Abstract
Notch/RBP-J signaling is required for generation of early T progenitors (ETP) and promotion of double-negative (DN) 4 cells from DN3 cells in thymocyte differentiation. However, whether Notch affects other steps during thymocyte differentiation remains unknown. Msx2-interacting nuclear target protein (Mint) is an endogenous inhibitor of Notch regulation. Concordantly, by ex vivo analyses of embryonic thymi and in vitro differentiation studies of fetal liver progenitors, we find that Mint deficiency enhances generation of ETP and DN4 cells. Unexpectedly, however, Mint deficiency impairs differentiation of ETP into DN2 cells, suggesting that Notch/RBP-J signaling negatively regulates DN1–DN2 transition.
Keywords: development, T cell
Notch/RBP-J signaling controls cell differentiation processes in a wide variety of tissues of multicellular organisms (1, 2), including lineage choice between T and B lymphocytes from hematopoietic progenitors (3, 4). Notch/RBP-J signaling is initiated by proteolytic processing of the Notch receptor on the membrane upon interaction with its ligands, such as Delta-like 1 (Dll1), expressed on neighboring cells (5). This processing releases the intracellular domain of Notch (NICD) from the membrane into the nucleus where NICD forms a complex with the DNA-binding protein RBP-J to transactivate target genes (6, 7). The mammalian genome encodes four Notch receptors that are expressed differentially and redundantly (8–10), whereas NICDs of all Notch receptors are targeted to ubiquitously expressed RBP-J (11, 12).
We and others have shown that Msx2-interacting nuclear target protein (Mint) acts as a suppressor of Notch/RBP-J signaling by competing RBP-J binding with NICD (13, 14). Overexpression of Mint suppresses transcriptional activation of Notch target genes in cultured cell lines (13, 14). Although mint−/− mice die in utero, displaying abnormalities in cardiac and pancreatic development, mint−/− fetal liver cells transplanted into irradiated rag2−/− mice produce more marginal zone B cells and fewer follicular B cells in spleen compared with wild-type cells (13). Inactivation of Notch/RBP-J signaling through genetic disruption of RBP-J, Notch2, or Dll1 results in phenotypes opposite those of Mint deficiency in spleen B cells (15–18). These results demonstrate that Mint negatively regulates Notch/RBP-J signaling in spleen, whereas it remains to be examined whether Mint has critical roles in other organs, such as thymus, brain, and testis of postnatal mice, where Mint is highly expressed (19).
Genetic disruption of Notch1 (20, 21) or RBP-J (22) in bone marrow progenitor cells leads to early developmental arrest of T cells and ectopic generation of B cells in the thymus. Consistently, activation of Notch/RBP-J signaling in hematopoietic stem cells by retroviral expression of NICD promotes the appearance of an immature T cell population in the bone marrow, and it suppresses B cell development (23). The most primitive T progenitors in adult thymus are found within a heterogeneous population, the double-negative (DN) 1 thymocytes (CD4−CD8−CD25−CD44+), and they are defined as the early T progenitors (ETPs) (c-KithiDN1) (24–26). ETPs are also heterogeneous, and some of them display the lineage potential for T, B, and natural killer (NK) cells but no long-term self-renewing activity, unlike the blood-circulating Lin−Sca-1hic-Kithi (LSK) cells (25, 27–29). Both LSK and DN1 cells cultured on the bone marrow stromal cell line OP9 with ectopic expression of Dll1 (OP9/Dll) generate ETP and T lineage cells, whereas they develop into B but not T cells on OP9 cells without expression of Dll1 (30). In addition, inhibition of Notch/RBP-J signaling impairs generation of ETPs and increases B lineage cells from LSK and DN1 cells (28, 31). Thus, the thymic environment, including Notch signaling, instructs bone marrow-derived progenitors such as LSK to differentiate into T cells, and it suppresses B cell differentiation (24, 32). Furthermore, Notch/RBP-J signaling controls T cell differentiation after the lineage commitment, DN3–DN4 transition in the thymus (33, 34), and the induction of Th2 with suppression of Th1 in the periphery (34–36). However, it is not known whether Notch signaling regulates the generation of DN2 and DN3 cells in the thymus. The major technical difficulty in tackling this problem stems from the fact that blockade of Notch/RBP-J signaling abolishes ETP generation completely, resulting in the absence of all thymocytes.
This work was designed to investigate where and how Notch regulates early T cell differentiation by using specific up-regulation of Notch signaling by Mint knockout. Transplantation and in vitro culture of mint−/− fetal liver cells revealed that Mint deficiency results in impaired DN1–DN2 transition, indicating that Notch/RBP-J signaling inhibits DN1–DN2 differentiation. In addition, Mint deficiency leads to enhanced production of ETP and DN4 cells, indicating that Mint negatively regulates Notch/RBP-J signaling at these steps.
Results
Mint Expression in Fetal Adult and Thymus.
