Abstract
In contrast to the deregulated hepatocellular division that is a feature of many hepatic diseases and malignancies, physiologic liver growth during embryonic development and after partial hepatectomy (PH) in adults is characterized by tightly controlled cell proliferation. We used forward genetic screening in zebrafish to test the hypothesis that a similar genetic program governs physiologic liver growth during hepatogenesis and regeneration after PH. We identified the uhrf1 gene, a cell cycle regulator and transcriptional activator of top2a expression, as required for hepatic outgrowth and embryonic survival. By developing a methodology to perform PH on adult zebrafish, we found that liver regeneration inuhrf1+/− adult animals is impaired.uhrf1 transcript levels dramatically increase after PH in both mice, and zebrafish and top2a is not up-regulated in uhrf1+/− livers after PH. This indicates that uhrf1 is required for physiologic liver growth in both embryos and adults and illustrates that zebrafish livers regenerate.
Keywords: hepatic outgrowth, hepatogenesis, partial hepatectomy
The liver's capacity to regenerate after acute injury allows for the full restoration of liver mass and function. In the most reliable model to study liver regeneration in rodents, ≈70% of the liver mass is removed with partial hepatectomy (PH), resulting in the reentry of the normally quiescent hepatocytes into the cell cycle (1). Within a week of this procedure, the presurgical liver mass is restored (2). Whereas pathologic liver growth is characterized by uncontrolled cell division, physiologic liver growth during PH-induced liver regeneration is a tightly regulated process. Hepatic outgrowth, the final stage of liver development during which the liver bud expands, is another example of physiologic liver growth. There is very little known regarding the process that controls hepatic outgrowth in the embryo, and with decades of research on liver regeneration, the genetic requirements of physiologic liver growth remains an active area of scientific inquiry.
Studies with knockout mice have identified a few genes that are essential for both hepatic outgrowth and regeneration; of these, none are liver-specific. For example, a liver specific knockout of c-jun results in defective liver regeneration (3), whereas homozygous c-jun deletion results in embryonic lethality and hypoplastic livers (4, 5). Similar studies have shown that the hepatocyte growth factor/c-met (6–8), β-catenin (9), and TNFα (10–12) pathways also regulate physiologic liver growth in embryos and adults. Comparison of the gene expression profiles in regenerating and embryonic livers has identified a handful of genes that are coregulated during both processes (13, 14); however, the functional significance of these findings has not yet been addressed.
Zebrafish present an excellent system for such genetic studies. The robust regenerative potential of adult zebrafish is well established (15), and PH-induced liver regeneration has been reported in trout (16), suggesting similar studies are possible in zebrafish. Zebrafish are renowned for developmental studies, and work on hepatogenesis has revealed that similar genes regulate hepatic patterning in zebrafish and mice (17, 18). Interestingly, studies in mice indicate that hepatic outgrowth requires both hepatomitogenic signal (18) combined with the active suppression of apoptotic signals (11, 19, 20). This dual-signaling mechanism regulating outgrowth has yet to be demonstrated in other species.
We have used the power of zebrafish-forward genetic screening to identify genes that regulate physiologic liver growth. First, embryonic mutants that fail to undergo hepatic outgrowth were identified. We then developed a PH model in zebrafish and demonstrate its utility to discover new genes that regulate liver regeneration. We show that the hi272 line, which bears an insertion in the ubiquitin-like protein containing PHD and ring finger domains-1 (uhrf1) gene (21), is defective in physiologic liver growth in embryos and adults. UHRF1 (also called Np95 in mice and ICBP90 in humans) has been shown to require cell cycle progression in mammalian tissue culture cells (22–25), and the expression of UHRF1 is up-regulated in cancer cells (24, 26–28). UHRF1 is a transcriptional activator of topoisomerase IIα (top2a, refs. 23 and 28; and an E3 ubiquitin ligase, refs. 24 and 29). Here, we report that zebrafish mutants bearing homozygous mutation of uhrf1 develop small-for-size livers and do not survive embryogenesis, whereas uhrf1+/− adults appear healthy but have impaired liver regeneration after PH. We describe liver regeneration in zebrafish and show the utility of this system for addressing whether the genetic program of liver development is recapitulated during regeneration.
Results
uhrf1 Is Required for Hepatic Outgrowth.
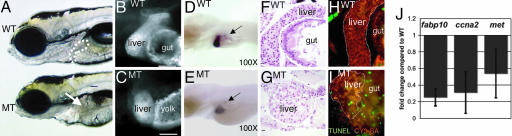
A screen in zebrafish by using retroviral insertional mutagenesis identified nearly 1/4 of the genes that are essential for embryogenesis, and the mutated gene has been identified in every line (21). Streptavidin linked to the fluorophore CY3 (CY3-SA) effectively labels the liver in fixed embryos and was used to screen 294 of these lines for mutants with liver size defects (30), and the hi272 was chosen for further analysis. Fig. 1 illustrates that, in addition to the liver phenotype, hi272 mutant (MT) embryos have a small head, eyes, and jaw, and an underdeveloped gut. Additionally, the diminished yolk consumption and uninflated swim bladder and embryonic lethality are phenotypes common to many of the mutants in this collection. The liver phenotype is revealed both by CY3-SA labeling and in situ hybridization with the liver-specific probe, fatty acid binding protein 10 (fabp10; Fig. 1 B–E). Histologically, the liver in day 5 WT embryos is a crescent-shaped organ composed primarily of hepatocytes (Fig. 1 F), whereas a ball of histologically normal hepatocytes is observed in MT embryos (Fig. 1 G).
