Abstract
The tyrosine phosphatase PTPN22 allele 1858T has been associated with rheumatoid arthritis (RA) and other autoimmune diseases. RA is the most frequent of those multifactorial diseases. The RA association was usually restricted to serum rheumatoid factor positive disease (RF+). No interaction was shown with HLA-DRB1, the first RA gene. Many case-control studies replicated the RA association, showing an allele frequency increase of ≈5% on average and large variations of population allele frequencies (2.1–15.5%). In multifactorial diseases, the final proof for a new susceptibility allele is provided by departure from Mendel's law (50% transmission from heterozygous parents). For PTPN22–1858T allele, convincing linkage proof was available only for type 1 diabetes. We aimed at providing this proof for RA. We analyzed 1,395 West European Caucasian individuals from 465 “trio” families. We replicated evidence for linkage, demonstrating departure from Mendel's law in this subset of early RA onset patients. We estimated the overtransmission of the 1858T allele in RF+ families: T = 63%, P < 0.0007. The 1858T allele frequency increased from 11.0% in controls to 17.4% in RF+ RA for the French Caucasian population and the susceptibility genotype (1858T/T or T/C) from 20.2% to 31.6% [odds ratio (OR) = 1.8 (1.2–2.8)]. In conclusion, we provided the linkage proof for the PTPN22–1858T allele and RF+ RA. With diabetes and RA, PTPN22 is therefore a “linkage-proven” autoimmunity gene. PTPN22 accounting for ≈1% of the RA familial aggregation, many new genes could be expected that are as many leads to definitive therapy for autoimmune diseases.
Keywords: linkage analysis, R620W polymorphism, rheumatoid factor, transmission disequilibrium, LYP protein
Rheumatoid arthritis (RA), the most frequent autoimmune human disease, affects ≈1% of the adult population worldwide. RA being a multifactorial disease, the identification of a new factor provides solid ground for further research, from pathophysiology to clinical applications, including therapy to definitely cure the patients. Environmental factors, including smoking and female hormones, are also involved, along with several RA susceptibility genes (1). The first RA gene, HLA-DRB1, established as early as in 1978, codes for the adaptative immune system HLA class II molecules (2). The RA susceptibility alleles encode a homologous “shared epitope” of the HLA molecule, the pathophysiological mechanism of which remains unknown (3). Both association and linkage to RA were repeatedly shown, definitely establishing HLA-DRB1 as the first “association and linkage proven” RA gene, for which modelization taken into account both association and linkage data are available (4–6). The HLA locus contributing approximately 1/5 to 1/3 of the RA familial aggregation, other factors including new RA genes remain to be established (7, 8). From the large number of recent suggestions in case-control studies, only PTPN22 was clearly replicated so far (9, 10). Indeed, the 1858T allele of the PTPN22-C1858T single nucleotide polymorphism, resulting in the R620W amino acid substitution, has been associated in Caucasian populations with RA and other autoimmune diseases, including type 1 diabetes, systemic lupus and thyroid diseases (refs. 9, 11, and 12; reviewed in ref. 13). A common feature of those diseases is the presence of autoantibodies in the serum. However, some diseases with autoantibodies are not associated with PTPN22, such as systemic scleroderma (14). In RA, the association is mostly restricted to the form positive for the most common autoantibody, rheumatoid factor (RF) (13).
The PTPN22 gene, located on chromosome 1p13, encodes the lymphoid-specific tyrosine phosphatase LYP, involved in the suppression of T cell activation, and thereby in T-dependent antibody production (13). The R620W polymorphism affects a proline-rich motif of LYP, involved in protein–protein interactions, although the molecular mechanism of the disease susceptibility remains to be clarified, as mutually exclusive gain and loss of function mechanisms have both been suggested (9, 13, 15, 16).
The large number of PTPN22 association reports in various autoimmune diseases provides compelling evidence for an implication of this gene in susceptibility to autoimmune diseases (13), but the “gold standard” for multifactorial diseases, i.e., the linkage proof, is convincingly met only for type 1 diabetes (reviewed in 13). The most straightforward linkage proof for a putative susceptibility allele, first used for establishing definitely the insulin gene as a type 1 diabetes gene (17), is the demonstration of a departure from the Mendel's first law, which states that the probability of transmission, for each allele of a heterozygous parent, is 50%. The linkage proof is particularly important to obtain when the putative susceptibility allele frequency (i) exhibits only a small patients controls difference and (ii) varies greatly between populations. Indeed, the overlap between the range of allele frequencies observed for patients and controls in different studies, might lead to some false-positive findings. In such a situation, the smaller the frequency difference between patients and controls, the higher the risk of false-positives and the rarer the reports of negative findings. The large sample size, required to reach significance for a small difference, does increase the difficulty of the matching task, aggravating the risk that some degree of mismatching would account for the small difference observed between patients and controls. The rarity of negative reports would be explained, in addition to the well known publication bias, by the difficulty in reaching adequate power for meaningful negative findings, because of the sample size required: that sample would need to be larger than that of the initial positive report. In the particular situation of RA and PTPN22, the overwhelming replication of positive cases-controls studies in Caucasian populations of European origin is very reassuring (13). The cases-controls allele frequency difference within a given study ranges from 2.8% to 7.1%, i.e., ≈5% on average, whereas the population frequency ranges from 2.1% to 15.5% (13). Therefore, the linkage proof remains important to fully establish the PTPN22-RA association. The PTPN22-1858T RA linkage data published so far being un-conclusive (9, 18, 19), we took advantage of the largest reported European family resource dedicated to RA linkage studies. Specifically, we aimed at providing the linkage proof for the PTPN22-RF+ RA association.
