Abstract
Accessions of a plant species can show considerable genetic differences that are analyzed effectively by using recombinant inbred line (RIL) populations. Here we describe the results of genome-wide expression variation analysis in an RIL population of Arabidopsis thaliana. For many genes, variation in expression could be explained by expression quantitative trait loci (eQTLs). The nature and consequences of this variation are discussed based on additional genetic parameters, such as heritability and transgression and by examining the genomic position of eQTLs versus gene position, polymorphism frequency, and gene ontology. Furthermore, we developed an approach for genetic regulatory network construction by combining eQTL mapping and regulator candidate gene selection. The power of our method was shown in a case study of genes associated with flowering time, a well studied regulatory network in Arabidopsis. Results that revealed clusters of coregulated genes and their most likely regulators were in agreement with published data, and unknown relationships could be predicted.
Keywords: natural variation
Analogous to classical traits, quantitative genetic variation is often observed for transcript levels of genes. Jansen and Nap (1), therefore, introduced the concept of genetical genomics, in which quantitative trait locus (QTL) analysis is applied to levels of transcript abundance and identifies genomic loci controlling the observed variation in expression (eQTLs). One of the best studied organisms with regard to gene expression regulation nowadays is yeast (2–8). However, in recent years several studies have demonstrated the feasibility of this approach in different organisms and diverse types of populations (5, 9–13). A logical next step would be the construction of genetic regulatory networks (14), which only a few studies have addressed up to now (2, 15). Although many studies on higher eukaryotes suffered from small populations or only analyzed a subset of genes present on the genome of the organism under study, the main reason holding back the identification of gene-by-gene regulation has been the lack of a reliable identification of candidate regulators. Although powerful in detecting loci controlling the observed variation for trait values, support intervals of QTLs are still of considerable width, often covering hundreds of genes. Consequently, the molecular dissection of quantitative trait regulation is still in its infancy and would greatly benefit from approaches that reduce the number of candidate genes in a QTL support interval. Promising results have been obtained by combining QTL analyses of physiological and gene expression traits, based on colocalization of (e)QTLs (10, 11, 16). However, when expression differences in genes are caused by differences in the expression of their regulator, it is likely that multiple functionally related genes show correlation in expression with the regulator, especially when their eQTLs colocate. We therefore developed an approach for the assignment of maximum-likelihood regulators by combining QTL analysis of gene expression profiling and iterative group analysis (iGA) (17) of functionally related genes with coinciding eQTLs. To apply the concept of genetical genomics to higher plants we analyzed genome-wide gene expression variation in a large, well studied recombinant inbred line (RIL) population of Arabidopsis thaliana. We show that for many genes the variation in transcript level can be explained by genetic factors. By integrating current knowledge of the genetics of a specific trait, we demonstrate the construction of genetic regulatory networks, which can serve to form hypotheses about as-yet-unknown regulatory steps.
Results
Genetic Control of Gene Expression in Plants Is Highly Complex.
To determine the effect of genetic factors involved in the regulation of expression, we analyzed genome-wide gene expression in the parents and an RIL population of a cross between the distinct accessions Landsberg erecta (Ler) and Cape Verde Islands (Cvi), consisting of 160 lines (18). Transcript levels of 24,065 genes were analyzed by DNA microarrays, of which 922 showed significant differential expression between the parents [P < 2.5 × 10−3; false-discovery rate (FDR) = 0.05]. Subsequent mapping resulted in 4,523 eQTLs detected for 4,066 genes (P < 5.29 × 10−5; FDR = 0.05, corresponding to a q value of 0.01) (19). Because the microarray probe set was designed on the sequenced accession Columbia (Col), we performed hybridizations of genomic DNA of the parental lines and found relatively few hybridization differences (supporting information (SI) Table 1). However, the low power to detect differences, due to the small number of replicates, might have led to an underestimation, as indicated by other studies (20).
Heritability values calculated from the parental data and the RIL population reached a median value of 28.6% and 74.7%, respectively (SI Figs. 3 and 4), which is in agreement with the discrepancy between the number of differentially expressed and mapped genes (i.e., genes for which an eQTL was found).
Although the fraction of mapped genes increased with higher heritability values, for many genes showing high heritability, no eQTL could be significantly detected (SI Fig. 4). These findings suggest that the regulation of expression of many genes is controlled by multiple eQTLs, of which many might not have passed the significance test because of their small effect. Likewise, only 65.6% of the genes differentially expressed between the parents could be mapped. However, for 15.0% of the genes for which the parents did not show a significant difference in expression levels, eQTLs could be detected. These observations and the much lower heritabilities calculated from the parental data, compared with those from the RIL population, indicate that eQTLs for a given gene might exert opposite additive effects, leading to a balanced expression in the parents but a transgressive expression pattern among the segregants of the population. To test this hypothesis, we tested each gene for significant transgression and found significant transgression of expression for 10,849 genes (45.1%). No relationship was found between the number of mapped genes and transgression (SI Fig. 6).
These data indicate that the regulation of gene expression in plants is largely under genetic control but is highly complex because of the involvement of multiple genes.
Distribution of eQTLs Identifies Regulatory Hot Spots.