Fig. 1.
hi272 mutant embryos have a small-for-size liver on day 5. (A) WT and MT liver day 5 embryos from hi272. The liver in phenotypically WT embryo is visible anterior to the intestinal bulb (white outline); the expected position of the liver in the MT is indicated by an arrow. (B and C) CY3-SA labeling of day 5 WT (B) and MT embryos (C). The gut in the mutant is malformed and does not stain with CY3-SA, and the yolk consumption is incomplete in the mutant by day 5, and thus is labeled with CY3-SA. (Scale bar: 50 μm.) (D and E) In situ hybridization with fabp10 and insulin probes on WT (D) and MT (E) embryos. Arrows point to pancreatic islets, labeled with insulin. (F and G) H&E-; stained sagittal sections of livers from WT(F) and MT (G) embryos. Images were taken through the widest section of the left liver lobe. (Scale bar: 10 μm.) (H and I) Apoptotic cells (green) are not seen in the CY3-SA labeled liver (white outline) and gut of WT (H) but are plentiful in MT (I) embryos. (J) Q-PCR on cDNA prepared from day 5 WT and MT embryos from hi272. Expression levels relative to tbp were calculated and shown as fold change compared with phenotypically WT siblings. The experiment was run in triplicate; bars indicate SD.
The failure to develop a full-size liver could be due to a defect in hepatic patterning or differentiation or from the inability to undergo hepatic outgrowth. To differentiate among these three options, we determined the expression of GATA6, hex, and prox1, genes important for endodermal and hepatic patterning (17), and assessed the pancreas in hi272 mutants. All of the patterning markers were expressed normally (data not shown), and there was not a difference in the pancreatic size, morphology (data not shown), or expression of insulin by islet cells (Fig. 1 D and E), suggesting that endodermal patterning is not affected. In addition, hepatocyte differentiation is also unaffected in hi272 mutants: (i) hepatocytes are histologically normal (Fig. 1 G), (ii) hepatic glycogen production and storage is unaffected (data not shown), and (iii) markers of liver function, fabp10 (Fig. 1E) and ceruloplasmin (data not shown), are expressed by MT hepatocytes. In contrast, hepatocellular apoptosis is increased in MT embryos (Fig. 1 H and I), proliferation appears decreased (data not shown), and cyclin A2 (ccna2) and the hepatocyte growth factor receptor, met, levels are decreased (Fig. 1 J). Taken together, these data suggest that the hepatic phenotype in hi272 MT embryos results from a defect in hepatic outgrowth.
The mutagenic viral insertion in the hi272 is in the first intron of the zebrafish uhrf1 gene (21). The hi3020 line has been identified as another allele, with an insertion 20 bp downstream of the hi272 insertion site (A. Amsterdam, personal communication). Both alleles are phenotypically identical with 100% penetrance (data not shown). uhrf1 mRNA levels in MTs from both lines are decreased by ≈75% compared with phenotypically WT siblings [supporting information (SI) Fig. 6A], confirming that both alleles are hypomorphic uhrf1 mutants. Mammals have two UHRF genes (UHRF1 and UHRF2), whereas zebrafish have only uhrf1, which is 66% identical to human UHRF1 at the amino acid level (SI Fig. 6AB). When referring to uhrf1, we will follow the nomenclature guidelines for the species under discussion.
uhrf1 Is Expressed in Proliferating Tissues.
We evaluated the expression pattern of uhrf1 in zebrafish embryos and adults. Using in situ hybridization, we found uhrf1 to be highly expressed at 24–48 h after fertilization (pf) in rapidly proliferating tissues, including the tectum, retina, and brachial arches. This pattern is similar to many genes that are markers of cell proliferation (zfin.org). Interestingly, during hepatic outgrowth (57 h pf, day 4), uhrf1 is preferentially expressed in the liver bud and expression is maintained in the fully developed liver (Fig. 2 E and F). Expression in the proximal gut also is observed at these times. Thus, the uhrf1 expression pattern in the embryo correlates with the tissues that are the most severely affected in the mutant. In adult zebrafish tissues, the highest expression of uhrf1 was detected in testis (Fig. 2 G), consistent with what is seen in mice (31) and humans (28) and is correlated with the high proliferation observed in this tissue; however, no fertility defects were noted for heterozygous animals. Importantly, low uhrf1 expression was detected in adult liver in zebrafish (Fig. 2 G), mice (31), and humans (28), suggesting a conserved function.
Fig. 2.
uhrf1 is expressed in zones of proliferation during organogenesis and in proliferating adult tissues. (A and B) In situ hybridization at 24 (A) and 34 (B) hours pf shows high levels of uhrf1 expression in rapidly proliferating cells. (C–F) At 48 h pf (C), expression has decreased in most tissues except the tectum, retina, and arches, and by 57 h (D), 4 days (E) and 5 days (F) pf, expression is evident in the liver bud and gut (arrowheads). No expression is seen in MT embryos at 4 days pf (E Inset). (G) uhrf1 message was detected in tissues from and adult male zebrafish by Q-PCR by using tbp as a reference.
Adult Livers Regenerate After PH.
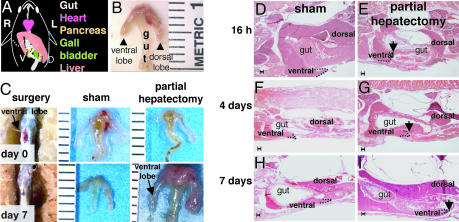
To establish a system in which to study physiologic liver growth in adults, we developed a procedure to carry out PH in zebrafish. The adult zebrafish liver is a bilobed organ positioned on the dorsal-ventral axis, slightly lateral to the gut and other organs of the gastrointestinal system (Fig. 3A). The dorsal lobe is ≈25% larger than the ventral lobe, and the two lobes are contiguous, lacking a pedicle between them. Thus, the anatomy of the zebrafish liver does not allow full removal of an entire lobe, as is performed in rodents through the ligation of individual pedicles. We opted for a small ventral-lateral incision to excise the caudal portion of the ventral lobe (Fig. 3 A and C). The procedure has a success rate of nearly 75% postoperative survival, and a median survival of 63% after overnight recovery. There is no significant difference in the survival after sham operation (SI Table 1). Survival is directly related to the size of the animal, and as technical expertise improved, the percent survival after overnight recovery averaged over 83–100% for PH animals.