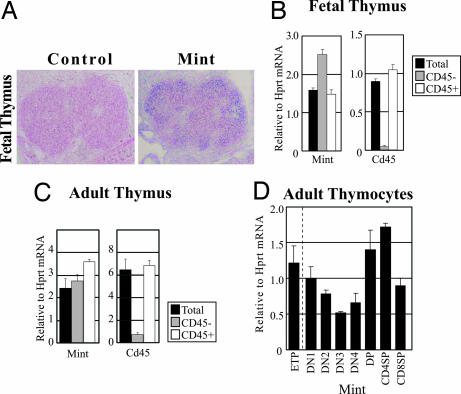
To investigate the role of Mint in thymus, we first examined expression of Mint mRNA. Thymic expression of Mint mRNA was detectable at E14.5 by in situ hybridization and quantitative RT-PCR (Fig. 1A and B). Interestingly, both CD45− nonlymphoid and CD45+ lymphoid cells derived from E14.5 fetal thymus as well as adult thymus (Fig. 1 B and C) expressed similar levels of Mint mRNA, which raises a striking possibility that Mint might control still uncharacterized functions of Notch/RBP-J signaling in thymic epithelial cells that also express Notch receptors and target genes (37). Mint is expressed in all subsets of thymocytes with varying levels (Fig. 1D). It is of note that ETP (c-KithiDN1) expressed Mint at a level similar to total DN1 cells.
Fig. 1.
Expression of Mint mRNA in developing fetal and adult thymus. (A) In situ hybridization analysis of E14.5 thymus using either Mint sense (Control) or anti-sense (Mint) probe. (B and C) Quantitative RT-PCR analysis for Mint expression in thymus from E14.5 embryos (B) and adult (C). Total RNA was prepared from unsorted cells (Total), nonlymphocyte (CD45−) fraction, or lymphocyte (CD45+) fraction, and then it was subjected to quantification. Each value represents the mean ± SEM of three individual thymi. (D) Quantitative RT-PCR analysis on adult thymocytes. Total RNA was prepared from indicated lymphocyte subsets, and then it was subjected to quantification. Each value represents the mean ± SEM of three individual thymi. Hprt, hypoxanthine phosphoribosyltransferase.
Increased ETP but Reduced Maturation of Thymocytes in Mint Deficiency.
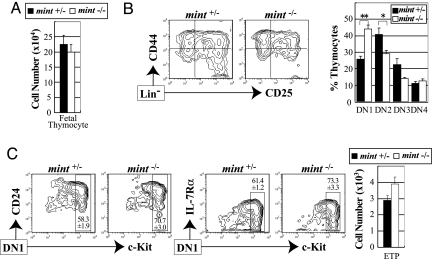
To address the Mint function in thymocyte development, we first analyzed the number and subset distribution of thymocytes in E14.5 mint−/− fetal thymi. Mint−/− mice used in the present study survive until E14.5, and they die soon after E16 because of cardiovascular defects with severe s.c. edema. Lymphoid progenitors begin to immigrate to the thymus around E11.5 (38, 39). Although few double-positive (DP) and single-positive (SP) cells were found at this stage of embryonic thymus, DN cells were clearly identified. The absolute number of mint−/− embryonic thymocytes did not differ from that of littermate mint+/− thymocytes (Fig. 2A). However, we noticed that the proportion of DN1 cells was increased in mint−/− thymocytes (Fig. 2B). The majority of DN1 cells were c-KithiCD24+IL-7R+/lo and indistinguishable between mint−/− and mint+/− thymocytes (Fig. 2C). Thus, c-KithiDN1 cells (ETPs) increased in mint−/− fetal thymi. Because Mint negatively regulates Notch/RBP-J signaling, the increased ETPs in mint−/− thymi agrees with recent reports that inhibition of Notch/RBP-J signaling impairs generation of ETPs (28, 31). By contrast, DN2 and DN3 cells were decreased in mint−/− fetal thymi compared with littermate controls (Fig. 2B), indicating a developmental defect at DN1–DN2 transition in mint−/− embryos. The results suggest that enhanced Notch/RBP-J signaling by Mint deficiency may inhibit generation of DN2 cells from DN1 ETP cells. Curiously, we have not observed any reduction of DN4 cells despite clear reduction of their immediate precursors (DN2 and DN3). This observation is consistent with the previous report that Notch/RBP-J signaling positively regulates the generation of DN4 cells from DN3 cells (33, 34), assuming that Mint inhibits Notch signaling at this step.
Fig. 2.
Enhanced ETP production and impaired DN1–DN2 transition in mint−/− fetal thymus. Thymocytes from E14.5 mint−/− embryos and mint+/− littermates were analyzed for their expression of lineage markers, CD44, CD25, c-Kit, CD24, and IL-7Rα. (A) Absolute cell numbers of total thymocytes. (B) Representative FACS profiles and percentage of indicated DN subsets among total thymocytes. Lineage-negative (DN) cells were subdivided into DN1–DN4 subsets as follows: DN1 (CD44+CD25−), DN2 (CD44+CD25+), DN3 (CD44−CD25+), and DN4 (CD44−CD25−). Each value represents the mean ± SEM of five to seven embryos. ∗∗, P < 0.01; ∗, P < 0.05, by Student's unpaired t test. (C) The percentages of ETPs within the DN1 subset (Left) and the absolute numbers of ETPs in embryonic thymi (Right). Each value represents the mean ± SEM of three embryos.
Defective DP T Cell Development in the Absence of Mint.