To characterize in more detail the genes whose expression showed significant linkage, we determined several features. We first analyzed the distribution of eQTLs along the genome of Arabidopsis and found a number of genomic regions containing numbers of eQTL significantly deviating from what can be expected by chance, as determined by permutation tests (SI Fig. 7). These hot spots may reflect local gene-dense regions, in contrast to cold spots, which may reflect low-gene-density regions such as centromers. Alternatively, hot spots may contain master regulators: genes controlling the expression of many other genes. The large number of genes mapping to the ERECTA gene, which was included as a phenotypic marker, illustrate this finding. Because 176 genes mapped to the ERECTA marker, this locus was considered to be an eQTL hot spot. Polymorphisms in ERECTA, a receptor protein kinase (21), are well known for their pleiotropic effect on many traits, including morphological differences (22).
Distant Gene Expression Regulation Occurs More Frequently but Local Regulation Is Stronger.
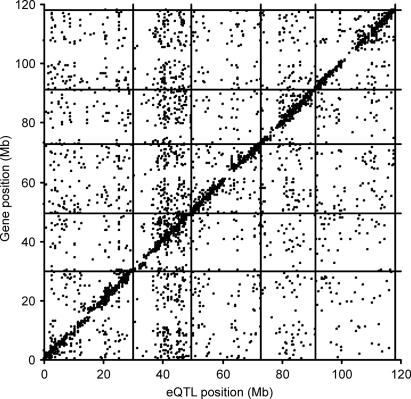
Genomic differences responsible for eQTLs occur either in regulatory genes affecting the transcript level of other genes (trans-regulation) or in the genes encoding the mRNA for which the eQTL was found (cis-regulation) (23). To compare the position of genes and their eQTLs, we anchored the genetic map to the physical map and found an almost linear genome-wide relation of 4.1 cM per Mbp (SI Fig. 8). When the position of each eQTL was plotted against the position of the gene for which that eQTL was found, a strong enrichment along the diagonal of the graph was observed (Fig. 1). This enrichment indicates that many genes, of which the majority are expected to be cis-regulated, map to their own physical position (6). To quantify this result, we defined local/distant regulation in terms of the positional coincidence of genes and their accompanying eQTL(s). Of 4,066 mapped genes, 1,875 (46.1%) colocated with the support interval of one of their eQTLs, corresponding to a region consistent with max{−log10 P} − 1.5 (where P expresses the significance of association) (24) and were therefore classified as locally regulated. Genes outside such intervals (1,958; 48.1%) were classified as distantly regulated. A minor number of 198 genes (4.9%) with multiple QTLs showed both local and distant regulation, whereas the physical position of 35 genes (0.9%) was unknown (SI Table 2). Because cis-regulation is often much stronger than trans-regulation (2), as also indicated by the median −log10 P values of 7.1 and 5.3 and the median explained variance of 30.3% and 22.6% for local and distant eQTLs, respectively, the ratio of detected local versus distant eQTL depends on the applied significance threshold (11–13). The stringent threshold applied here, corrected for multiple testing, might therefore have underestimated distant regulation. When the threshold was decreased from 5.29 × 10−5 to 6.5 × 10−4 (FDR = 0.25, q = 0.05), 7,604 transcripts showed at least one linkage, with 2,167 (28.5%) being locally regulated, 4,587 (60.3%) being distantly regulated, and 794 (10.4%) being both locally and distantly regulated. Based on their P value distributions (19), the overall proportion of locally and distantly regulated genes were estimated at 40.5% and 15.3%, respectively. A second parameter affecting the assignment of locally versus distantly regulated transcripts is the setting of the eQTL support interval. However, when a wider interval of max{−log10 P} − 2.0 was used at P < 5.29 × 10−5, results were similar with 2,007 (49.4%), 1,832 (45.1%), and 192 (4.7%) genes classified as locally, distantly, and both locally and distantly regulated, respectively.
Fig. 1.
Distribution of mapped genes versus the position of their accompanying eQTL. Positions of detected eQTL are plotted against the position of the gene for which that eQTL was found. Chromosomal borders are depicted as horizontal and vertical lines. Mb, megabase.
Local Regulation Correlates with SNP Frequency and Is Less Frequent in Regulatory Genes.
To determine whether a relationship exists between SNP or gene density and the number of mapped genes, we performed a sliding-window regression analysis. A strong correlation was observed between gene density and the number of locally and distantly regulated genes (r2 = 0.88, P < 0.0001 and r2 = 0.91, P < 0.0001, respectively) (SI Fig. 9). A weaker but significant correlation was also found between gene and SNP frequency (r2 = 0.34, P < 0.0001). Even when the number of mapped genes in a window was corrected for gene density, a significant correlation was still found between SNP frequency and the number of locally regulated genes (r2 = 0.32, P < 0.0001), although incidental differences in hybridization efficiency might have contributed to an overestimation. Such a relationship was not found for distantly regulated genes (r2 = −0.003, P = 0.89) (SI Fig. 9).
To assess whether there was a functional enrichment for genes whose variation in expression could be genetically explained, we computed the proportion of these genes for each Gene Ontology molecular function and biological process category (The Arabidopsis Information Resource; www.arabidopsis.org) (SI Fig. 10). Genes involved in regulatory processes showed significantly less genetically explainable variation in expression (25) (SI Table 3). However, small changes in expression level, which may be more frequent in regulatory genes, are more difficult to detect but can nevertheless be very relevant biologically, because they may result in large changes in expression of target genes. Furthermore, many regulatory genes often display pleiotropic effects. A change in expression of such key regulators can affect the expression of many more target genes, which may skew the distribution of differently expressed genes in favor of classes containing predominantly target genes.