Results
We analyzed in total 1,395 European Caucasian individuals from 465 trio families (one RA case and both parents): 319 French families and 146 from other continental West-European countries: 54 Italian, 40 Spanish, 24 Belgium, 14 Dutch, and 14 Portuguese (Table 1). We had observed preliminary linkage evidence in the RF+ subgroup in a sample of 200 French families: 61% transmission of the 1858T allele (P < 0.04) (18). We analyzed 265 additional families, which provided a 99% power to replicate true linkage, based on the results from the initial family set. We first checked, in the replication set, the absence of significant deviation from the Hardy-Weinberg equilibrium in controls, using the two untransmitted chromosomes of each trio family as one “virtual control” (data not shown). We replicated linkage evidence with, compared with the Mendel's expectation of 50%, an observed overtransmission of 66% in the 188 RF+ families of the replication sample: T (transmission testing Mendel's law) = 66%, P < 0.007) (Table 2). This replication demonstrated linkage to RF+ RA for the PTPN22–1858T allele.
Table 1.
Characteristics of rheumatoid arthritis index cases
| Index characteristics | Initial set 200 RA index | Replication set 265 RA index | Global sample 465 RA index | French Caucasian families 319 RA index | RF+ French Caucasian families with unaffected parents 218 RA index | Other continental West European Caucasian families 146 RA index |
|---|---|---|---|---|---|---|
| Female, n (%) | 177 (88.5) | 228 (86.0) | 405 (87.1) | 281 (88.1) | 191 (87.6) | 124 (84.9) |
| Mean age at RA onset, years ± SD | 31.7 ± 9.4 | 30.3 ± 9.4 | 30.9 ± 9.5 | 30.9 ± 9.5 | 31.1 ± 9.6 | 30.9 ± 9.3 |
| Mean disease duration, years ± SD | 15.3 ± 7.8 | 8.1 ± 7.0 | 11.2 ± 8.2 | 13.4 ± 8.2 | 12.4 ± 8.3 | 6.4 ± 5.8 |
| Bone erosions, n (%) | 169 (84.5) | 187 (72.2) | 356 (77.5) | 257 (80.6) | 182 (83.5) | 99 (70.2) |
| RF+, n (%) | 157 (78.5) | 188 (73.4) | 345 (75.6) | 239 (75.0) | 218 (100.0) | 106 (77.4) |
| Nodules, n (%) | 50 (25) | 42 (16.3) | 92 (20.1) | 74 (23.2) | 59 (27.3) | 18 (13.0) |
Table 2.
RF+ RA linkage proof for the PTPN22–1858T allele
| Samples | RF+ subgroup (n = 345) |
RF− subgroup (n = 120) |
All families (n = 465) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trans. | Un. | T | P | Trans. | Un. | T | P | Trans. | Un. | T | P | |
| Initial set (n = 200) | 55 | 35 | 61% | 0.02 | 11 | 11 | 50% | 0.6 | 66 | 46 | 59% | 0.04 |
| Replication set (n = 265) | 44 | 23 | 66% | 0.007 | 15 | 12 | 55% | 0.3 | 59 | 35 | 63% | 0.009 |
| Global sample (n = 465) | 99 | 58 | 63% | 0.0007 | 26 | 23 | 53% | 0.4 | 125 | 81 | 61% | 0.001 |
| French Caucasian subgroup (n = 319) | 80 | 45 | 64% | 0.001 | 20 | 15 | 57% | 0.2 | 100 | 60 | 62% | 0.001 |
| Other continental West European Caucasian subgroup (n = 146) | 19 | 13 | 59% | 0.2 | 6 | 8 | 43% | 0.4 | 25 | 21 | 54% | 0.3 |
Trans., transmitted 1858T allele; Un., untransmitted; T, percentage of transmission of the 1858T allele from heterozygous parents, compared with Mendel's expectation of 50%, using the exact test with calculation of the significance level (see Statistical Analysis).
In the RF− subgroup, we observed no significant linkage (P = 0.3). In the global sample (RF+ and RF− families), linkage remained significant (T = 63%, P < 0.009).