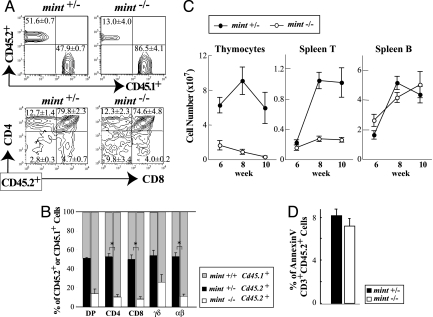
To analyze further the effects of Mint deficiency on thymocyte development at later differentiation stages in adult, we investigated thymocyte differentiation of mint−/− fetal liver cells transplanted into sublethally irradiated rag2−/− mice, which are devoid of thymocytes except for DN cells (40). To monitor the differentiation efficiency of mint−/− or mint+/− E14.5 fetal liver cells (CD45.2+), an equal number of CD45.1+ fetal liver cells from wild-type B6.SJL embryos was added as an internal control of transplantation. The proportion of CD45.2+ thymocytes was 52% in mice receiving mint+/− fetal liver cells (mint+/− recipients) 6 weeks after transplantation, whereas only 13% of thymocytes were CD45.2+ in mice receiving mint−/− fetal liver cells (mint−/− recipients), indicating the defect of thymocyte differentiation in Mint deficiency (Fig. 3A). To de-termine the stage at which the defect occurred in Mint defi-ciency, thymocytes were further analyzed for their expression of CD4 and CD8 in the CD45.2+ cells. The proportions of DP, CD4+, and CD8+ subsets in CD45.2+ cells were similar between mint−/− and mint+/− recipients (Fig. 3A). Consequently, the proportion of CD45.2+ cells was almost equally reduced in DP, CD4, and CD8 subsets in mint−/− recipients, suggesting that thymocyte differentiation is blocked before DP cells (Fig. 3B). This interpretation is in accord with normal development of DP and SP T cells in RBP-Jf/f × CD4-Cre mice (34). In mint−/− recipients, the proportions of CD45.2+ αβT and γδT cells were both severely reduced. Because rag2−/− thymus contains the background CD45.2 DN cells, analyses of the subsets of CD45.2 DN cells of mint−/− recipient are not useful. Nonetheless, it is worth noting that the proportion of DN cells in CD45.2+ mint−/− recipients (9.8%) is higher than that (2.8%) in CD45.2+ mint+/− recipients (Fig. 3A). This observation is in agreement with the above conclusion that mint−/− thymocyte differentiation is blocked somewhere in DN cells. The number of CD45.2+ thymocytes further decreased along with time in mint−/− recipients, whereas that in mint+/− recipients showed no significant reduction (Fig. 3C). Consistent with this observation, the number of CD45.2+CD3+ splenocytes of mint−/− recipient remained low, whereas the mint+/− counterpart increased dramatically. By contrast, almost no change for B220+ splenocytes was seen between mint−/− and mint+/− recipients.
Fig. 3.
Mint deficiency affects early T cell development before the DP stage in fetal liver transplantation assay. (A) Representative FACS profiles 6 weeks after transplantation. Each value represents the mean ± SEM of three recipient mice per group. (B) Percentages of CD45.2+ cells among the indicated thymocyte subsets at 6 weeks after transplantation. Each value represents the mean ± SEM of three recipient mice per group. ∗, P < 0.05 by Student's unpaired t test. (C) absolute number of CD45.2+ cells among thymocytes, CD3+ spleen T cells, and B220+ spleen B cells at various time points after transplantation. Each value represents the mean ± SEM of 6–10 recipient mice per group. (D) Percentages of apoptotic annexin V+ cells among CD45.2+CD3+ thymocytes at 8 weeks after transplantation. Each value represents the mean ± SEM of three to five mice per group.
Given the severe reduction in the number of CD45.2+ thymocytes in mint−/− recipients, we analyzed potential cell death of CD45.2+ cells among CD3+ thymocytes by annexin V staining, an early marker for apoptosis, which showed no change between mint−/− and mint+/− recipients (Fig. 3D). All of these results suggest that Mint deficiency causes a developmental defect in the DN stage but shows indifference to B lineage commitment.
Impaired DN1–DN2 Transition by Mint Deficiency.
To address which step of DN stages is affected by Mint deficiency, we used the in vitro T cell differentiation assay that was originally developed by Schmitt and Zuniga-Pflucker (41). Lin−ScaIhic-Kithi (LSK) cells were sorted from E14.5 fetal livers, and their T cell differentiation was recapitulated on Tst-4/Dll1, a thymic stromal cell line Tst-4 expressing Dll1 (42, 43). Differentiation of cultured cells was analyzed for their expression of Thy1.2 and CD19. Mint−/− and mint+/− LSK cells generated a similar number of Thy1.2+ T cells at day 6 on Tst-4/Dll1 cells (Fig. 4A Upper Left). The number of T cells from mint−/− LSK cells became far less than that from mint+/− LSK cells as coculture continued. The reduced number of mint−/− Thy1.2+ T cells at day 14 is mostly the result of reduction of DN cells (Fig. 4A Lower Left). Under this condition, no CD4+ or CD8+ SP T cells were generated, whereas DP cells began to appear at day 10 (Fig. 4A Lower Right). Generation of DP cells was severely impaired in mint−/− LSK cell coculture compared with mint+/− LSK cell coculture at day 14. The reduction of T cells by Mint deficiency affected both αβT and γδT cells (Fig. 4B). On Tst-4/Dll1, neither mint−/− nor mint+/− LSK cells were able to differentiate into CD19+ B cells, whereas both efficiently differentiated into CD19+ B cells but not Thy1.2+ T cells on Tst-4. Importantly, the same numbers of B cells were generated from mint−/− and mint+/− LSK cells by coculture with Tst-4 (Fig. 4A Upper Right), suggesting that multipotential capacity of LSK in not affected by Mint deficiency.