Interestingly, when these analyses were performed separately for locally and distantly regulated genes, regulatory categories showed a comparable proportion of distantly regulated genes with other classes but a much smaller proportion of locally regulated genes (SI Fig. 10). Comparing locally to distantly regulated genes (25) resulted in significant overrepresentation of distantly regulated genes in 10 Gene Ontology biological process categories, all involved in regulation. This finding agrees with the general assumption that regulatory genes are much more strongly conserved than other genes because of their often pleiotropic effects.
A Dual Approach for the Construction of Regulatory Networks Reveals Regulatory Steps for Flowering Time.
Genetic regulatory networks consist of a collection of genes, which are interconnected because one gene regulates the transcription of another directly or indirectly. The analysis of gene expression in a mapping population can greatly enhance the construction of such networks. If an eQTL results from differences in expression of a regulator, this regulator is likely to show correlation in expression levels with the gene that mapped to its position (2). Multiple genes involved in the same biological process mapping to the same position indicates that many of them might be under the control of the same gene. We reasoned that the best candidate within an eQTL interval is the gene whose expression best correlates with multiple genes mapping to the position of that gene. We therefore combined expression trait profiling with eQTL mapping, gene annotation, and extended iGA (17) to sort candidate regulators based on their PC (possibility of change) value, which tells how likely a given regulator is to observe a strong correlation with multiple members of a selected group of genes. This approach enabled us to drastically narrow down the number of candidate genes in an eQTL interval and select the best candidate for the construction of genetic regulatory networks.
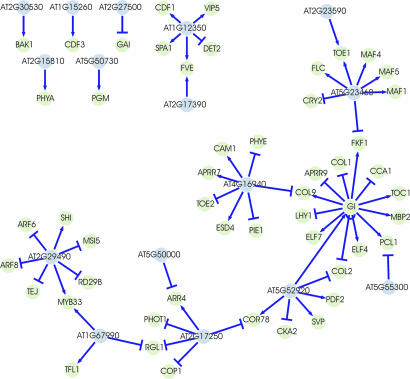
To verify our approach, we focused on one of the best studied and most complete genetic regulatory networks available in plants: the regulation of flowering in Arabidopsis. Flowering time is highly variable between accessions of Arabidopsis (22). Variation in flowering time also exists between Ler and Cvi, and several studies have reported QTLs for this trait (26–28). Although flowering starts much later, the expression of genes that indicate commitment to flowering are already apparent at a very early stage and find their transcription peak in the seedling stage (29, 30). We selected a set of 192 genes known to be involved in the control of flowering from recent literature (see SI Table 5 for a full list) and keyword searching in the The Arabidopsis Information Resource database; 175 of these genes were analyzed in our study. Analysis of their expression level in the parental accessions assigned eight of them as being differentially expressed. However, 83 genes showed at least one eQTL at a genome-wide threshold of 2.23 × 10−3. We calculated PC values for correlation in expression profiles, using the group of 83 mapped flower genes and all candidate genes within their eQTL support intervals. We then selected the genes within the eQTL support interval of a given flower gene with significant PC values (FDR = 0.05) as candidates for this eQTL (SI Table 5). Regulators were predicted for 51 genes, whereas for 32 genes no significant PC value was obtained. Fig. 2 shows a network of flower genes and their most likely regulators. The most significant regulator detected was GIGANTEA (GI) with a PC value of 1.01 × 10−12. Thirteen genes mapped to GI, including GI itself, and all of them contributed to the lowest PC value. GI is the first member of an output pathway of the circadian clock that controls flowering time and has been shown to regulate circadian rhythms in Arabidopsis (31). At the position of GI, a minor flowering-time QTL (26) and a circadian period length QTL (32, 33) were identified, which indicates the physiological consequences of this complex pattern of gene expression variation. Indeed, many of the genes, such as CCA1 (see SI Table 5 for details), LHY1, ELF4, and TOC1, for which GI was identified as their most likely regulator, belong to the core circadian oscillator (34). Others are involved in the regulation of the circadian clock, such as PCL1, APRR9, and FKF1 (32, 35), or play a role in floral transition, such as ELF7 and the CONSTANS-LIKE family COL1, COL2, and COL9 (36–38). A second cluster of coregulated genes is involved in floral repression and mapped to FLG, another major QTL for flowering time. Where the floral repressors FLC, MAF1, MAF4, MAF5, and TOE1 (34) are up-regulated, the floral promoter CRY2 (34) is down-regulated by this locus, in agreement with findings that FLC expression negatively correlates with CRY2 (39). In addition to FLG, CRY2 and FLC are major-effect QTLs for flowering time in the Ler × Cvi population, and significant epistasis has been found between CRY2 and FLC (39) and between the FLC region and the FLG locus (26). Although HUA2 was previously suggested as a candidate for the FLG locus (40), we did not identify it as such and found a gene with unknown function (At5g23460) to be the most likely candidate. Other clusters are predominantly involved in hormonal pathways (MYB33, ARF6, ARF8, RD29B, and SHI) (41, 42) and the photoperiod pathway (PIE1, CAM1, PHYE, and ESD4) (34, 43) of flowering.
Fig. 2.
Regulatory network of genes involved in the transition to flowering. Flower genes (green dots) are connected to their most likely regulator (blue dots) by directional edges. Arrows, stimulative regulation; bars, repressive regulation.