As the replication sample of 265 families showed no significant difference with the initial sample of 200 families for T, we pooled both samples to provide an improved estimation of Mendel's departure. We obtained T = 63% for RF+ RA (P < 0.0007). There was no significant difference between the paternal T and the maternal T (67% vs. 61%, P = 0.4). We observed no significant linkage in RF− families (T = 53%; P = 0.4). The linkage remained significant in the global sample (T = 61%; P < 0.001) (Table 2).
T for RF+ RA was similar in French and in other European families [T = 64% and 59%, respectively, P = 0.6 (Table 2)], suggesting that T might be a disease parameter independent from the population investigated. For RF− RA, T varied around 50% in those two populations, with no significant difference (T = 57% and 43%, respectively, P = 0.4), in keeping with the interpretation of the absence of linkage in this subgroup (Table 2).
Having established linkage, we obtained from those data an unbiased estimation of the PTPN22 association in RA. Indeed, such family data provide, for each case investigated, perfectly matched controls, as far as the population of origin is concerned: for each case, a “virtual control” is obtained from the two untransmitted parental chromosomes. Each case's chromosome, transmitted by a given parent, is perfectly matched for the population of origin with the untransmitted chromosome of that very same parent: both chromosomes come from the same individual. To get “disease-free controls,” families with one parent possibly affected with the disease need to be excluded.
To obtain reliable estimates for the association between RF+ RA and the PTPN22–1858T allele, we focused on the French Caucasian population, the sample size for other populations being too small. The criteria used for each of the trio families investigated, was the European Caucasian origin for each of the four parental chromosomes, i.e., each of the four grandparents. We selected RF+ RA and, to obtain true “disease-free” controls, we excluded families with one parent affected with RA or undefined inflammatory arthritis (n = 21), resulting in a 218 families sample (Table 1). We observed in controls a 1858T allele frequency of 11.0% and a susceptibility genotype (1858T/T or T/C) of 20.2%, increasing to 17.4%, [odds ratio (OR) = 1.7, 95% confidence interval (C.I.) = 1.1–2.5, P = 0.007] and 31.6% (OR = 1.8, 95% C.I. = 1.2–2.8, P = 0.006), respectively (Table 3).
Table 3.
Association of PTPN22-1858T allele and RF+ RA in the continental European Caucasian population
| Subgroups | Cases |
Controls* |
ORs (95% C.I.) |
Allele |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TT | TC | CC | TT | TC | CC | TT vs. CC | TC vs. CC | TTCT vs. CC | OR | P value | |
| Global sample (n = 465) | 12 | 114 | 339 | 7 | 80 | 378 | 1.86 (0.72–4.77) | 1.58 (1.15–2.19) | 1.61 (1.18–2.20) | 1.55 (1.17–2.04) | 0.002 |
| RF+ sample (n = 345) | 9 | 89 | 247 | 4 | 58 | 283 | 2.42 (0.73–7.95) | 1.75 (1.21–2.54) | 1.80 (1.26–2.59) | 1.73 (1.25–2.40) | 0.0008 |
| French global (n = 319) | 11 | 89 | 219 | 6 | 59 | 254 | 2.05 (0.75–5.64) | 1.74 (1.20–2.54) | 1.78 (1.24–2.55) | 1.68 (1.22–2.31) | 0.001 |
| RF+ French (n = 239) | 8 | 70 | 161 | 4 | 43 | 192 | 2.25 (0.66–7.61) | 1.93 (1.25–2.98) | 1.97 (1.30–3.00) | 1.83 (1.26–2.65) | 0.001 |
| RF+ French with unaffected parents (n = 218) | 7 | 62 | 149 | 4 | 40 | 174 | 1.94 (0.56–6.77) | 1.80 (1.14–2.83) | 1.82 (1.17–2.82) | 1.70 (1.15–2.51) | 0.007 |
*Controls are “virtual controls” derived from untransmitted alleles for each trio family.
There was no practical interest in describing the absence of association in the French rheumatoid factor negative RA subgroup, as no linkage was shown for the rheumatoid factor negative subgroup, nor in describing association data in the remaining sample, too heterogeneous globally in population of origin for allowing extrapolation and with too small a sample size per country for useful individual estimations.
Moreover, to confirm the absence of interaction between the two confirmed RA genes, HLA-DRB1 and PTPN22, we used the refined shared epitope classification, a successful modelization of HLA-DRB1 in RA (5) that we recently validated (20) and that is now in use (6). No significant difference was observed for the frequency of the 2 PTPN22 genotypes (1858T/T or T/C; 1858C/C) in the first fully HLA-DRB1-typed 200 French families between the 6 HLA-DRB1 genotypes recently described (4, 5) (data not shown).
