), and FITC-conjugated anti-IgE (clone F23.1; eBioscience, San Diego, CA). At least 10,000 live cells were analyzed on a FACSCalibur flow cytometer with CELLQuest software (BD Biosciences, San Jose, CA). Dead cells were excluded from the analysis by forward-scatter and side-scatter intensity and propidium-iodide gating.
Histology and Immunohistochemistry.
Tissue sections were fixed overnight in Mildform (Wako, Osaka, Japan). The sections were stained with hematoxylin and eosin or immunostained as described in ref. 28 by using the following primary antibodies: rabbit anti-bcl6 (clone N3; Santa Cruz Biotechnology, Santa Cruz, CA), anti-mouse IgG (clone R11–89; BD Biosciences) anti-mouse IgM (clone II/41; BD Biosciences), anti-mouse Bp-1 (clone 6C3; eBioscience), and anti-mouse CD43 (clone S7; BD PharMingen, San Diego, CA).
Real-Time PCR.
Expression of AID was measured with real-time PCR by using IQ SYBR green supermix and iCycler iQ (Bio-Rad, Hercules, CA) (46). Expression levels were normalized to that of GAPDH mRNA. The oligonucleotide sequences used were as follows: AID-F, 5′-CGTGGTGAAGAGGAGAGATAGTG-3′; AID-R, 5′-CAGTCTGAGATGTAGCGTAGGAA-3′; GAPDH-F, 5′-TGTGTCCGTCGTGGATCTGA-3′; and GAPDH-R, 5′-CCTGCTTCACCACCTTCTTGAT-3′. The PCR conditions were 40 cycles of 95°C for 15 sec and 60°C for 1 min. All reactions were performed in duplicate.
Preparation of Genomic DNA, Sequence, and SHM Analysis.
Genomic DNA from the spleens and lymph nodes of Emu-cmyc Tg mice was extracted according to standard methods. The Pim1 and IgH genes were amplified by genomic PCR using Pyrobest polymerase (Takara, Shiga, Japan). The oligonucleotide sequences used for genomic PCR were as follows: Pim1–1-F, 5′-GCAACGCCACCCGCAGTCTGAG-3′ and Pim1–1-R, 5′-CCAGCACCTGCCAGAAGAAT-3′ (for Pim1 exon 1–4 fragment); and VH J558-F, 5′-CAGCCTGACATCTGAGGACTC-3′ and JH4 intron-R, 5′-CTCCACCAGACCTCTCTAGAC-3′. The PCR conditions were 35 cycles of 94°C for 30 sec, 66°C for 30 sec, and 72°C for 1 min for Pim1 and 35 cycles of 94°C for 30 sec, 63°C for 30 sec, and 72°C for 2 min for VH. The Ig gene cDNA templates were amplified with 3′ primer corresponding to constant regions of expressed Ig genes and degenerate 5′ primers to variable regions. Primers were as follows: Cmm-R, 5′-CCCGAATTCGCTCTCGCAGGAGAC-3′; VH1-F, 5′-CCCGAATTCGAGGTGAAGCTGGTGGAGWC-3′; and VH2-F, 5′-CCCGAATTCCAGGTCCAGTTGCAGCAGWC-3′. PCR conditions were 30 cycles of 94°C for 30 sec, 60°C for 30 sec, and 72°C for 1 min. PCR products were gel-purified by using a PCR purification kit (Promega, Madison, WI) and sequenced directly on an ABI3700 sequencer (Applied Biosystems, Foster City, CA).
Supplementary Material
Acknowledgments
We thank R. Shinkura for useful suggestions and K. Fukui, Y. Shiraki, and T. Nishikawa for excellent secretarial help. This study was supported by the Takeda Science Foundation, the Japan Society for the Promotion of Science, and Centers of Excellence (COE) Grant 12CE2006 from the Ministry of Education, Culture, Sports, Science, and Technology.
Abbreviations
- AID
activation-induced cytidine deaminase
- SHM
somatic hypermutation
- CSR
class switch recombination
- GC
germinal center
- Tg
transgenic.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610732104/DC1.