Fig. 3.
Partial hepatectomy results in regeneration within 7 days. (A) Schematic of the gastrointestinal organs, viewed from the animals ventral side. The animal's left (L) and right (R) are labeled as are the dorsal (D) and ventral (V) liver lobes. The dashed line marks the site of resection of the ventral lobe. (B) Dissected gut and liver with a piece of spleen (s) attached. Ruler marks are millimeters. (C) PH in zebrafish is carried out on day 0 (Upper) by creating a small incision through the ventral body wall and the ventral lobe of the liver is resected. By day 7 (Lower), the wound has healed and a ventral lobe is evident in both sham and animals subject to PH. (D–I) H&E stained sagittal sections of fish taken at 16 h (D and E) and 4 (F and G) and 7 (H and I) days after PH or sham operation. The tip of the dorsal lobe is outlined with a dotted line and indicated with an arrow in PH samples. (Scale bars: 100 μm.)
To assess the amount of tissue removed by this procedure, 15 animals (n = 4 at 16 h, n = 11 at 24 h) were collected during the first day after PH, and the size and shape of the liver was analyzed histologically on step-sagittal sections through the whole fish (representative example shown in Fig. 3 E). In sham-operated fish, the dorsal lobe extends caudal to the bowel, whereas the tip of the ventral lobe is more rostral (Fig. 3 D, F, and H). In 100% of the fish that underwent PH, extensive inflammation was observed at the site of resection and, in all cases, the entire caudal region of the ventral lobe was missing (Fig. 3 E), indicating that nearly half of the ventral lobe, or ≈20–40% of the total liver mass, had been successfully removed.
To characterize the regeneration process, we collected animals at 16, 24, 48, 60, and 72 h and 4 and 7 days after PH, and carried out histological analysis to determine liver morphology during regeneration. Fig. 4 illustrates the phases of regeneration at 16 h and 4 and 7 days after PH. The unperturbed zebrafish liver is falciform, tapering to a rounded point at the tip (Fig. 3 D, F, and H). This architecture is disrupted during the initial phase of regeneration (Fig. 3 E). By day 4 of recovery, the tip of the liver lobe extended farther caudally and the rounded tip structure was nearly restored (Fig. 3 G). Seven days after PH, the liver is indistinguishable from sham-operated animals (Fig. 3 H and I). This demonstrates that PH can lead to complete liver regeneration in zebrafish within a week after PH. We did not find any difference in the pattern or extent of regeneration if the dorsal lobe instead of the ventral lobe was resected (data not shown). Animals from age 6 to 36 months were indistinguishable in procedure survival and regeneration (data not shown), indicating that animal age does not affect liver regeneration as it does in rodents (32).
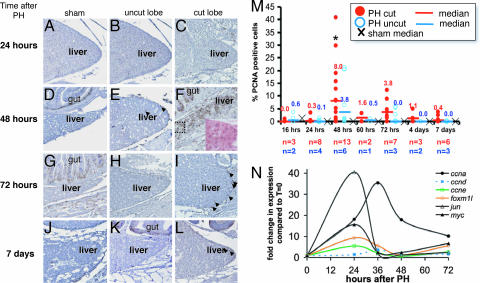
Fig. 4.
PCNA expression peaks at 48 h after PH. (A–L) PCNA immunostaining is indicated by brown nuclei. The gut lies medial to the dorsal liver lobe, serving as a positive control (see D, F, G, and K). Samples taken from sham operated animals (A, D, G, and J), and both the uncut (ventral) lobe (B, E, H, and K) and cut (dorsal) lobe (C, F, I, and L) of animals that underwent PH at 24 h (A–C), 48 h (D–F), 72 h (G–I), and 7 days (J–L) after surgery. Arrows indicate PCNA-positive hepatocyte nuclei. (F Inset) A magnification of the boxed region, stained with H&E. (M) The PCNA index was determined from the cut (red circles) and uncut (blue circles) lobes of PH animals, and the median is plotted as a bar with the value indicated above. The median PCNA index for sham animals is illustrated with an ×. ∗, a significant difference in the PCNA index in the cut lobe at 48 h compared with the cut lobe at 24 h (P < 0.0035) and 72 h (P < 0.095). (N) The remaining ventral lobe of zebrafish livers were collected at the indicated times after PH. The expression levels were determined in two fish by using Q-PCR with ef1a as a reference, the values were averaged and are plotted as fold change compared with expression levels in quiescent livers (t = 0) from five fish.
Cell Proliferation After PH.
We used immunohistochemical nuclear staining of proliferating cell nuclear antigen (PCNA) as a proliferating cell marker in regenerating livers. Liver regeneration in mammals is accomplished by hepatocyte proliferation, and only under certain circumstances are progenitor cells thought to play a part in this process (33). Aside from a few infiltrating inflammatory cells and, in some cases, the cells of Glisson's capsule, virtually all of the PCNA-positive cells in the regenerating liver are hepatocytes (see Fig. 4 F Inset). Although contribution of hepatic progenitor cells to regeneration in zebrafish could not be ruled out because of the lack of specific markers for these cells, we surmise that hepatocyte proliferation accounts for the restoration of liver mass.