To identify other possible target genes of the most significant regulator (GI), we calculated the correlation coefficient between the genes of the GI regulatory cluster and all other genes. Strong correlation was observed for 280 transcripts at an empirical correlation coefficient cutoff of 0.55, corresponding to a FDR of 9.5 × 10−5 (SI Table 6). Many of these genes showed no significant linkage at the position of GI but several displayed a suggestive QTL. Although correlation can be a result of linked genetic effect, only 32 locally regulated genes were located within 2.5 Mbp of GI. The highest correlation coefficient (0.75) was found for a CONSTANS-LIKE PROTEIN encoding gene (At1g07050). The long day integrator CONSTANS (CO) has been shown to be a direct target of GI (31), although it was not identified as such in our study. Two other genes associated with circadian rhythms, APRR5 and WNK1, were detected, and both showed a suggestive QTL at the position of GI. APRR genes are paralogs of TOC1 and have been shown to be regulated by the protein kinase WNK1 (44). These results suggest that the feedback regulation of the circadian clock by GI acts, at least partly, through WNK1 and APRR5.
Discussion
Genetic Variation in Gene Expression Is Abundant and Complex.
We determined differences in gene expression between two distinct accessions of Arabidopsis and within an RIL population derived from these accessions.
Our data suggest that variation in gene expression among genetically different plants of the same species is for a large part genetically controlled and highly complex. Although eQTLs were detected for >4,000 genes, only 922 were differentially expressed between the parents, which suggests that the expression of many genes is controlled by multiple loci with opposing effects, avoiding large differences between natural accessions but generating strong transgression in a segregating population. This suggestion is supported by the differences in heritability, as calculated from the parental and population expression analyses. This difference between the two heritability estimates might have several reasons. First, statistical issues might bias the outcome of the analyses. False negatives might bias the number of genes differentially expressed between the parents downwards, because statistical power was limited to 10 replicate measurements of each parent. On the other hand, false positives due to low signal-to-noise ratios for low-expressed genes might bias the number of mapped genes upwards. However, most mapped genes had medium-to-high expression levels (SI Fig. 5).
A second and more likely reason why mapped genes were not significantly differently expressed between the parents might be the complex genetic inheritance of gene expression. Illustrating this finding is that although the median heritability of mapped genes was 82.4%, only a median 28.4% of the variation observed for mapped genes could be explained by significant eQTLs. Furthermore, although the proportion of mapped genes increased with higher heritability values, many genes with a high heritability could not be mapped significantly. Together with the strong transgression observed for many genes, these data imply that regulation of expression often occurs through the added effect of numerous small effect loci, each of which fail to pass the significance threshold.
Because two color arrays were used in this study, a dye effect can be expected in subsequent analyses. In our experiment, dye effect was controlled and corrected at two levels. At the level of the experimental design, we balanced the dye effect between two alleles by optimizing for the number of Ler–Cvi and Cvi–Ler comparisons at each marker position (45). At the analysis level we included the gene-specific differential effect between the two dyes in the QTL analysis model (46).
Molecular Background of Expression Variation.
Factors ranging from abiotic external influences to direct active control of transcriptional activity influence the level of transcript abundance of a given gene. Here, we focused on genetic factors contributing to whole-genome transcript levels. Our data showed that genes whose transcript variation could be mapped are not equally distributed over the Arabidopsis genome. Although a strong correlation between the total number of genes per unit of chromosome and those that could be mapped was observed, other explanations, such as differences in chromatin structure or SNP frequency, cannot be excluded. The correlation observed between SNP frequency and the proportion of mapped genes illustrates this result.
Anchoring of the genetic map enabled us to define local versus distant regulation. Although, in general, local regulation seems stronger, distant regulation occurs more frequently. This distant regulation was demonstrated by decreasing the significance threshold; only a minor number of additional locally regulated genes were detected, whereas the number of distantly regulated genes increased >2-fold. Because the vast majority of genes showing local linkage are expected to be cis-regulated (6), this difference in increase can be explained by the direct influence of cis-polymorphisms on expression, whereas trans-polymorphisms exert their effect indirectly through a change in expression or coding sequence of a second gene. Taking together the strong transgression observed for many genes and the number of distantly versus locally regulated genes, it is conceivable that many cis-regulated genes exert pleiotropic effects on the expression of other genes and are causal for many of the eQTLs acting in trans.
Regulatory Networks.
For many biological processes, the genes contributing to a certain phenotype are often well known. However, in many cases, little is known about the regulation and interaction of these genes. We combined expression information with eQTL mapping, gene annotation, and iGA to identify likely regulators. This approach enabled the construction of maximum-likelihood genetic regulatory networks from a genome-wide genetical genomics experiment. A case study that used genes involved in the well known process of transition from a vegetative state to a flowering state confirmed many of the interactions identified previously. Moreover, numerous interactions that can serve to form hypotheses for future studies were predicted. It must be noted, however, that analyses were performed on data from a single time point. It is not unlikely that regulation occurs differently at other developmental stages or diurnal phase or even organ, specifically. Especially for pathways influenced by the circadian clock, such as flowering time, expression differences at one time point can be caused by differences in circadian phase (32, 47). Accuracy and reliability would therefore benefit from gene expression analysis at multiple developmental stages and time points. Nevertheless, confidence in the followed approach was gained, because many functionally related genes grouped together indicating common and simultaneous regulation. We assigned the gene with the lowest PC value as the most likely candidate responsible for this regulation although other genes with significant PC values cannot be ruled out a priori. Subsequent in-depth analysis should be performed to unambiguously identify genes underlying eQTLs, but the number of candidate genes decreased substantially with the described method.