Fig. 4.
Mint deficiency enhances ETP production and impairs the DN1–DN2 transition in in vitro T cell differentiation. (A) The cells cultured on Tst-4/Dll1 (+ Dll1) or Tst-4 (No Dll1) were analyzed for their expression of Thy1.2 and CD19 at various time points. The cells generated on Tst-4/Dll1 were analyzed for their expression of lineage markers or CD4 and CD8 at various time points, and the absolute cell numbers of Lineage-negative (DN) cells and DP cells are calculated. (B) The cells generated in A were analyzed for their expression of TCRβ and TCRγδ at day 14, and the absolute cell numbers of αβT and γδT cells are calculated. (C) The Lin− cells among the cells generated in A were analyzed for their expression of CD44 and CD25 at indicated day in vitro, and they were subdivided into DN1–DN4 subsets as in Fig. 2B. (Upper) Representative FACS profiles and (Lower) absolute cell numbers of indicated thymocyte subsets. (D) The Lin− cells among the cells generated in A were analyzed for their expression of CD44, CD25, and c-Kit at day 6, and the absolute cell numbers of ETPs are calculated. Each value represents the mean ± SEM of three independent samples per group. ∗, P < 0.05 by Student's unpaired t test.
To determine the exact step of the DN stages affected by Mint deficiency, we examined DN cells generated by culturing mint−/− and mint+/− LSK cells on Tst-4/Dll1 for their expression of CD44 and CD25 (Fig. 4C). At day 6, the number of mint−/− DN1 cells was >2-fold higher than that of mint+/− DN1 cells. Because the fraction and absolute number of c-Kithi cells increased in mint−/− DN1 cells compared with mint+/− DN1 cells, the increase of DN1 cells is primarily caused by more efficient generation of c-KithiDN1 (ETP) cells from mint−/− LSK cells (Fig. 4D). c-KithiDN1 cells were confirmed to have surface phenotypes similar to those identified in fetal thymus (data not shown). Previous experiments using the OP9/Dll1 system with exogenous cytokines showed that the majority of LSK or common lymphoid progenitors differentiate into DN3 or later stage cells by 7–8 days (25, 41). In the present system without exogenous cytokines, enhanced generation of ETP from mint−/− LSK cells persisted until day 10 and slowed down at day 14. At these time points, the differentiation profile is probably skewed to the terminal stages partially because of limitation of stem cells. Kinetics of differentiation regulation is likely to be best represented at day 6. The in vitro observations at days 6–10 are probably similar to thymocyte differentiation stages in E14.5 thymus 3 days after immigration of lymphocyte progenitors (Fig. 2B). The increase of ETPs in Mint deficiency also agrees with the previous reports that Notch/RBP-J signaling facilitates ETP production (28, 31).
Despite more abundant ETP at day 6, we found no clear increase but rather reduction of subsequent stage cells (DN2 and DN3) in mint−/− cells compared with mint+/− cells (Fig. 4C). No obvious increase in mint−/− DN2 cells persisted at days 10 and 14. These in vitro findings agree with the observations in the mint−/− fetal thymus, showing reduced DN2 cells despite increased ETP generation (Fig. 2B). Taken together, these results demonstrate that Mint deficiency inhibits DN1–DN2 transition. It is therefore likely that Notch/RBP-J signaling negatively regulates the DN1–DN2 transition.
It is worth noting that mint−/− DN4 cells increased significantly compared with mint+/− DN4 cells at day 6. A similar increase of DN4 cells was observed in mint−/− fetal thymus as shown above. The results are in good agreement with the previous finding that the DN3–DN4 transition is impaired by the defect of Notch/RBP-J signaling (33, 34).
Increased Expression of Nrarp in Mint-Deficient DN1 Cells.