PCNA-positive hepatocytes were rare, observed in sham-operated animals (Fig. 4 A, D, G, and J) despite the robust PCNA staining of the adjacent gut or inflammatory cells in the healing wound in the same section (Fig. 4 D and G), indicating that hepatocytes are quiescent in unperturbed zebrafish livers, as in mammals (2). After PH, PCNA-positive cells were concentrated at the site of resection (Fig. 4 F and I), indicating that the lobe that was removed indeed was growing back. We noted that in some cases, PCNA-positive hepatocytes were present at the tip of the unexcised lobe (Fig. 4 E), suggesting that some compensatory regeneration also occurs. Typically, after 72 h after PH, <1% of the cells were PCNA-positive (the sample containing the highest percent of PCNA-positive cells at day 7 is shown in Fig. 4 L).
To quantitatively evaluate hepatocyte proliferation after PH and to determine the region of the liver that is responsible for regeneration, we calculated the PCNA index (no. of PCNA-positive nuclei/total no. of hepatocytes) at the tips of both lobes in 42 PH and 21 sham-operated animals as a function of time after surgery. Fig. 4 M illustrates both the range of PCNA indices for the cut (red dots) and uncut (blue dots) lobes, as well as the median values (horizontal lines with corresponding numerical values). The PCNA indices for the dorsal and ventral lobes of sham animals were pooled, as there was no significant difference between these; the median sham PCNA index is <1 at all time points and is denoted by an ×. In both the cut and uncut lobes of animals that underwent PH, the PCNA index peaks at 48 h after PH; however, the increase in the cut lobe is significant, with P < 0.003 compared with the cut lobe PCNA index at 24 h, and P < 0.09 compared with the cut lobe at 72 h. The PCNA index returned to baseline on days 4 and 7 after PH, consistent with the observation that liver size is reestablished at these times (Fig. 4 G and I).
Many studies have defined the changes in gene expression as a function of time after PH in mammals. Among the up-regulated genes are c-jun and c-myc, which are activated in the first several hours after PH (34). These and other transcription factors, such as Foxm1b (35), activate the transcription of the G1 cyclins, such as cyclin D1, E, and A2, which drive the hepatocytes from G1 into S phase of the cell cycle (2). To investigate the genes that are up-regulated in regenerating zebrafish livers, we performed PH on 18-month-old WT animals. We collected samples from the tip of the regenerating lobe at 24, 36, 48, and 72 h from two animals and analyzed the RNA expression levels of jun, myc, foxm1l (the zebrafish Foxm1b homolog), ccnd1, ccne, and ccna2 by using quantitative PCR (Q-PCR). The expression levels for each gene was averaged between the two samples and plotted as the fold change relative to the average expression level in five quiescent livers (Fig. 4 N). jun and myc are up-regulated at 24 h, likely reflecting a more dramatic up-regulation at an earlier time point, as is reported in rodents (36). foxm1l and ccne are also up-regulated at 24 h, and ccnd1 and ccna2 show peak expression at 36 h after PH.
uhrf1 and ccna2 Are Coregulated After PH in Mouse and Zebrafish.
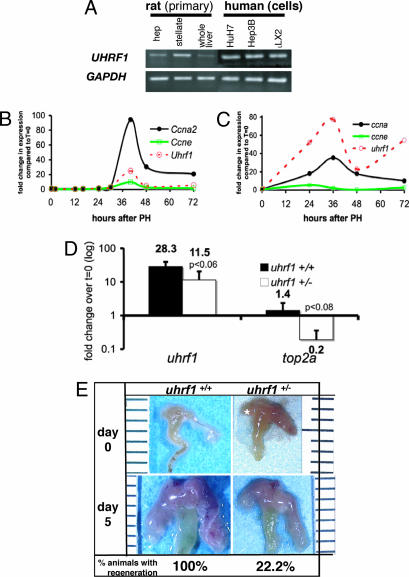
The Uhrf1 transcript is present in mammalian primary and immortalized liver cell lines. In Fig. 5 A, we detect the transcript by PCR in primary rat hepatocytes and stellate cells as well as immortalized cell lines Huh7, Hep3b (hepatocyte), and LX2 (stellate cells). Because many of the genes that have been shown to be critical for liver regeneration are up-regulated after PH (37) and we reasoned that if uhrf1 were required for physiologic liver growth, it too would be up-regulated during recovery from PH. Using Q-PCR to detect uhrf1 message in samples collected from regenerating mouse (Fig. 5 B) and zebrafish (Fig. 5 C) livers, we found that expression was drastically up-regulated during regeneration, in parallel with ccna2 expression. Ccne expression peaks at 24 h in zebrafish regenerating livers and at 40 h in mice. Transcription of TOP2A is directly regulated by UHRF1 (23, 28), and Top2a is up-regulated during mouse liver regeneration (38) with the same kinetics that we observe for Uhrf1. To determine whether heterozygous mutation of uhrf1 hinders its capacity to activate top2a during liver regeneration, we used Q-PCR to compare its expression in livers from uhrf1+/+ and uhrf1+/− zebrafish at the peak time of uhrf1 expression after PH (36 h). Fig. 5 D illustrates that there is a nearly 30-fold increase in uhrf1 and a 1.4-fold increase in top2a expression in WT livers at 36 h after PH. The difference in magnitude in the uhrf1 levels in WT fish at 36 h in Fig. 5 C and D reflects the natural variation from animal to animal that is seen in this system: we observed that the absolute expression levels vary with different samples, but that the pattern observed is consistent across the experiments in which we analyzed expression in >12 animals. As expected, uhrf1 transcript levels in uhrf1+/− animals are roughly one-half that found in animals carrying both copies of the gene, and expression of top2a after PH is markedly suppressed. This demonstrates that activation of top2a depends on uhrf1 in vivo and suggests that cell cycle activation during zebrafish liver regeneration may depend on Uhrf1 activity.