Methods
Plant Material and Tissue Collection.
Aerial parts of seedlings from the accessions Ler and Cvi and a population of 160 RILs derived from a cross between these parents (18, 24) were grown and collected as described in ref. 24.
Linkage Map Construction and Anchoring to the Physical Map.
The genetic map was constructed from a subset of the markers available, at http://nasc.nott.ac.uk, with a few new markers added. The computer program JoinMap 4 (48) was used for the calculation of linkage groups and genetic distances. In total, 144 markers were used, with an average spacing of 3.5 cM. The largest distance between two markers was 10.8 cM.
To anchor the genetic map to the physical map of Arabidopsis, the total set of 291 available markers was analyzed. First, a genetic map that comprised all 291 markers was constructed. Physical positions of molecular PCR markers were obtained from The Arabidopsis Information Resource, release 6.0 (www.arabidopsis.org). Sequences of amplified fragment length polymorphism markers were obtained by in silico amplification of Col markers that were polymorphic between Ler and Cvi (49) or by sequencing fragments polymorphic between Ler and Cvi but absent in Col. The retrieved marker sequences were then blasted against the completely sequenced Col genome, and center positions of positive hits were taken as the physical position. Physical positions could be established for 179 markers; positions of remaining markers were inferred from interpolation by using the closest nearby markers for which a physical position was known. The largest gap between two markers with confirmed physical position comprised 3.5 Mb, which corresponded to a genetic distance of ≈15 cM.
Sample Preparation.
Total RNA of each line was isolated from two biological replicates by using phenol–chloroform extraction (50). Extracts were then combined and purified with RNeasy (Qiagen, Valencia, CA), amplified with the MessageAmp aRNA kit (Ambion, Austin, TX) incorporating 5-(3-aminoallyl)-UTP, and labeled with Cy3 or Cy5 mono-reactive dye (Amersham, Piscataway, NJ). All RNA products were purified by using the Rneasy kit (Qiagen). Labeled RNA was fragmented for 15 min before hybridization (fragmentation reagent obtained from Ambion).
Microarray Analyses.
Arabidopsis DNA microarrays were provide by the Galbraith laboratory (University of Arizona, Tucson, AZ) and were produced from a set of 70-mer oligonucleotides, representing 24,065 unique genes (Array-Ready Oligo Set, version 1.0, Qiagen-Operon).
DNA probe immobilization and hybridization was performed according to instructions from the Galbraith laboratory. Arrays were scanned by using a ScanArray Express HT (PerkinElmer, Wellesley, MA) and quantified by using Imagene 6.0 (BioDiscovery, El Segundo, CA).
Experimental Design.
Genome-wide gene expression analysis was carried out for Ler and Cvi and an RIL population derived from a cross between these two accessions. Ten replicates of the parental lines were compared in direct hybridizations by using a dye swap design. The 160 RILs were analyzed by direct hybridization of two genetically distant lines on each array, leading to a total of 80 slides. A distant pair design, which was proposed specifically for genetic studies on gene expression (45) was used. An optimal design was obtained through simulated annealing, in which pairs of genetically distant lines were hybridized to maximize the direct comparisons between two different alleles at each marker. The numbers of Ler–Cvi and Cvi–Ler comparisons at each marker were optimized for equal ratio to balance dye effects, and their total number was optimized for minimal extra variation across other markers. The observed signal intensities on the arrays were subjected to general normalization procedures (51, 52). Resulting log signal intensities and log ratios between cohybridized RILs were used for further analyses.
Statistical Analyses.
Differential expression of genes between the two parents was tested for significance. For each gene, the P value of a t test and the corresponding q values (19) were computed (51). The P value significance threshold was 2.5 × 10−3 at a q value cutoff of 0.05.
Log signal intensities of gene expression were used to test for genetic variance of expression traits. Spot effects were removed by treating it as a random effect in a linear mixed model.
Heritability of expression in the parental accessions was calculated as follows (53):
where Vg and Ve represent the components of variance among and within accessions respectively. The factor 0.5 was applied to adjust for the 2-fold overestimation of additive genetic variance among inbred strains.
Heritability of expression within the RIL population was calculated by using the pooled variance of the parents as an estimate of the within line variance:
where VRIL and Ve are the variance among adjusted expression intensities in the segregants and the pooled variance within parental measurements, respectively. To prevent overestimation, we removed outliers more than three standard deviations away from the mean values. We discarded 1,470 (6.1%) negative heritability values.
Transgressive segregation was determined in terms of the pooled standard deviation of the parents (3). We calculated the number of RILs, n, whose expression level lay beyond the region μ ± 2 × SD, where μ and SD are the mean and the standard deviation of parental phenotypic values, respectively. To determine significance, phenotype values of parents and segregants were reassigned at random to null parents and segregants for each transcript. The number of transgressive individuals, n0, was then recorded. The total number of transcripts with n0 greater than a given threshold m represented the genome-wide false-positive count at m. The FDR was computed as the ratio between the estimated false-positive count at m and the number of nonpermuted transcripts with n > m. Results were averaged over 20 permutations. The FDR = 0.05 cutoff corresponded to m = 33.