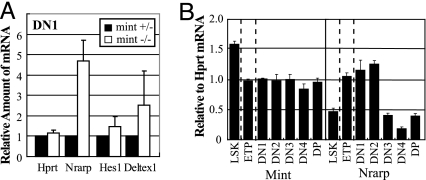
Because Mint acts as a negative regulator of Notch/RBP-J signaling in lymphocyte differentiation steps, we assumed that Mint deficiency impairs DN1–DN2 transition through up-regulation of Notch target genes. Among the limited number of direct Notch target genes known to date, Nrarp has been reported to block DN1–DN2 transition when it is overexpressed (44), serving as a good candidate for our analysis. To test whether Mint deficiency leads to increased expression of Nrarp, we prepared total RNA from DN1 cells generated from mint+/− and mint−/− LSK cells cultured on Tst-4/Dll1 for 10 days and subjected to quantitative RT-PCR (Fig. 5A). Nrarp expression is increased by >4-fold in mint−/− DN1 cells compared with control mint+/− DN cells. The increase of other known Notch candidates (Deltex1 and Hes1) was less significant (Fig. 5A). The relative level of Hprt, an invariant control, was not affected between mint−/− and mint+/− DN1 cells. We also studied expression of Nrarp mRNA in various thymocyte subsets generated in vitro at day 10, and we found that expression of Nrarp is initially high in ETP, DN1, and DN2 stages and attenuated in later stages, whereas that of Mint does not vary much among each subset (Fig. 5B). These results are consistent with the assumption that Notch negatively regulates the DN1–DN2 transition through activation of Nrarp.
Fig. 5.
Mint deficiency leads to induction of Notch target genes. (A) Cells generated from mint−/− or mint+/− cells cultured on Tst-4/Dll1 for 10 days were subjected to FACS sorting of DN1 cells. Total RNA was isolated from the sorted DN1 cells and analyzed by quantitative RT-PCR. Each value represents the mean ± SEM of the amount of mRNA relative to that of the mint+− group in three independent experiments. (B) Expression of Mint and Nrarp in the indicated lymphocyte subsets generated from wild-type cells cultured on Tst-4/Dll1 for 10 days. Each value represents the mean ± SEM of three independent samples.
Discussion
We have demonstrated that Mint regulates three steps in early thymocyte development: generation of the ETP, differentiation of DN1 to DN2 cells, and differentiation of DN3 to DN4 cells (Fig. 6). First, Mint deficiency leads to the increase of ETP in fetal thymus and in vitro T cell differentiation. The recent reports that genetic disruption of Notch (31) or Notch inhibitors (i.e., Lunatic fringe and a DN form of Mastermind-like 1) induces defective ETP production and increased generation of intrathymic B cells (28, 31) provide evidence that notch positively regulates ETP generation in thymus. Thus, the increase of mint−/− ETP shows that Mint acts as a negative regulator of Notch/RBP-J signaling that positively regulates ETP generation.
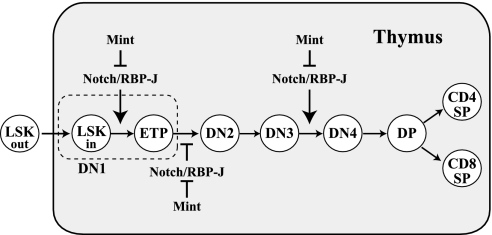
Fig. 6.
Regulation of thymocyte development by Notch/RBP-J signaling. It has been demonstrated that Notch/RBP-J signaling is required for intrathymic T cell development at three steps: (i) ETP production, (ii) γδΤ segregation, and (iii) DN3–DN4 transition. This work reveals that Mint negatively regulates Notch/RBP-J signaling at ETP production and DN3–DN4 transition. Furthermore, this work identified DN1–DN2 transition as a regulatory step controlled by Notch/RBP-J signaling, and it showed that Mint negatively regulates Notch/RBP-J signaling at this step. DN1 cells include ETP and recent immigrants from bone marrow which are represented by LSKin. Circulating LSK are shown by LSKout.
Second, Mint deficiency leads to impaired DN1–DN2 transition in fetal thymus and in vitro T differentiation. In view of the negative regulatory role of Mint in Notch/RBP-J signaling, this finding indicates that the DN1–DN2 transition is negatively regulated by Notch signaling. Earlier studies failed to identify the step and mode of regulation by Notch in early T thymocyte development after ETP generation (24). The negative regulation of the DN1–DN2 transition by Notch is supported by the present and previous findings that the impaired DN1–DN2 transition is associated with the induction of Nrarp, one of direct Notch target genes that blocks DN1–DN2 transition when it is overexpressed (44, 45). Taken together, Mint deficiency up-regulates Notch signaling at DN1, resulting in enhanced expression of Nrarp, which blocks DN1 transition to DN2. Although we also observed slight up-regulation of Deltex1, another direct Notch target in mint−/− DN1 cells, Deltex1 does not seem to be involved in control of DN1–DN2 transition (44). It is possible that still-undiscovered Notch target(s) is up-regulated and involved in negative regulation of DN1–DN2 transition in cooperation with Nrarp. It has been demonstrated that E2A is critical for DN1–DN2 transition (46, 47) and that NICD overexpression induces degradation of E2A by RBP-J-dependent transcription in splenic T and B cells (48). However, we did not detect any decrease of E2A protein in mint−/− DN1 cells nor any increase in expression of Id protein that inactivates E2A (data not shown), suggesting that the E2A pathway may not be involved in the negative regulation of DN1–DN2 transition by Notch/RBP-J signaling.
Third, studies on embryonic thymus and LSK cell coculture with stromal cells indicate that mint−/− DN4 cells were not reduced despite the clear decrease in the number of mint−/− DN2 and DN3 cells compared with mint+/− cells. Because conditional deletion of Notch1 or RBP-J at the DN2–DN3 stage using lck-Cre drastically reduces DN4 but not DN3 cells in adult thymus, Notch/RBP-J signaling positively regulates the generation of DN4 cells from DN3 cells (33, 34). It is therefore reasonable to assume that Mint also negatively regulates Notch/RBP-J signaling at the DN3–DN4 transition.