Finally, the relative contribution of PTPN22 to RA genetics was estimated by using the conceptual frame developed by Risch (21, 22). Indeed, T could be used to estimate the effect of the deviation from Mendel's law on affected sib-pair analysis. For the PTPN22 locus, assuming that all of the genetic effect is brought by the 1858T allele, by using the estimates from the global French sample T = 62%, together with the parental 1858T allele frequency of 14% in this population, the excess of allele sharing identical by descent (IBD) at the PTPN22 locus for RA would be IBD = 51%, in very small excess to the Mendel's expectation of IBD = 50%. The affected sib pairs sharing no PTPN22 alleles identical by descent would be IBD_0 = 24%, just below the Mendel's expectation of IBD_0 = 25%. We would then obtain a PTPN22 locus contribution to RA familial aggregation of 1%, by using Risch's parameters lambdaPTPN22 = 1.02 and lambdasib = 11 (23).
Discussion
We demonstrated here linkage between RF+ RA and the PTPN22–1858T allele, by replication in a West European sample. We provided the linkage estimate T = 63% (P < 0.0007) for the largest European trio RA family resource published to date, measuring the over-transmission of the PTPN22–1858T allele compared with the 50% transmission expected from Mendel's law. We observed no significant difference for T among the French families belonging either to the initial set or to the 119 additional French families. So, the very significant P value in the French Caucasian RF+ subgroup, in comparison with those of the RF+ initial set alone should not be explained by particular RA characteristics of this subset (see Materials and Methods) nor by an increased transmission disequilibrium. We also observed no significant difference for T between the French and the mixed European sample, suggesting that T could be used as a disease parameter, here for RF+ RA, independent from the susceptibility allele frequency in the underlying population. Showing clear absence of linkage in the RF− subgroup, we provided strong evidence in favor of a restriction of this genetic effect to RF+ RA. This evidence implies that the PTPN22–1858T allele would be involved in one of the mechanisms underlying RA clinical heterogeneity, which it could contribute to identify.
Given the perfect population match between patients and disease-free virtual controls, we obtained an accurate estimation of the association for one population, the Caucasian French: the allele frequency increases from 11.0% in controls to 17.4% in patients (OR = 1.7, 95% C.I. = 1.1–2.5, P = 0.007) and the susceptible genotype 1858T/Tor T/C from 20.2% to 31.6% (OR = 1.8, 95% C.I. = 1.2–2.8, P = 0.006). Of course, this estimation could be reliably applied only to the same population and for similar disease, i.e., the French Caucasian population for relatively early onset RF+ RA.
We confirmed the lack of genotype correlation with the first RA gene, HLA-DRB1, by using the recently validated new classification of HLA-DRB1 RA genotypes. The contribution of PTPN22–1858T allele to RA genetics, if of major importance in quality, seems to be small in quantity compared with the strongest RA-associated HLA-DRB1 allele: T for the HLA-DRB1 “S2” allele, essentially *0401, is 77% (n = 200 families, P < 1.4 × 10−9) (5, 20). By using the conceptual frame developed by Risch (21, 22), we obtained a PTPN22 locus contribution to RA familial aggregation of 1%, lambdaPTPN22 = 1.02 and lambdasib = 11 (23). By comparison, the HLA locus contribution would be 19% (using the data obtained from the same population: lambdaHLA = 1.60) (7). This small locus contribution clearly shows that the PTPN22 gene is extremely difficult to detect by using affected sib pair analysis genome scan, because the difference between IBD = 51% and 50% is so small. It is not surprising that it remained indeed undetected at the French genome scan, although we used the first available refined genetic map (24, 25). RA genetics might therefore involve a large number of genes with contributions equivalent to that of PTPN22, together with a small number of genes detectable by affected sib pair analysis, the major one being HLA-DRB1.
These results are consistent with the literature. The evidence from cases-controls studies in European populations is compelling (13). There is only one recent publication of a negative report in a population partly of European origin, from Columbia, that could be clearly explained by the low 1858T allele frequency in that population (26). The few published linkage data are compatible with linkage. As explained above, the absence of consistent linkage suggestion observed in affected sib-pair analysis is expected (8, 24, 27). Begovich et al. were unsure of their linkage evidence (9) and we considered our first evidence as only preliminary (18). Seldin et al. provided linkage evidence only for the very isolated Finnish population, with parents missing in their family material so that no direct estimate of T could be provided, the authors requesting further linkage studies for confirmation (28).
A limitation of our study is to provide reliable association estimates only for one European Caucasian population and in a specific subset of RA patients with young age at RA onset. In addition, if the “control” frequencies can be considered reliable for that French population, the patient frequencies are representative only for early onset RA, given the requisite for parents participation in trio families. Also, the frequency of the homozygous genotype 1858T/T was too rare in the population investigated, to test for the dose effect on disease risk that has been reported (13, 29). The 1858T-negative PTPN22 haplotype that has been recently suggested to be an independent RA susceptibility allele was not investigated (30).