Fig. 5.
uhrf1 is required for liver regeneration. (A) uhrf1 message was detected by standard PCR in rat primary hepatocytes, stellate cells, and whole liver tissue, in human hepatocyte cell lines (Huh7 and Hep3B), and in the stellate cell line (LX2) compared with GAPDH. (B and C) Mouse (B) and zebrafish (C) livers were collected at the indicated times after PH. The expression levels of uhrf1, ccne, and ccna2 were determined by using Q-PCR with cyclophilin as a reference for mouse and ef1a as a reference for zebrafish. Graphs represent fold change compared with quiescent livers. For zebrafish, n = 2 for each PH time point and n = 5 for quiescent liver samples. (D) The level of uhrf1 and top2a transcripts in quiescent (t = 0; n = 3) and regenerating (t = 36 h; n = 3) livers were determined by Q-PCR with ef1a as a reference. Numbers are expressed as the average fold change at 36 h compared with the t = 0 samples. Error bars indicate SD, and P values are labeled. (E) Seven uhrf1+/+ fish and nine uhrf1+/− fish were subject to PH. After 5 days of recovery, the livers were dissected and assessed for regrowth. Representative animals are shown. One hundred percent (7/7) of the uhrf1+/+ animals had substantial regeneration, whereas only 22% (2/7) of the uhrf1+/− animals had any regrowth 5 days after PH.
uhrf1 Is Required for Liver Regeneration After PH.
To test whether uhrf1 is required for liver regeneration, we performed PH on uhrf1+/− adults. One hundred percent (9/9) of the uhrf1+/− animals survived surgery and overnight recovery, compared with 88% (7/8) of the uhrf1+/+. Fig. 5 D shows a representative example of livers from animals of both genotypes on days 0 and day 5 after PH: whereas all (7/7) of the WT animals had full regrowth of the ventral lobe by this time, 78% (7/9) of the uhrf1+/− animals showed no significant regrowth. In the 22% of the uhrf1+/− animals that showed some ventral lobe regeneration, the size of the lobe was significantly smaller than WT (data not shown), and no mutant animal demonstrated full regeneration by day 5. A similar pattern was observed at day 7 (data not shown). Importantly, fin regeneration was not affected in uhrf1+/− animals (SI Fig. 7). These data illustrate that uhrf1 is required for efficient liver regeneration.
Discussion
We have reasoned that a similar genetic program governs physiologic liver growth in embryos and adults and used forward genetic screening in zebrafish to address this. We discovered that a loss-of-function homozygous mutation in the uhrf1 gene results in a small-for-size liver and embryonic lethality on day 5–6 pf and that heterozygous mutation of this gene in adult zebrafish impedes liver regeneration. This demonstrates the utility of the zebrafish PH model for identifying genes that are required for liver growth.
In comparison to the advances in understanding of hepatic patterning (17, 18, 39), hepatic outgrowth is understudied. Mouse knockout models have revealed a handful of genes that are required for outgrowth and have demonstrated that disruption of a number of these genes results in hepatic hypoplasia and apoptosis (4, 5, 10, 11, 12, 40). In zebrafish, we identified a similar pattern in uhrf1 embryos. They undergoes normal hepatic patterning, and several pieces of evidence indicate that mutation of uhrf1 blocks hepatic outgrowth: (i) the size and morphology of uhrf1 livers is that of WT livers at the commencement of hepatic outgrowth (day 3.5), (ii) uhrf1 is expressed in WT livers during outgrowth, (iii) met and ccna2 transcripts are decreased, and (iv) hepatocyte apoptosis is increased in uhrf1 embryos. This finding is significant, for it implies that positive signals driving proliferation as well as suppression of apoptosis is the mechanism for regulating hepatic outgrowth that is conserved across species. Given that UHRF1 is highly expressed in human fetal livers (28), it may play similar role in humans.
Is uhrf1 a liver-specific proliferation regulator? Our data points to a requirement for uhrf1 in a defined number of tissues and suggest that it is one of several important regulators of hepatocyte proliferation. Indeed, the requirement for uhrf1 has been demonstrated in a number of cell types in vitro suggests against this (23–25, 28, 29, 41). In the zebrafish embryo, it is possible that uhrf1 expression is required for proliferation of many tissues, but that maternally derived uhrf1 mRNA produces sufficient Uhrf1 to advance the embryos through the early stages of development. Then, when the maternal message is depleted, the organs that proliferate later in development, such as liver and gut, may be more severely affected. Should this be the case, it would highlight the use of the zebrafish mutant for these studies, because maternal mRNA in mouse embryos is much less than is found in zebrafish, germ-line Uhrf1 mutation in mice would result in early embryonic lethality. Moreover, as the majority of genes previously identified as essential for hepatic outgrowth in mice are not liver specific, c-jun, for example (4, 5), but are required preferentially in the liver for outgrowth and regeneration.