References
- 1.Rajewsky K. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 2.Migliazza A, Martinotti S, Chen W, Fusco C, Ye BH, Knowles DM, Offit K, Chaganti RS, Dalla-Favera R. Proc Natl Acad Sci USA. 1995;92:12520–12524. doi: 10.1073/pnas.92.26.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RS, Kuppers R, Dalla-Favera R. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 4.Shen HM, Peters A, Baron B, Zhu X, Storb U. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- 5.Arakawa H, Hauschild J, Buerstedde JM. Science. 2002;295:1301–1306. doi: 10.1126/science.1067308. [DOI] [PubMed] [Google Scholar]
- 6.Harris RS, Sale JE, Petersen-Mahrt SK, Neuberger MS. Curr Biol. 2002;12:435–438. doi: 10.1016/s0960-9822(02)00717-0. [DOI] [PubMed] [Google Scholar]
- 7.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 8.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 9.Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 10.Greeve J, Philipsen A, Krause K, Klapper W, Heidorn K, Castle BE, Janda J, Marcu KB, Parwaresch R. Blood. 2003;101:3574–3580. doi: 10.1182/blood-2002-08-2424. [DOI] [PubMed] [Google Scholar]
- 11.Smit LA, Bende RJ, Aten J, Guikema JE, Aarts WM, van Noesel CJ. Cancer Res. 2003;63:3894–3898. [PubMed] [Google Scholar]
- 12.Cerutti A, Zan H, Kim EC, Shah S, Schattner EJ, Schaffer A, Casali P. J Immunol. 2002;169:6594–6603. doi: 10.4049/jimmunol.169.11.6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forconi F, Sahota SS, Raspadori D, Ippoliti M, Babbage G, Lauria F, Stevenson FK. Blood. 2004;104:3312–3317. doi: 10.1182/blood-2004-03-0950. [DOI] [PubMed] [Google Scholar]
- 14.Greiner A, Tobollik S, Buettner M, Jungnickel B, Herrmann K, Kremmer E, Niedobitek G. J Pathol. 2005;205:541–547. doi: 10.1002/path.1746. [DOI] [PubMed] [Google Scholar]
- 15.Hardianti MS, Tatsumi E, Syampurnawati M, Furuta K, Suzuki A, Saigo K, Kawano S, Takenokuchi M, Kumagai S, Matsuo Y, et al. Eur J Haematol. 2005;74:11–19. doi: 10.1111/j.1600-0609.2004.00338.x. [DOI] [PubMed] [Google Scholar]
- 16.Oppezzo P, Vuillier F, Vasconcelos Y, Dumas G, Magnac C, Payelle-Brogard B, Pritsch O, Dighiero G. Blood. 2003;101:4029–4032. doi: 10.1182/blood-2002-10-3175. [DOI] [PubMed] [Google Scholar]
- 17.Okazaki IM, Hiai H, Kakazu N, Yamada S, Muramatsu M, Kinoshita K, Honjo T. J Exp Med. 2003;197:1173–1181. doi: 10.1084/jem.20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotani A, Okazaki IM, Muramatsu M, Kinoshita K, Begum NA, Nakajima T, Saito H, Honjo T. Proc Natl Acad Sci USA. 2005;102:4506–4511. doi: 10.1073/pnas.0500830102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, Honjo T, Nussenzweig A, Nussenzweig MC. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 20.Ramiro AR, Jankovic M, Callen E, Difilippantonio S, Chen HT, McBride KM, Eisenreich TR, Chen J, Dickins RA, Lowe SW, et al. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Unniraman S, Zhou S, Schatz DG. Nat Immunol. 2004;5:1117–1123. doi: 10.1038/ni1127. [DOI] [PubMed] [Google Scholar]
- 22.Langdon WY, Harris AW, Cory S, Adams JM. Cell. 1986;47:11–18. doi: 10.1016/0092-8674(86)90361-2. [DOI] [PubMed] [Google Scholar]
- 23.Alt JR, Greiner TC, Cleveland JL, Eischen CM. EMBO J. 2003;22:1442–1450. doi: 10.1093/emboj/cdg133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 25.Egle A, Harris AW, Bouillet P, Cory S. Proc Natl Acad Sci USA. 2004;101:6164–6169. doi: 10.1073/pnas.0401471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grawunder U, Leu TM, Schatz DG, Werner A, Rolink AG, Melchers F, Winkler TH. Immunity. 1995;3:601–608. doi: 10.1016/1074-7613(95)90131-0. [DOI] [PubMed] [Google Scholar]