A number of new research studies are warranted. The involvement in RA heterogeneity is to be refined, in particular the correlation with other autoantibodies, such as anti-citrullinated antibodies (10). Taking advantage of the statistical independence between the PTPN22 and the HLA-DRB1 genotypes, a combined genetic test should be investigated for its potential contribution to early diagnosis of RA, along with rheumatoid factor and anti-citrullinated antibodies (31). However, evaluating the impact in terms of OR of the combination of these two genes by using a modelization approach should be of great interest. A role for PTPN22 as RA severity gene is to be thoroughly explored, although first reports are negative (32). A role in treatment response, especially in B cells directed treatment such as rituximab, is to be investigated (13). A model integrating both PTPN22 and HLA-DRB1 genetic contribution is warranted. The pathophysiology of PTPN22 susceptibility genotype needs to be clarified, to open new leads toward etiological treatment of RA (13). Rare mutations that might have high penetrance should be searched for and investigated, as the same gene can contribute to a disease not only with a common susceptibility allele of low penetrance, but also with rare deleterious mutations of high penetrance, such as RET gene in Hirschprung disease (33). Such a mutation was reported and needs to be investigated (30). This gene is to be further explored for various populations, to provide accurate genotype association estimates for RA as well as for other autoimmune diseases. It would be particularly interesting to determine why PTPN22 is clearly involved in some diseases with autoantibodies, but not with all, such as systemic scleroderma (14). Family based association studies will help to validate associations. The few positive reports of an association with RF− RA, in view of the vast majority of negative reports, might be false positives, as suggested by the clear negative linkage findings in our study (13). There were association reports for one form of idiopathic juvenile arthritis, but a well powered negative linkage study raised serious doubts on the reality of the association (28, 34–36).
This success stimulates the search for new RA and autoimmunity genes, keeping in mind that some genes could be population specific, such as PTPN22, which 1858T allele is very rare or absent in Asian and African populations. As far as RA is concerned, a large number of susceptibility genes could be expected, many of which being virtually undetectable at affected sib pair analysis genome scan. The superior power of the trio families based linkage studies, compared with affected sib-pair studies, for such genes as PTPN22, has long been demonstrated (37). With one additional recent success for the IFN regulatory factor IRF5 gene in systemic lupus erythematosus, the field of multifactorial diseases genetics is now increasingly successful (38).
In conclusion, together with the literature, this study provides the final piece of the “association and linkage proof” for PTPN22 as the second RA gene, 28 years after the detection of HLA-DRB1 (2) and, together with type 1 diabetes, for PTPN22–1858T as an autoimmunity susceptibility allele (13, 16). This success of multifactorial diseases genetics lays solid grounds for research on those frequent and severe autoimmune diseases, from pathophysiology to clinical applications, including definitive therapy.
Materials and Methods
Patients and Families.
The initial sample was made of the DNA from 200 trio families from French Caucasian origin. The replication sample was made of the DNA from 265 trio families with one RA patient and both parents, from West European Caucasian origin as recorded for each of the four grandparents, that had been recruited through ECRAF (the European Consortium on Rheumatoid Arthritis Families). RA fulfilled the 1987 American College of Rheumatology [formerly, the American Rheumatism Association) criteria (39)]. Clinical data were reviewed by three rheumatologists, all former university fellows (S.L., P. Fritz, and A.-C.R.). All individuals provided informed written consent, and the study was approved by ethics committees in each country.
For each index, characteristics collected were sex, age at RA onset, disease duration in years, presence of bone erosions at x-ray examination, presence of rheumatoid nodules, and seropositivity for rheumatoid factor (ever, as determined by Latex fixation, by Waaler-Rose assay or by Laser Nephelometry). The anti-citrullinated antibodies status was not available.
Clinical characteristics of the RA index cases are presented in Table 1. The relatively young mean age of RA onset was explained by the inclusion criteria, with the request for two living parents. The characteristics of the 119 RA French Caucasian index, not presented in Table 1, did not differ significantly from those of the 200 RA index from the initial set, in terms of bone erosions (73.9%), rheumatoid factor positivity (68.9%), nodules (20.5%), and mean age at RA onset (29.5 ± 9.5 years).
Genotyping.
Blood samples were collected for DNA extraction and genotyping. PTPN22 C1858T polymorphism was genotyped by PCR-restriction fragment length polymorphism (RFLP). The forward and reverse primers were, respectively, 5′-GATAATGTTGCTTCAACGGAATTT-3′ and 5′-CCATCCCACACTTTATTTTATACT-3′. The presence of the 1858T allele created a restriction site of XcmI enzyme. The reliability of the genotypes was established by genotyping 200 samples by the RsaI PCR-RFLP, RsaI cutting the 1858C allele, with 100% consistency. HLA-DRB1 genotyping has been described (20).
Statistical Analysis.
The Hardy–Weinberg equilibrium was checked in controls, by using a χ2 test with one degree of freedom. The linkage analysis relied on the transmission disequilibrium test, which compares, for a given allele, the transmission of that allele from heterozygous parents to RA patients, with the transmission expected from Mendel's first law (i.e., 50%) (40, 41). The statistical analysis of the transmission disequilibrium test used the exact test with the calculation of the significance level (called P) as follows:
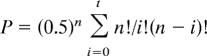 |
where t = number of alleles transmitted [t < (n − t)]; i = different values taken by t; and n = number of heterozygous parents. The transmission disequilibrium test was performed in the initial sample and in the replication sample separately, then after pooling in a global sample. Because Mendel's law is universal, the fact the global sample was heterogeneous in population had no effect on the validity of the analysis. Transmission disequilibrium was also investigated in RF+ and RF− subgroups.