To determine whether uhrf1 is required for liver regeneration, we have developed a model of PH-induced regeneration in zebrafish. We show that livers from adult zebrafish can regenerate fully after PH and that the regenerated livers appear grossly, architecturally, histologically, and functionally normal. Unlike regeneration in rodent models of PH, where remnant lobes become proliferative after excision of adjoining lobes (i.e., compensatory regeneration), zebrafish liver regeneration is a true regenerative process in that the liver mass increase is contiguous with the resection site. We do observe some compensatory proliferation of hepatocytes at a site distant to the resection, indicating that a circulating factor may play a part in this process, as has been determined in rodents (42). Our data demonstrate that liver cell proliferation after PH peaks at 48 h, preceded by a up-regulation of several cell cycle regulatory genes. Liver regrowth is complete within 7 days. We observe that the pattern of gene expression that is consistent with the finding that PCNA immunostaining peaks at 48 h in the regenerating ventral lobe and illustrate that the pattern of hepatocyte cell cycle reentry after PH is similar to what is observed in mammals, although the precise time and magnitude in which a set of genes become activated may vary by species. We postulate that growth is due to hepatocyte proliferation, however, development of specific markers for the different populations of zebrafish liver cells will allow studies addressing the identity of the proliferative cells. Interestingly, preliminary data imply that hepatocytes in uhrf1+/− livers after PH may be hypoplastic and apoptotic (data not shown), suggesting that parallel processes might regulate hepatic outgrowth and regeneration.
We tested the hypothesis that uhrf1 is required for regeneration by performing PH and observing liver regrowth by gross analysis. Most uhrf1+/− animals have a stunted knob 5 days after PH, and there is no significant difference at day 7 (data not shown). Moreover, mRNA levels of uhrf1 after PH are approximately half of that in WT fish. These data indicate that heterozygous mutation of uhrf1 results in a haploinsufficiency phenotype that is revealed in the liver only when significant proliferation is required, similar to the example of hepatic outgrowth.
TOP2A is required for cell cycle progression, and as such has been exploited as a target for chemotherapies (43). UHRF1 directly activates TOP2A transcription in vitro (25, 27, 28), and Top2a up-regulation after PH has been reported in mice (38, 44) and, shown here, in zebrafish. Interestingly, top2a expression after PH in zebrafish requires uhrf1, providing an in vivo example of this relationship and suggests a mechanism by which uhrf1 mutation hampers liver regeneration. In contrast, it is not likely that the uhrf1 embryonic phenotype is mediated exclusively through regulation of top2a. First, mutation of top2a in zebrafish (hi3635) results in a severe and early embryonic lethal phenotype (21), whereas the uhrf1 mutants are relatively healthy on day 5 pf. Second, significant down-regulation of top2a message in uhrf1 embryos was not observed; however, if Uhrf1 regulates top2a only in the liver, then our assays with whole embryo mRNA may underestimate grossly the level of top2a suppression. In situ hybridization analysis of the hepatic changes in top2a expression are required before conclusions can be made. Indeed, top2a expression is regulated by a number of other genes (45), raising the possibility that Uhrf1 mediates additional, top2a-independent, cell cycle effects in hepatocytes. There has been a report that Uhrf1 can negatively regulate expression of the retinoblastoma gene at the transcriptional and posttranscriptional levels (23), and this has been proposed as one mechanism by which alterations in UHRF1 expression may contribute to hyperproliferation. However, we were unable to detect a similar pattern of rb1 mRNA expression in uhrf1 mutant tissue (data not shown). Establishing a link between uhrf1 and rb1 in zebrafish awaits development of suitable reagents for interrogating Rb1 protein.
In addition to regulating the transcription of TOP2A (28) and RB1 (23), UHRF1 has been shown to possess E3 ubiquitin ligase activity targeting histone H3 (29) and itself (24), yet the functional relevance of monoubiquitinization of these targets is unknown. Loss of UHRF1 function abrogates S phase entry, and its overexpression shortens the cell cycle (22–24, 46), in agreement with our finding that uhrf1 is up-regulated during regeneration in parallel with ccna2 and is required for hepatocyte proliferation in vivo. We postulate that this role is liver specific in adults, because fin regeneration (SI Fig. 7) and wound healing are unaffected in uhrf1+/− animals. It is of interest to dissect how Uhrf1 acts to control proliferation in vivo, because it has been proposed as a prognostic marker for some human tumors (26).
Materials and Methods
Screen.
Mutants were generated as described in ref. 21. CY3-SA staining was performed as described (30). Mutants that were healthy on day 5 and had little or no liver tissue staining with CY3-SA were scored as small-for-size liver mutants. The phenotype had 100% penetrance in more than five clutches of hi272 and hi3020 embryos.
In Situ Hybridization.
Day 5 embryos were processed for in situ hybridization by using a probe mixture containing dioxygenin-labeled anti-sense fabp10 (30) and insulin, prepared as described in ref. 47. Experiments were carried out in duplicate on at least 10 embryos.
Histology.
TUNEL assays were performed on three WT and MT embryos. For adult zebrafish, tissue was fixed and processed for PCNA immunohistochemistry.
Between 150 and 250 PCNA-positive cells at the tips of both the ventral (cut) and dorsal (uncut) liver lobes were counted on one- to two-step sections. The PCNA index was calculated as (no. of PCNA positive cells)/(total no. of cells counted). See SI Text for detailed methods.
Partial Hepatectomy.
Before surgery, adult zebrafish aged 6–30 months were deprived of food for 16–24 h. They were anesthetized in 1% MESAB for 40 seconds and placed on a sponge immersed in MESAB, with continual bathing of the gills during the 90-second surgery. A 1- to 2-mm V-shaped incision was made through the ventral body wall, caudal to the heart and ≈1 mm medial to the left pectoral fin. The ventral liver lobe was resected by using forceps and fine-spring scissors. Animals recovered in fish water at room temperature for 2–4 h and then maintained at 28°C. Sham-operated animals were subject to the same anesthetic and body wall incision without liver resection. Animals were monitored daily and euthanized upon signs of distress. PH was performed on 8- to 26-month-old uhrf1+/− adults and on age-matched controls that did not bear an insertion in the uhrf1 gene. Samples from regenerating livers were collected at the distal tip of the resected lobe.