It is important to know which of the Notch receptors and ligands are involved in the regulation of the DN1–DN2 transition. Although the thymic subcapsular region containing most of DN cells expresses Notch1, 2, and 3 (37), conditional disruption of Notch2 or germ-line disruption of Notch3 has not been shown to affect thymocyte development (16, 49). It has been demonstrated that genetic disruption of Notch1 causes impaired ETP production and DN3–DN4 transition (31, 33). Although Notch1 is a most likely candidate to regulate the DN1–DN2 transition, this regulation has not been adequately addressed because the absence of Notch1 abrogates ETP and subsequent T lineage cells from thymus. The direct confirmation of this conclusion depends on availability of Cre Tg mice that appropriately delete a floxed Notch1 allele in ETPs.
It is puzzling why Notch negatively regulates the DN1–DN2 transition. Interestingly, the regulation of this step by Notch is not strict or absolute because the DN2, DN3, and DN4 cells were produced in the absence of the endogenous Notch inhibitor Mint. It is therefore likely that the DN1–DN2 transition is modulated so that the numbers of DN2 and DN3 cells are relatively limited. One possible explanation is that abundant DN2/DN3 cells may disturb the balance between γδ and αβ lineage (34). This model can be tested in the future with conditional knockout strategy for Mint (e.g., lck-Cre transgene-mediated deletion) that can bypass the impaired DN1–DN2 transition observed in this work. Although we did not address potential roles of Mint in thymic epithelial cells in this work, the conditional knockout strategy for RBP-J and Mint will also shed light on the functions of Notch/RBP-J signaling in thymic epithelial development and function in the future.
It is worth noting that fetal liver progenitors have the potential to generate B cells in the absence of Notch signaling almost 10 times stronger than that of T cells in the presence of Notch signaling as assessed by coculture with thymic stromal cells (Fig. 4). Nonetheless, Mint deficiency has almost no effect on the number of CD19+ B cells in in vitro coculture with Tst-4. These results indicate that Mint deficiency does not disturb the generation of B cell progenitors from LSK. By analyzing differentiation of mint−/− and mint+/− fetal liver cells injected into rag2−/− mice, we found few B220+ cells in thymus (data not shown), and we did not observe a significant difference in the number of B220+ spleen cells (Fig. 3C). Similar observations are reported by analyses of peripheral B220+ cells from the MX-Cre transgene-mediated Notch1 conditional knockout mice (20, 50). The results indicate that T and B lineage segregation in thymus has little contribution in the number of B cells in the periphery. Although the final conclusion should wait for characterization of B progenitors in thymus, it is likely that Notch is involved in blockade of B differentiation from either precommitted or multipotential progenitors only in thymus.
In summary, we demonstrate the roles of Mint, the Notch inhibitor, at three steps of early T cell development: ETP generation, the DN1–DN2 transition, and the DN3–DN4 transition. The finding revealed the negative regulation of the DN1–DN2 transition by Notch/RBP-J signaling. It is particularly striking that Notch regulates the three steps of early thymocyte differentiation in different directions. The biological significance of this complex regulation remains to be seen.
Materials and Methods
Mice.
Mint-deficient mice have been described in ref. 13. These mice were backcrossed to C57BL/6J >10 times, and they were maintained as heterozygotes for the mutant mint allele. Mint−/− and mint+/− embryos used in the current study were generated by crossing heterozygous males and females. Rag2−/− mice were described in ref. 40, and B6.SJL mice were obtained from the Jackson Laboratory (Bar Harbor, ME). All mouse protocols were approved by the Institute of Laboratory Animals, Kyoto University Graduate School of Medicine.
In Situ Hybridization.
Cryosections (8–12 μm) of E14.5 fetal thymus were fixed in 4% paraformaldehyde. In situ hybridization was performed as described in ref. 51. Antisense and sense probes were transcribed in vitro by using a fragment of Mint cDNA (GenBank accession no. NM_019763; nucleotide positions 1–670 of the ORF) in pGEM-T Easy vector as a template.
Flow Cytometry and Antibodies.