For the association analysis, we used the genotypes relative risk, which compares the affected offspring's genotype and the control genotype derived from untransmitted parental chromosomes (42). For each case genotype, we had therefore a control genotype perfectly matched for the population of origin. As the frequency of the homozygous genotype T/T has been reported to be very low in Caucasian population, the T/T genotype was pooled to the C/T genotype to calculate the genotype relative risk, by using the method proposed by Lathrop (43). The OR and 95% C.I. were estimated by using the method of Woolf (44) as modified by Haldane (45). The T allele frequency in the controls was estimated in the French Caucasian subgroup of trio families where only trio families with both parents without RA or undefined inflammatory arthritis were studied. The study of the interaction between HLA-DRB1 and PTPN22 was performed in the first 200 French Caucasian trio families, relying on a χ2 test with two degrees of freedom.
The PTPN22 locus contribution to RA genetics was estimated by using the conceptual frame developed by Risch (21, 22). Indeed, T could be used to estimate the effect of the deviation from Mendel's law on affected sib-pair analysis. For the PTPN22 locus, assuming that all of the genetic effect is brought by the 1858T allele, by using the estimates from the global French sample, together with the parental 1858T allele frequency in this population, the excess of allele sharing identical by descent (IBD) at the PTPN22 locus for RA would be estimated and compared with the Mendel's expectation of IBD = 50%. The affected sib pairs sharing no PTPN22 alleles identical by descent would also be estimated and compared with the Mendel's expectation of IBD_0 = 25%. Then, the PTPN22 locus contribution to RA familial aggregation should be calculated, by using Risch's parameters lambdaPTPN22 and lambdasib (23).
Power Calculation.
Taking into account the transmission disequilibrium of 61% in RF+ RA observed in the French Caucasian population (18), the replication trio sample of 188 RF+ RA families provided a 93% power to show a significant transmission disequilibrium, with P < 0.05.
Acknowledgments
We thank the RA family members for their participation, Dr. Pierre Fritz for reviewing the clinical data, Dr. J. F. Prud'homme, Dr. C Bouchier, Prof. J. Weissenbach (Généthon), Mrs. M. F. Legrand, and Prof. G. Thomas (Fondation Jean-Dausset-CEPH) for technical help with the DNA samples. This work was supported by Association Française des Polyarthritiques, Société Française de Rhumatologie, Association Rhumatisme et Travail, Association Polyarctique, Groupe Taitbout, Académie de Médecine, Association de Recherche sur la Polyarthrite, Genopole, Conseil Régional Ile de France, Fondation pour la Recherche Médicale, Université Evry-Val d'Essonne, and unrestricted institutional support from Wyeth, Schering-Plough, Pfizer, and Amgen.
Abbreviations
- RA
rheumatoid arthritis
- RF+/RF−
rheumatoid factor positive/negative
- T
transmission testing Mendel's law
- IBD
identical by descent
- C.I.
confidence interval
- OR
odds ratio.
Footnotes
The authors declare no conflict of interest.
*GenHotel-EA 3886, University Evry-Paris 7 Medical School, Member of the AutoCure European Consortium, CP5727, 91057 Evry-Genopole Cedex, France
†Fédération de Rhumatologie, Pôle de l'Appareil Locomoteur, Lariboisière Hospital, AP-HP, 2 Rue Ambroise Paré, 75010 Paris, France
‡Institut National de la Santé et de la Recherche Médicale, U 396, Hôpital Saint-Louis, 75010 Paris, France
§Unité de Génétique Clinique, Pôle des Laboratoires Médicaux-Imagerie-Pharmacie, Lariboisière Hospital, AP-HP, 2 Rue Ambroise Paré, 75010 Paris, France
¶Consultation de Génétique Adulte, Centre Hospitalier Sud Francilien, 59 Boulevard H. Dunant, 91106 Evry-Corbeil, France
‖Généthon, 91002 Evry, France
††Institut National de la Santé et de la Recherche Médicale, U794, 91034 Evry, France
‡‡Centre National de Sequençage-Génoscope, F-91057 Evry, France
§§Katholieke Universiteit Leuven, BE-3000 Leuven, Belgium
¶¶La Paz Hospital, 28046 Madrid, Spain
***National Tissue Typing Center, 11527 Athens, Greece
†††Pisa University, 56126 Pisa, Italy
‡‡‡Nijmegen University, 6500HB Nijmegen, The Netherlands
§§§Porto San Joao Hospital, 4200 Porto, Portugal.