RNA Extraction, cDNA Preparation, and PCR.
RNA samples from zebrafish embryos and adult tissue were isolated by standard protocols. Mouse liver samples were provided by L. Greenbaum (University of Pennsylvania, Philadelphia, PA), and collected as described (38). Rat liver samples were provided by Natalie Nieto (Mount Sinai) and collected as described (48), except that pronase was not used in the hepatocyte isolation protocol. See SI Text for additional experimental details. See SI Table 2 for primer details.
Supplementary Material
Acknowledgments
We are indebted to Nancy Hopkins for support; Adam Amsterdam and Sarah Farrington for animal care, tireless help, and lively discussions; Smita Gopinath and Valeriy Demchev for technical assistance; Scott Friedman, Meena Bansal, David Cohen, and Jerry Trier provided helpful comments on the manuscript; Christian Lawrence for zebrafish care; Charlie Whittaker for bioinformatics analysis; Linda Greenbaum for mouse liver mRNA; and Natalia Nieto for rat liver samples. C.U. was supported by National Institutes of Health Grant K08DK067240.
Abbreviations
- MT
mutant
- PCNA
proliferating cell nuclear antigen
- pf
after fertilization
- PH
partial hepatectomy.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610774104/DC1.
References
- 1.Bucher NL, Swaffield MN. Cancer Res. 1964;24:1611–1625. [PubMed] [Google Scholar]
- 2.Taub R. Nat Rev Mol Cell Biol. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 3.Behrens A, Sibilia M, David JP, Mohle-Steinlein U, Tronche F, Schutz G, Wagner EF. EMBO J. 2002;21:1782–1790. doi: 10.1093/emboj/21.7.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hilberg F, Aguzzi A, Howells N, Wagner EF. Nature. 1993;365:179–181. doi: 10.1038/365179a0. [DOI] [PubMed] [Google Scholar]
- 5.Johnson RS, van Lingen B, Papaioannou VE, Spiegelman BM. Genes Dev. 1993;7:1309–1317. doi: 10.1101/gad.7.7b.1309. [DOI] [PubMed] [Google Scholar]
- 6.Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C. Nature. 1995;376:768–771. doi: 10.1038/376768a0. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt C, Bladt F, Goedecke S, Brinkmann V, Zschiesche W, Sharpe M, Gherardi E, Birchmeier C. Nature. 1995;373:699–702. doi: 10.1038/373699a0. [DOI] [PubMed] [Google Scholar]
- 8.Huh C-G, Factor VM, Sanchez A, Uchida K, Conner EA, Thorgeirsson SS. Proc Natl Acad Sci USA. 2004;101:4477–4482. doi: 10.1073/pnas.0306068101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan X, Behari J, Cieply B, Michalopoulos GK, Monga SP. Gastroenterology. 2006;131:1561–1572. doi: 10.1053/j.gastro.2006.08.042. [DOI] [PubMed] [Google Scholar]
- 10.Bonnard M, Mirtsos C, Suzuki S, Graham K, Huang J, Ng M, Itie A, Wakeham A, Shahinian A, Henzel WJ, et al. EMBO J. 2000;19:4976–4985. doi: 10.1093/emboj/19.18.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rudolph D, Yeh WC, Wakeham A, Rudolph B, Nallainathan D, Potter J, Elia AJ, Mak TW. Genes Dev. 2000;14:854–862. [PMC free article] [PubMed] [Google Scholar]
- 12.Yamada Y, Kirillova I, Peschon JJ, Fausto N. Proc Natl Acad Sci USA. 1997;94:1441–1446. doi: 10.1073/pnas.94.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jochheim-Richter A, Rudrich U, Koczan D, Hillemann T, Tewes S, Petry M, Kispert A, Sharma AD, Attaran F, Manns MP, Ott M. Differentiation. 2006;74:167–173. doi: 10.1111/j.1432-0436.2006.00066.x. [DOI] [PubMed] [Google Scholar]
- 14.Kelley-Loughnane N, Sabla GE, Ley-Ebert C, Aronow BJ, Bezerra JA. Hepatology. 2002;35:525–534. doi: 10.1053/jhep.2002.31351. [DOI] [PubMed] [Google Scholar]
- 15.Poss KD, Keating MT, Nechiporuk A. Dev Dyn. 2003;226:202–210. doi: 10.1002/dvdy.10220. [DOI] [PubMed] [Google Scholar]
- 16.Okihiro MS, Hinton DE. Toxicol Pathol. 2000;28:342–356. doi: 10.1177/019262330002800215. [DOI] [PubMed] [Google Scholar]
- 17.Field HA, Ober EA, Roeser T, Stainier DY. Dev Biol. 2003;253:279–290. doi: 10.1016/s0012-1606(02)00017-9. [DOI] [PubMed] [Google Scholar]
- 18.Zaret KS. Nat Rev Genet. 2002;3:499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- 19.Li Q, Van Antwerp D, Mercurio F, Lee KF, Verma IM. Science. 1999;284:321–325. doi: 10.1126/science.284.5412.321. [DOI] [PubMed] [Google Scholar]