Multiple QTL Analysis.
Gene expression in the mapping population was analyzed for significant eQTLs. For each gene, the log-ratios of signal intensities were subjected to multiple QTL mapping. Cofactors were selected by using a backward elimination process (54) (see SI Text). For every marker-by-gene combination, the multiple QTL mapping mapping model can be given as
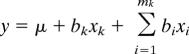 |
where y is the expression ratio of a transcript, μ is the gene-specific differential effect between Cy3 and Cy5 dyes (characterized as consistent across samples) (46), x denotes the genotype comparison and takes the following values: 1 for Ler–Cvi, −1 for Cvi–Ler, and 0 for Ler–Ler and Cvi–Cvi; b is the substitution effect; k is the kth marker under study; and i denotes the cofactors from 1 to mk, outside a 30-cM interval of the kth marker. The P value from a t test that tested the hypothesis that bk = 0 was used as a measure of significance of the association.
A genome-wide P-value threshold of 2.23 × 10−3 at α = 0.05 for a single trait was estimated by a 10,000-permutation test (55). But for a study with 24,065 gene transcripts, we controlled the FDR based on the pool of P values for all markers and all transcripts. Because the P values are correlated when markers are linked, the FDR increases depending on the number of markers on a chromosome (56). In our experiment, the maximum number of markers reached 35 (chromosome 5), and a simulation analysis (data not shown) that used Storey's algorithm to control the FDR (57) at a desired level showed a 4.4-fold increase of the actual FDR. To account for this increase, we corrected the FDR by a factor of 5 and calculated the genome wide P-value threshold at Storey's FDR of 0.01 for all gene-marker P values, to make sure that the real FDR rate is <0.05 (corrected FDR = 0.05). The estimated P-value threshold then corresponded to 5.29 × 10−5, and this threshold was used as a significance threshold for the detection of eQTL.
Explained variance of detected eQTLs was estimated by fitting expression ratios of all detected eQTLs and their interactions in a linear model. We used ANOVA to estimate the fraction of variance explained by each eQTL and eQTL interactions.
Local and Distant Regulation.
We determined the physical position of each eQTL by anchoring the genetic map of the Ler × Cvi population to the physical map of the sequenced accession Col. Support intervals were then calculated by setting left and right border positions associated with max{−log10 P} − 1.5, where P represents the significance value for linkage (24).
The physical positions of genes (The Arabidopsis Information Resource, version 2005.12.8) showing significant linkage of expression values were then compared with the positions of their respective eQTL(s); a gene was classified as locally regulated when its position coincided with the support interval and as distantly regulated when it did not.
Distribution of Hot Spots.
eQTL hot spots are shown by the frequency distribution of the number of significant eQTLs detected. Each eQTL is presented by the marker showing the most significant linkage. The frequency distribution of eQTL by chance was empirically estimated by 250 permutations (58). The 95th percentile, corresponding to 43 eQTLs, was used as a confidence threshold for the occurrence of a hot spot.
Sliding-Window Analyses.
All 24,065 genes analyzed were positioned on the Arabidopsis physical map, and the ATG start codon was used as the start of each gene. Each gene was classified as locally regulated, distantly regulated, or nonregulated. The frequency of the total number of genes and the number of locally and distantly regulated genes along each chromosome was determined in a 5-Mbp sliding window by using a 50-Kbp step size. Polymorphisms between Ler and Cvi in 875 sequenced loci (59) were downloaded from the MSQT website (http://msqt.weigelworld.org) and filtered for unique positions. INDELs were recorded as a single polymorphism by using the physical position of the first nucleotide difference. A total number of 4,032 polymorphisms were subjected to further analysis. A sliding-window analysis for SNP frequency was then carried out as described above. Observed gene and SNP frequencies per window were standardized by using the genome-wide average and standard deviation, and resulting z scores were plotted at the physical position of the center of each window.
Genetic Network Construction.
A group of 83 functionally related genes and their potential regulators were used for the construction of a genetic regulatory network. All of the genes that were physically located in an eQTL interval were assigned as a regulator candidate for the gene for which that eQTL was detected. The candidates were sorted by using iGA (17). We postulated that, among all possible regulators, the best candidates are those that correlate particularly well to a large number of their potential target genes. To test that postulation, we calculated all pair-wise Spearman rank correlations on expression profiles (80 log ratios of cohybridized RILs) between each of the 83 functionally related genes and all potential regulators in their eQTL intervals. These values were then rank-ordered so that the strongly correlated gene-candidate pairs were at the top of the list. For each given candidate, we determined the iGA possibility of change value (PC value; SI Table 5). The PC-value threshold was Bonferroni-adjusted as 0.05/m, where m is the total number of candidate genes. Any candidate with a significant PC value is a putative regulator, and all genes contributing to this value are putative target genes. We defined the regulatory relation in terms of the sign of the correlation coefficient. If the correlation coefficient is negative, regulation is repressive; otherwise, it is stimulative.