References
- 1.Klareskog L, Stolt P, Lundberg K, Kallberg H, Bengtsson C, Grunewald J, Ronnelid J, Harris HE, Ulfgren AK, Rantapaa-Dahlqvist S, et al. Arthritis Rheum. 2006;54:38–46. doi: 10.1002/art.21575. [DOI] [PubMed] [Google Scholar]
- 2.Stastny P. N Engl J Med. 1978;298:869–871. doi: 10.1056/NEJM197804202981602. [DOI] [PubMed] [Google Scholar]
- 3.Gregersen PK, Silver J, Winchester RJ. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 4.Seldin MF, Amos CI, Ward R, Gregersen PK. Arthritis Rheum. 1999;42:1071–1079. doi: 10.1002/1529-0131(199906)42:6<1071::AID-ANR1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 5.Tezenas du Montcel S, Michou L, Petit-Teixeira E, Osorio J, Lemaire I, Lasbleiz S, Pierlot C, Quillet P, Bardin T, Prum B, et al. Arthritis Rheum. 2005;52:1063–1068. doi: 10.1002/art.20989. [DOI] [PubMed] [Google Scholar]
- 6.Gourraud PA, Boyer JF, Barnetche T, Abbal M, Cambon-Thomsen A, Cantagrel A, Constantin A. Arthritis Rheum. 2006;54:593–599. doi: 10.1002/art.21630. [DOI] [PubMed] [Google Scholar]
- 7.Cornelis F, Faure S, Martinez M, Prud'homme JF, Fritz P, Dib C, Alves H, Barrera P, de Vries N, Pascual-Salcedo D, et al. Proc Natl Acad Sci USA. 1998;95:10746–10750. doi: 10.1073/pnas.95.18.10746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amos CI, Chen WV, Lee A, Li W, Kern M, Lundsten R, Batliwalla F, Wener M, Remmers E, Kastner DA, et al. Genes Immun. 2006;7:277–286. doi: 10.1038/sj.gene.6364295. [DOI] [PubMed] [Google Scholar]
- 9.Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, Ardlie KG, Huang Q, Smith AM, Spoerke JM, et al. Am J Hum Genet. 2004;75:330–337. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plenge RM, Padyukov L, Remmers EF, Purcell S, Lee AT, Karlson EW, Wolfe F, Kastner DL, Alfredsson L, Altshuler D, et al. Am J Hum Genet. 2005;77:1044–1060. doi: 10.1086/498651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, MacMurray J, Meloni GF, Lucarelli P, Pellecchia M, et al. Nat Genet. 2004;36:337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 12.Kyogoku C, Langefeld CD, Ortmann WA, Lee A, Selby S, Carlton VE, Chang M, Ramos P, Baechler EC, Batliwalla FM, et al. Am J Hum Genet. 2004;75:504–507. doi: 10.1086/423790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregersen PK, Lee HS, Batliwalla F, Begovich AB. Semin Immunol. 2006;18:214–223. doi: 10.1016/j.smim.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 14.Wipff J, Allanore Y, Kahan A, Meyer O, Mouthon L, Guillevin L, Pierlot C, Glikmans E, Bardin T, Boileau C, et al. Ann Rheum Dis. 2006;65:1230–1232. doi: 10.1136/ard.2005.048181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vang T, Congia M, Macis MD, Musumeci L, Orru V, Zavattari P, Nika K, Tautz L, Tasken K, Cucca F, et al. Nat Genet. 2005;37:1317–1319. doi: 10.1038/ng1673. [DOI] [PubMed] [Google Scholar]
- 16.Bottini N, Vang T, Cucca F, Mustelin T. Semin Immunol. 2006;18:207–213. doi: 10.1016/j.smim.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 17.Julier C, Hyer RN, Davies J, Merlin F, Soularue P, Briant L, Cathelineau G, Deschamps I, Rotter JI, Froguel P, et al. Nat Genet. 1991;354:155–159. doi: 10.1038/354155a0. [DOI] [PubMed] [Google Scholar]
- 18.Dieude P, Garnier S, Michou L, Petit-Teixeira E, Glikmans E, Pierlot C, Lasbleiz S, Bardin T, Prum B, Cornelis F. Arthritis Res Ther. 2005;7:R1200–R1207. doi: 10.1186/ar1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seldin MF, Shigeta R, Laiho K, Li H, Saila H, Savolainen A, Leirisalo-Repo M, Aho K, Tuomilehto-Wolf E, Kaarela K, et al. Genes Immun. 2005;6:720–722. doi: 10.1038/sj.gene.6364255. [DOI] [PubMed] [Google Scholar]
- 20.Michou L, Croiseau P, Petit-Teixeira E, Tezenas du Montcel S, Lemaire I, Pierlot C, Osorio J, Frigui W, Lasbleiz S, Quillet P, et al. Arthritis Res Ther. 2006;8:R79. doi: 10.1186/ar1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Risch N. Am J Hum Genet. 1987;40:1–14. [PMC free article] [PubMed] [Google Scholar]