- 20.Rosenfeld ME, Prichard L, Shiojiri N, Fausto N. Am J Pathol. 2000;156:997–1007. doi: 10.1016/S0002-9440(10)64967-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Proc Natl Acad Sci USA. 2004;101:12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arima Y, Hirota T, Bronner C, Mousli M, Fujiwara T, Niwa S, Ishikawa H, Saya H. Genes Cells. 2004;9:131–142. doi: 10.1111/j.1356-9597.2004.00710.x. [DOI] [PubMed] [Google Scholar]
- 23.Jeanblanc M, Mousli M, Hopfner R, Bathami K, Martinet N, Abbady AQ, Siffert JC, Mathieu E, Muller CD, Bronner C. Oncogene. 2005;24:7337–7345. doi: 10.1038/sj.onc.1208878. [DOI] [PubMed] [Google Scholar]
- 24.Jenkins Y, Markovtsov V, Lang W, Sharma P, Pearsall D, Warner J, Franci C, Huang B, Huang J, Yam GC, et al. Mol Biol Cell. 2005;16:5621–5629. doi: 10.1091/mbc.E05-03-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hopfner R, Mousli M, Oudet P, Bronner C. Anticancer Res. 2002;22:3165–3170. [PubMed] [Google Scholar]
- 26.Crnogorac-Jurcevic T, Gangeswaran R, Bhakta V, Capurso G, Lattimore S, Akada M, Sunamura M, Prime W, Campbell F, Brentnall TA, et al. Gastroenterology. 2005;129:1454–1463. doi: 10.1053/j.gastro.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 27.Mousli M, Hopfner R, Abbady AQ, Monte D, Jeanblanc M, Oudet P, Louis B, Bronner C. Br J Cancer. 2003;89:120–127. doi: 10.1038/sj.bjc.6601068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hopfner R, Mousli M, Jeltsch JM, Voulgaris A, Lutz Y, Marin C, Bellocq JP, Oudet P, Bronner C. Cancer Res. 2000;60:121–128. [PubMed] [Google Scholar]
- 29.Citterio E, Papait R, Nicassio F, Vecchi M, Gomiero P, Mantovani R, Di Fiore PP, Bonapace IM. Mol Cell Biol. 2004;24:2526–2535. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sadler KC, Amsterdam A, Soroka C, Boyer J, Hopkins N. Development (Cambridge, UK) 2005;132:3561–3572. doi: 10.1242/dev.01918. [DOI] [PubMed] [Google Scholar]
- 31.Fujimori A, Matsuda Y, Takemoto Y, Hashimoto Y, Kubo E, Araki R, Fukumura R, Mita K, Tatsumi K, Muto M. Mamm Genome. 1998;9:1032–1035. doi: 10.1007/s003359900920. [DOI] [PubMed] [Google Scholar]
- 32.Bucher NL, Swaffield MN, Ditroia JF. Cancer Res. 1964;24:509–512. [PubMed] [Google Scholar]
- 33.Fausto N. Hepatology. 2004;39:1477–1487. doi: 10.1002/hep.20214. [DOI] [PubMed] [Google Scholar]
- 34.Morello D, Fitzgerald MJ, Babinet C, Fausto N. Mol Cell Biol. 1990;10:3185–3193. doi: 10.1128/mcb.10.6.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Kiyokawa H, Dennewitz MB, Costa RH. Proc Natl Acad Sci USA. 2002;99:16881–16886. doi: 10.1073/pnas.252570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morello D, Lavenu A, Babinet C. Oncogene. 1990;5:1511–1519. [PubMed] [Google Scholar]
- 37.Fausto N. Am J Physiol. 1999;277:G917–G921. doi: 10.1152/ajpgi.1999.277.5.G917. [DOI] [PubMed] [Google Scholar]
- 38.White P, Brestelli JE, Kaestner KH, Greenbaum LE. J Biol Chem. 2005;280:3715–3722. doi: 10.1074/jbc.M410844200. [DOI] [PubMed] [Google Scholar]
- 39.Ober EA, Verkade H, Field HA, Stainier DY. Nature. 2006;442:688–691. doi: 10.1038/nature04888. [DOI] [PubMed] [Google Scholar]
- 40.Nishina H, Vaz C, Billia P, Nghiem M, Sasaki T, De la Pompa JL, Furlonger K, Paige C, Hui C, Fischer KD, et al. Development (Cambridge, UK) 1999;126:505–516. doi: 10.1242/dev.126.3.505. [DOI] [PubMed] [Google Scholar]
- 41.Muto M, Kanari Y, Kubo E, Takabe T, Kurihara T, Fujimori A, Tatsumi K. J Biol Chem. 2002;277:34549–34555. doi: 10.1074/jbc.M205189200. [DOI] [PubMed] [Google Scholar]
- 42.Bucher NL. N Engl J Med. 1967;696:738–746. doi: 10.1056/NEJM196710052771405. [DOI] [PubMed] [Google Scholar]
- 43.Giles GI, Sharma RP. Med Chem. 2005;1:383–394. doi: 10.2174/1573406054368738. [DOI] [PubMed] [Google Scholar]
- 44.Fan G, Xu R, Wessendorf MW, Ma X, Kren BT, Steer CJ. Cell Growth Differ. 1995;6:1463–1476. [PubMed] [Google Scholar]
- 45.Bakshi RP, Galande S, Muniyappa K. Crit Rev Biochem Mol Biol. 2001;36:1–37. doi: 10.1080/20014091074165. [DOI] [PubMed] [Google Scholar]
- 46.Bonapace IM, Latella L, Papait R, Nicassio F, Sacco A, Muto M, Crescenzi M, Di Fiore PP. J Cell Biol. 2002;157:909–914. doi: 10.1083/jcb.200201025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun Z, Hopkins N. Genes Dev. 2001;15:3217–3229. doi: 10.1101/gad946701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nieto N, Friedman SL, Cederbaum AI. Hepatology. 2002;35:62–73. doi: 10.1053/jhep.2002.30362. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.