Potential target genes outside the initial group of functionally related genes were identified by using expression trait correlations (60), for which we used the regulators and target genes obtained from the iGA study as seed transcripts. We then split the log ratio gene-expression profile matrix (axb) into two parts: a1xb and a2 xb, where a is the total number of gene transcripts (a = 24,065 in our case); a1 is the number of seed transcripts; a2 is the number of other genes (a1+a2 = a) and b is the number of arrays (b = 80 in our case). We then computed the Spearman correlation coefficient and its corresponding P value between each a1 seed gene and a2 transcript. A 95 percentile empirical threshold (r = 0.55) and its corresponding FDR (19) (FDR = 9.5 × 10−5) were estimated by performing 1,000 permutations.
Supplementary Material
Acknowledgments
We thank David Galbraith for providing microarrays and protocols, Janny Peters for providing data of in silico amplified fragment length polymorphism analysis, and Linus van der Plas for critical reading of the manuscript. This work was supported by Netherlands Organization for Scientific Research, Program Genomics Grant 050-10-029.
Abbreviations
- QTL
quantitative trait locus
- eQTL
expression quantitative trait locus
- iGA
iterative group analysis
- RIL
recombinant inbred line
- Ler
Landsberg erecta
- Cvi
Cape Verde Islands
- FDR
false-discovery rate
- Col
Columbia
- PC
possibility of change.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610429104/DC1.
References
- 1.Jansen RC, Nap JP. Trends Genet. 2001;17:388–391. doi: 10.1016/s0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- 2.Bing N, Hoeschele I. Genetics. 2005;170:533–542. doi: 10.1534/genetics.105.041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brem RB, Kruglyak L. Proc Natl Acad Sci USA. 2005;102:1572–1577. doi: 10.1073/pnas.0408709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brem RB, Storey JD, Whittle J, Kruglyak L. Nature. 2005;436:701–703. doi: 10.1038/nature03865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brem RB, Yvert G, Clinton R, Kruglyak L. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 6.Ronald J, Brem RB, Whittle J, Kruglyak L. PLoS Genet. 2005;1:e25. doi: 10.1371/journal.pgen.0010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Storey JD, Akey JM, Kruglyak L. PLoS Biol. 2005;3:e267. doi: 10.1371/journal.pbio.0030267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yvert G, Brem RB, Whittle J, Akey JM, Foss E, Smith EN, Mackelprang R, Kruglyak L. Nat Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- 9.Bystrykh L, Weersing E, Dontje B, Sutton S, Pletcher MT, Wiltshire T, Su AI, Vellenga E, Wang J, Manly KF, et al. Nat Genet. 2005;37:225–232. doi: 10.1038/ng1497. [DOI] [PubMed] [Google Scholar]
- 10.DeCook R, Lall S, Nettleton D, Howell SH. Genetics. 2006;172:1155–1164. doi: 10.1534/genetics.105.042275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hubner N, Wallace CA, Zimdahl H, Petretto E, Schulz H, Maciver F, Mueller M, Hummel O, Monti J, Zidek V, et al. Nat Genet. 2005;37:243–253. doi: 10.1038/ng1522. [DOI] [PubMed] [Google Scholar]
- 12.Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, et al. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 14.Kendziorski C, Wang P. Mamm Genome. 2006;17:509–517. doi: 10.1007/s00335-005-0189-6. [DOI] [PubMed] [Google Scholar]
- 15.Kliebenstein DJ, West MA, van Leeuwen H, Loudet O, Doerge RW, St Clair DA. BMC Bioinformatics. 2006;7:308. doi: 10.1186/1471-2105-7-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wayne ML, McIntyre LM. Proc Natl Acad Sci USA. 2002;99:14903–14906. doi: 10.1073/pnas.222549199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breitling R, Amtmann A, Herzyk P. BMC Bioinformatics. 2004;5:34. doi: 10.1186/1471-2105-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alonso-Blanco C, Peeters AJ, Koornneef M, Lister C, Dean C, van den Bosch N, Pot J, Kuiper MT. Plant J. 1998;14:259–271. doi: 10.1046/j.1365-313x.1998.00115.x. [DOI] [PubMed] [Google Scholar]
- 19.Storey JD, Tibshirani R. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borevitz J. Methods Mol Biol. 2006;323:137–145. doi: 10.1385/1-59745-003-0:137. [DOI] [PubMed] [Google Scholar]
- 21.Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y. Plant Cell. 1996;8:735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koornneef M, Alonso-Blanco C, Vreugdenhil D. Annu Rev Plant Biol. 2004;55:141–172. doi: 10.1146/annurev.arplant.55.031903.141605. [DOI] [PubMed] [Google Scholar]
- 23.Rockman MV, Kruglyak L. Nat Rev Genet. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- 24.Keurentjes JJB, Fu J, de Vos CH, Lommen A, Hall RD, Bino RJ, van der Plas LHW, Jansen RC, Vreugdenhil D, Koornneef M. Nat Genet. 2006;38:842–849. doi: 10.1038/ng1815. [DOI] [PubMed] [Google Scholar]