- 22.Risch N. Am J Hum Genet. 1990;46:222–228. [PMC free article] [PubMed] [Google Scholar]
- 23.Seldin MF, Amos CI, Ward R, Gregersen PK. Arthritis Rheum. 1999;42:1071–1079. doi: 10.1002/1529-0131(199906)42:6<1071::AID-ANR1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 24.Osorio y Fortea J, Bukulmez H, Petit-Teixeira E, Michou L, Pierlot C, Cailleau-Moindrault S, Lemaire I, Lasbleiz S, Alibert O, Quillet P, et al. Arthritis Rheum. 2004;50:2757–2765. doi: 10.1002/art.20458. [DOI] [PubMed] [Google Scholar]
- 25.Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, et al. Nature. 1996;380:152–154. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- 26.Gomez LM, Anaya JM, Gonzalez CI, Pineda-Tamayo R, Otero W, Arango A, Martin J. Genes Immun. 2005;6:628–631. doi: 10.1038/sj.gene.6364261. [DOI] [PubMed] [Google Scholar]
- 27.Etzel CJ, Chen WV, Shepard N, Jawaheer D, Cornelis F, Seldin MF, Gregersen PK, Amos CI. Hum Genet. 2006;119:634–641. doi: 10.1007/s00439-006-0171-8. [DOI] [PubMed] [Google Scholar]
- 28.Seldin MF, Shigeta R, Laiho K, Li H, Saila H, Savolainen A, Leirisalo-Repo M, Aho K, Tuomilehto-Wolf E, Kaarela K, et al. Genes Immun. 2005;6:720–722. doi: 10.1038/sj.gene.6364255. [DOI] [PubMed] [Google Scholar]
- 29.Lee AT, Li W, Liew A, Bombardier C, Weisman M, Massarotti EM, Kent J, Wolfe F, Begovich AB, Gregersen PK. Genes Immun. 2005;6:129–133. doi: 10.1038/sj.gene.6364159. [DOI] [PubMed] [Google Scholar]
- 30.Carlton VE, Hu X, Chokkalingam AP, Schrodi SJ, Brandon R, Alexander HC, Chang M, Catanese JJ, Leong DU, Ardlie KG, et al. Am J Hum Genet. 2005;77:567–581. doi: 10.1086/468189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johansson M, Arlestig L, Hallmans G, Rantapaa-Dahlqvist S. Arthritis Res Ther. 2005;8:R19. doi: 10.1186/ar1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wesoly J, van der Helm-van Mil AH, Toes RE, Chollalingam AP, Carlton VE, Begovich AB, Huizinga TW. Arthritis Rheum. 2005;52:2948–2950. doi: 10.1002/art.21294. [DOI] [PubMed] [Google Scholar]
- 33.de Pontual L, Pelet A, Trochet D, Jaubert F, Espinosa-Parrilla Y, Munnich A, Brunet JF, Goridis C, Feingold J, Lyonnet S, et al. J Med Genet. 2006;43:419–423. doi: 10.1136/jmg.2005.040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hinks A, Worthington J, Thomson W. Rheumatology (Oxford) 2006;45:365–368. doi: 10.1093/rheumatology/kel005. [DOI] [PubMed] [Google Scholar]
- 35.Hinks A, Barton A, John S, Bruce I, Hawkins C, Griffiths CE, Donn R, Thomson W, Silman A, Worthington J. Arthritis Rheum. 2005;52:1694–1699. doi: 10.1002/art.21049. [DOI] [PubMed] [Google Scholar]
- 36.Viken MK, Amundsen SS, Kvien TK, Boberg KM, Gilboe IM, Lilleby V, Sollid LM, Forre OT, Thorsby E, Smerdel A, et al. Genes Immun. 2005;6:271–273. doi: 10.1038/sj.gene.6364178. [DOI] [PubMed] [Google Scholar]
- 37.Risch N, Merikangas K. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 38.Graham RR, Kozyrev SV, Baechler EC, Reddy MV, Plenge RM, Bauer JW, Ortmann WA, Koeuth T, Gonzalez Escribano MF, Argentine Spanish Collaborative Groups et al. Nat Genet. 2006;38:550–555. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- 39.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS, et al. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 40.Spielman RS, McGinnis RE, Ewens WJ. Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- 41.Schaid DJ. Am J Hum Genet. 1998;63:935–941. doi: 10.1086/302077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terwilliger JD, Ott J. Hum Hered. 1992;42:337–346. doi: 10.1159/000154096. [DOI] [PubMed] [Google Scholar]
- 43.Lathrop GM. Tissue Antigens. 1983;22:160–166. doi: 10.1111/j.1399-0039.1983.tb01183.x. [DOI] [PubMed] [Google Scholar]
- 44.Woolf B. Ann Hum Genet. 1955;19:251–253. doi: 10.1111/j.1469-1809.1955.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 45.Haldane JBS. Ann Hum Genet. 1955;20:309–311. doi: 10.1111/j.1469-1809.1955.tb01285.x. [DOI] [PubMed] [Google Scholar]