The following mAbs were purchased from BD Biosciences (San Diego, CA): FITC-conjugated anti-CD24 (M1/69), anti-NK1.1 (PK136), anti-Gr1 (RB6-8C5), and anti-CD19 (1D3); phycoerythrin (PE)-conjugated anti-TCRγδ (GL3), anti-CD19 (1D3), anti-ScaI (E13-161.7), and anti-CD45.1 (A20); and biotin-conjugated anti-Mac1 (M1/70), anti-CD25 (7D4), and anti-nerve growth factor receptor (C40-1457). The following mAbs were purchased from eBiosciences (San Diego, CA): FITC-conjugated anti-TER119 (TER-119), anti-Mac1 (M1/70), anti-CD4 (RM4-5), anti-CD8 (53-6.7), anti-CD3ε (145-2C11), anti-CD25 (7D4), and anti-CD45.2 (104); PE-conjugated anti-CD4 (RM4-5) and anti-CD44 (IM7); allophycocyanin-conjugated anti-CD3ε (145-2C11), anti-CD25 (7D4), anti-CD8α (53-6.7), anti-TCRβ (H57-597), anti-Thy1.2 (53-2.1), and anti-c-Kit (2B8); and biotin-conjugated anti-IL-7Rα (A7R34), anti-CD3ε (145-2C11), anti-NK1.1 (PK136), anti-TER119 (TER-119), anti-Gr1 (RB6-8C5), anti-B220 (RA3-6B2), anti-CD19 (MB19-1), anti-CD4 (RM4-5), anti-CD8 (53-6.7), and anti-CD44 (IM7). For annexin V staining, the annexin V staining kit (BD Biosciences) was used. All FACS analyses were performed on FACSCalibur (BD Biosciences), and data were analyzed by using CellQuest software (BD Biosciences).
Fetal Liver Transplantation into rag2−/− Mice.
E14.5 fetal liver cells from mint+/− and mint−/− embryos carrying the CD45.2 allotype (5 × 105 cells) were mixed with an equal amount of those from B6.SJL embryos carrying the CD45.1 allotype (5 × 105 cells), and then they were injected i.v. into 4-Gy irradiated rag2−/− mice as described in ref. 13. At 6–10 weeks after transplantation, single-cell suspensions were prepared from thymi and spleens of the recipient rag2−/− mice, and then they were subjected to FACS analysis.
Quantitative RT-PCR.
The quantitative RT-PCR was performed as described in ref. 52 with minor modifications. Total RNA was prepared from FACS-sorted cells by using an RNeasy Mini Kit (Qiagen, Valencia, CA), and total RNA was treated with DNase I (DNA-free; Ambion, Austin, TX). First-strand cDNA was synthesized from DNase I-treated total RNA with random hexamers by using ABI cDNA Synthesis Kit (Applied Biosystems, Foster City, CA). Synthesized cDNA was subsequently mixed with 2× iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) and various sets of gene-specific forward and reverse primers [supporting information (SI) Table 1], and it was then subjected to real-time PCR quantification using an iCycler iQ detection system (Bio-Rad). All reactions were performed in duplicate. The relative amounts of mRNAs were calculated by using the standard curve method and normalized by invariant controls Gapdh or Hprt.
In Vitro T Cell Differentiation Assay.
LSK cells were sorted from E14.5 fetal livers of mint−/− or mint+/− embryos, seeded on a monolayer of Tst-4/Dll1, a thymus-derived stromal cell line expressing Dll1, or a parental cell line Tst-4 (42, 43). Six to 14 days after in vitro culture, cells generated from LSK cells were harvested and subjected to FACS analysis. Contaminating Tst-4/Dll1 cells and Tst-4 cells were gated out by staining nerve growth factor receptor artificially introduced into these stromal cell lines.
Supplementary Material
Acknowledgments
We thank H. Kawamoto (RIKEN, Yokohama, Kanagawa, Japan) for providing Tst-4 and Tst-4/Dll1 cells; S. Fagarasan (RIKEN), T. Okazaki, M. Muramatsu, N. Minato, H. Nagaoka, N. Yamamoto (Kyoto University, Kyoto, Japan), and K. Tanigaki (Shiga Medical Center for Adult Diseases, Shiga, Japan) for helpful discussions; Y. Sasaki, H. Hijikata, M. Nakata, E. Inoue, A. Noguchi for excellent technical assistance; and Y. Shiraki for the secretarial support. This work was supported in part by the Ministry of Education, Science, Sports, and Culture, Grants-in-Aid for Specially Promoted Research, 17002015, 2005–2010 (to T.H.), Scientific Research on Priority Areas, 18019018, 2006–2007 (to D.Y.), and Young Scientists (A), 17689013, 2005–2006 (to D.Y.).
Abbreviations
- Dll1
Delta-like 1
- DN
double-negative
- DP
double-positive
- E
embryonic day
- ETP
early T cell progenitor
- Hprt
hypoxanthine phosphoribosyltransferase
- LSK
Lineage −Sca-1hic-Kithi
- Mint
Msx2-interacting nuclear target protein
- NICD
Notch intracellular domain
- NK
natural killer
- SP
single-positive.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610520104/DC1.