- 25.Al-Shahrour F, Diaz-Uriarte R, Dopazo J. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 26.Alonso-Blanco C, El-Assal SED, Coupland G, Koornneef M. Genetics. 1998;149:749–764. doi: 10.1093/genetics/149.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juenger TE, Sen S, Stowe KA, Simms EL. Genetica. 2005;123:87–105. doi: 10.1007/s10709-003-2717-1. [DOI] [PubMed] [Google Scholar]
- 28.Ungerer MC, Halldorsdottir SS, Modliszewski JL, Mackay TF, Purugganan MD. Genetics. 2002;160:1133–1151. doi: 10.1093/genetics/160.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- 30.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizoguchi T, Wright L, Fujiwara S, Cremer F, Lee K, Onouchi H, Mouradov A, Fowler S, Kamada H, Putterill J, Coupland G. Plant Cell. 2005;17:2255–2270. doi: 10.1105/tpc.105.033464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michael TP, Salome PA, Yu HJ, Spencer TR, Sharp EL, McPeek MA, Alonso JM, Ecker JR, McClung CR. Science. 2003;302:1049–1053. doi: 10.1126/science.1082971. [DOI] [PubMed] [Google Scholar]
- 33.Swarup K, Alonso-Blanco C, Lynn JR, Michaels SD, Amasino RM, Koornneef M, Millar AJ. Plant J. 1999;20:67–77. doi: 10.1046/j.1365-313x.1999.00577.x. [DOI] [PubMed] [Google Scholar]
- 34.Boss PK, Bastow RM, Mylne JS, Dean C. Plant Cell. 2004;16:S18–S31. doi: 10.1105/tpc.015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Onai K, Ishiura M. Genes Cells. 2005;10:963–972. doi: 10.1111/j.1365-2443.2005.00892.x. [DOI] [PubMed] [Google Scholar]
- 36.Cheng XF, Wang ZY. Plant J. 2005;43:758–768. doi: 10.1111/j.1365-313X.2005.02491.x. [DOI] [PubMed] [Google Scholar]
- 37.He Y, Doyle MR, Amasino RM. Genes Dev. 2004;18:2774–2784. doi: 10.1101/gad.1244504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ledger S, Strayer C, Ashton F, Kay SA, Putterill J. Plant J. 2001;26:15–22. doi: 10.1046/j.1365-313x.2001.01003.x. [DOI] [PubMed] [Google Scholar]
- 39.El-Assal SED, Alonso-Blanco C, Peeters AJ, Wagemaker C, Weller JL, Koornneef M. Plant Physiol. 2003;133:1504–1516. doi: 10.1104/pp.103.029819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doyle MR, Bizzell CM, Keller MR, Michaels SD, Song J, Noh YS, Amasino RM. Plant J. 2005;41:376–385. doi: 10.1111/j.1365-313X.2004.02300.x. [DOI] [PubMed] [Google Scholar]
- 41.Mouradov A, Cremer F, Coupland G. Plant Cell. 2002;14:S111–S130. doi: 10.1105/tpc.001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, Guilfoyle TJ, Hagen G, Alonso JM, Cohen JD, Farmer EE, et al. Development (Cambridge, UK) 2005;132:4107–4118. doi: 10.1242/dev.01955. [DOI] [PubMed] [Google Scholar]
- 43.Levy YY, Dean C. Plant Cell. 1998;10:1973–1990. doi: 10.1105/tpc.10.12.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakamichi N, Murakami-Kojima M, Sato E, Kishi Y, Yamashino T, Mizuno T. Biosci Biotechnol Biochem. 2002;66:2429–2436. doi: 10.1271/bbb.66.2429. [DOI] [PubMed] [Google Scholar]
- 45.Fu J, Jansen RC. Genetics. 2006;172:1993–1999. doi: 10.1534/genetics.105.047001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dobbin KK, Kawasaki ES, Petersen DW, Simon RM. Bioinformatics. 2005;21:2430–2437. doi: 10.1093/bioinformatics/bti378. [DOI] [PubMed] [Google Scholar]
- 47.Darrah C, Taylor BL, Edwards KD, Brown PE, Hall A, McWatters HG. Plant Physiol. 2006;140:1464–1474. doi: 10.1104/pp.105.074518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Ooijen JW. JoinMap, Software for the Calculation of Genetic Linkage Maps. Wageningen, The Netherlands: Kyazma BV; 2006. Version 4. [Google Scholar]
- 49.Peters JL, Constandt H, Neyt P, Cnops G, Zethof J, Zabeau M, Gerats T. Plant Physiol. 2001;127:1579–1589. [PMC free article] [PubMed] [Google Scholar]
- 50.Jones JD, Dunsmuir P, Bedbrook J. EMBO J. 1985;4:2411–2418. doi: 10.1002/j.1460-2075.1985.tb03949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smyth GK. Stat Appl Genet Mol Biol. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 52.Yang YH, Buckley MJ, Dudoit S, Speed TP. J Comput Graph Stat. 2002;11:108–136. [Google Scholar]
- 53.Hegmann JP, Possidente B. Behav Genet. 1981;11:103–114. doi: 10.1007/BF01065621. [DOI] [PubMed] [Google Scholar]
- 54.Jansen RC. Genetics. 1993;135:205–211. doi: 10.1093/genetics/135.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Churchill GA, Doerge RW. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benjamini Y, Yekutieli D. Ann Stat. 2001;29:1165–1188. [Google Scholar]
- 57.Storey JD. J R Statist Soc B. 2002;64:479–498. [Google Scholar]
- 58.de Koning DJ, Haley CS. Trends Genet. 2005;21:377–381. doi: 10.1016/j.tig.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 59.Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, et al. PLoS Biol. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lan H, Chen M, Flowers JB, Yandell BS, Stapleton DS, Mata CM, Mui ET, Flowers MT, Schueler KL, Manly KF, et al. PLoS Genet. 2006;2:e6. doi: 10.1371/journal.pgen.0020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.