- 27.Lu LM, Ogawa M, Kamoto T, Yamada Y, Pataer A, Hiai H. Leuk Res. 1997;21:337–342. doi: 10.1016/s0145-2126(96)00124-5. [DOI] [PubMed] [Google Scholar]
- 28.Tsuruyama T, Nakamura T, Jin G, Ozeki M, Yamada Y, Hiai H. Proc Natl Acad Sci USA. 2002;99:8253–8258. doi: 10.1073/pnas.112202899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hardy RR, Hayakawa K. Annu Rev Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 30.He B, Raab-Traub N, Casali P, Cerutti A. J Immunol. 2003;171:5215–5224. doi: 10.4049/jimmunol.171.10.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muto T, Muramatsu M, Taniwaki M, Kinoshita K, Honjo T. Genomics. 2000;68:85–88. doi: 10.1006/geno.2000.6268. [DOI] [PubMed] [Google Scholar]
- 32.Poltoratsky V, Woo CJ, Tippin B, Martin A, Goodman MF, Scharff MD. Proc Natl Acad Sci USA. 2001;98:7976–7981. doi: 10.1073/pnas.141222198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cattoretti G, Chang CC, Cechova K, Zhang J, Ye BH, Falini B, Louie DC, Offit K, Chaganti RS, Dalla-Favera R. Blood. 1995;86:45–53. [PubMed] [Google Scholar]
- 34.Honjo T, Muramatsu M, Fagarasan S. Immunity. 2004;20:659–668. doi: 10.1016/j.immuni.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 35.Stevenson FK, Sahota SS, Ottensmeier CH, Zhu D, Forconi F, Hamblin TJ. Adv Cancer Res. 2001;83:81–116. doi: 10.1016/s0065-230x(01)83004-9. [DOI] [PubMed] [Google Scholar]
- 36.Ye BH, Cattoretti G, Shen Q, Zhang J, Hawe N, de Waard R, Leung C, Nouri-Shirazi M, Orazi A, Chaganti RS, et al. Nat Genet. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 37.MacLennan IC, Toellner KM, Cunningham AF, Serre K, Sze DM, Zuniga E, Cook MC, Vinuesa CG. Immunol Rev. 2003;194:8–18. doi: 10.1034/j.1600-065x.2003.00058.x. [DOI] [PubMed] [Google Scholar]
- 38.Zhu D, Qi CF, Morse HC, III, Janz S, Stevenson FK. Blood. 2005;105:2135–2137. doi: 10.1182/blood-2004-07-2573. [DOI] [PubMed] [Google Scholar]
- 39.Cattoretti G, Büttner M, Shaknovich R, Kremmer E, Alobeid B, Niedobitek G. Blood. 2006;107:3967–3975. doi: 10.1182/blood-2005-10-4170. [DOI] [PubMed] [Google Scholar]
- 40.Gordon MS, Kanegai CM, Doerr JR, Wall R. Proc Natl Acad Sci USA. 2003;100:4126–4131. doi: 10.1073/pnas.0735266100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shirogane T, Fukada T, Muller JM, Shima DT, Hibi M, Hirano T. Immunity. 1999;11:709–719. doi: 10.1016/s1074-7613(00)80145-4. [DOI] [PubMed] [Google Scholar]
- 42.Sahota SS, Davis Z, Hamblin TJ, Stevenson FK. Blood. 2000;95:3534–3540. [PubMed] [Google Scholar]
- 43.Harris AW, Pinkert CA, Crawford M, Langdon WY, Brinster RL, Adams JM. J Exp Med. 1988;167:353–371. doi: 10.1084/jem.167.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mao C, Jiang L, Melo-Jorge M, Puthenveetil M, Zhang X, Carroll MC, Imanishi-Kari T. Immunity. 2004;20:133–144. doi: 10.1016/s1074-7613(04)00019-6. [DOI] [PubMed] [Google Scholar]
- 45.Rolink AG, Andersson J, Melchers F. Eur J Immunol. 1998;28:3738–3748. doi: 10.1002/(SICI)1521-4141(199811)28:11<3738::AID-IMMU3738>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 46.Yabe D, Komuro R, Liang G, Goldstein JL, Brown MS. Proc Natl Acad Sci USA. 2003;100:3155–3160. doi: 10.1073/pnas.0130116100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.