References
- 1.Artavanis-Tsakonas S, Rand MD, Lake RJ. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Lai EC. Development (Cambridge, UK) 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 3.Maillard I, Fang T, Pear WS. Annu Rev Immunol. 2005;23:945–974. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- 4.Radtke F, Wilson A, Mancini SJC, MacDonald HR. Nat Immunol. 2004;5:247–253. doi: 10.1038/ni1045. [DOI] [PubMed] [Google Scholar]
- 5.Schroeter EH, Kisslinger JA, Kopan R. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 6.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 7.Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Curr Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- 8.Lardelli M, Dahlstrand J, Lendahl U. Mech Dev. 1994;46:123–136. doi: 10.1016/0925-4773(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 9.Lardelli M, Lendahl U. Exp Cell Res. 1993;204:364–372. doi: 10.1006/excr.1993.1044. [DOI] [PubMed] [Google Scholar]
- 10.Uyttendaele H, Marazzi G, Wu G, Yan Q, Sassoon D, Kitajewski J. Development (Cambridge, UK) 1996;122:2251–2259. doi: 10.1242/dev.122.7.2251. [DOI] [PubMed] [Google Scholar]
- 11.Kato H, Sakai T, Tamura K, Minoguchi S, Shirayoshi Y, Hamada Y, Tsujimoto Y, Honjo T. FEBS Lett. 1996;395:221–224. doi: 10.1016/0014-5793(96)01046-0. [DOI] [PubMed] [Google Scholar]
- 12.Mizutani T, Taniguchi Y, Aoki T, Hashimoto N, Honjo T. Proc Natl Acad Sci USA. 2001;98:9026–9031. doi: 10.1073/pnas.161269998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuroda K, Han H, Tani S, Tanigaki K, Tun T, Furukawa T, Taniguchi Y, Kurooka H, Hamada Y, Toyokuni S, Honjo T. Immunity. 2003;18:301–312. doi: 10.1016/s1074-7613(03)00029-3. [DOI] [PubMed] [Google Scholar]
- 14.Oswald F, Kostezka U, Astrahantseff K, Bourteele S, Dillinger K, Zechner U, Ludwig L, Wilda M, Hameister H, Knochel W, et al. EMBO J. 2002;21:5417–5426. doi: 10.1093/emboj/cdf549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hozumi K, Negishi N, Suzuki D, Abe N, Sotomaru Y, Tamaoki N, Mailhos C, Ish-Horowicz D, Habu S, Owen MJ. Nat Immunol. 2004;5:638–644. doi: 10.1038/ni1075. [DOI] [PubMed] [Google Scholar]
- 16.Saito T, Chiba S, Ichikawa M, Kunisato A, Asai T, Shimizu K, Yamaguchi T, Yamamoto G, Seo S, Kumano K, et al. Immunity. 2003;18:675–685. doi: 10.1016/s1074-7613(03)00111-0. [DOI] [PubMed] [Google Scholar]
- 17.Tanigaki K, Han H, Yamamoto N, Tashiro K, Ikegawa M, Kuroda K, Suzuki A, Nakano T, Honjo T. Nat Immunol. 2002;3:443–450. doi: 10.1038/ni793. [DOI] [PubMed] [Google Scholar]
- 18.Witt CM, Won WJ, Hurez V, Klug CA. J Immunol. 2003;171:2783–2788. doi: 10.4049/jimmunol.171.6.2783. [DOI] [PubMed] [Google Scholar]
- 19.Shi Y, Downes M, Xie W, Kao HY, Ordentlich P, Tsai CC, Hon M, Evans RM. Genes Dev. 2001;15:1140–1151. doi: 10.1101/gad.871201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, Aguet M. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 21.Wilson A, MacDonald HR, Radtke F. J Exp Med. 2001;194:1003–1012. doi: 10.1084/jem.194.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han H, Tanigaki K, Yamamoto N, Kuroda K, Yoshimoto M, Nakahata T, Ikuta K, Honjo T. Int Immunol. 2002;14:637–645. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- 23.Pui JC, Allman D, Xu L, DeRocco S, Karnell FG, Bakkour S, Lee JY, Kadesch T, Hardy RR, Aster JC, Pear WS. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 24.Wu L. Curr Opin Immunol. 2006;18:121–126. doi: 10.1016/j.coi.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 25.Porritt HE, Rumfelt LL, Tabrizifard S, Schmitt TM, Zuniga-Pflucker JC, Petrie HT. Immunity. 2004;20:735–745. doi: 10.1016/j.immuni.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Allman D, Sambandam A, Kim S, Miller JP, Pagan A, Well D, Meraz A, Bhandoola A. Nat Immunol. 2003;4:168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 27.Benz C, Bleul CC. J Exp Med. 2005;202:21–31. doi: 10.1084/jem.20050146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambandam A, Maillard I, Zediak VP, Xu L, Gerstein RM, Aster JC, Pear WS, Bhandoola A. Nat Immunol. 2005;6:663–670. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- 29.Schwarz BA, Bhandoola A. Nat Immunol. 2004;5:953–960. doi: 10.1038/ni1101. [DOI] [PubMed] [Google Scholar]
- 30.Schmitt TM, Zuniga-Pflucker JC. Crit Rev Immunol. 2005;25:141–160. doi: 10.1615/critrevimmunol.v25.i2.40. [DOI] [PubMed] [Google Scholar]
- 31.Tan JB, Visan I, Yuan JS, Guidos CJ. Nat Immunol. 2005;6:671–679. doi: 10.1038/ni1217. [DOI] [PubMed] [Google Scholar]
- 32.Rothenberg EV, Taghon T. Annu Rev Immunol. 2005;23:601–649. doi: 10.1146/annurev.immunol.23.021704.115737. [DOI] [PubMed] [Google Scholar]
- 33.Wolfer A, Wilson A, Nemir M, MacDonald HR, Radtke F. Immunity. 2002;16:869–879. doi: 10.1016/s1074-7613(02)00330-8. [DOI] [PubMed] [Google Scholar]
- 34.Tanigaki K, Tsuji M, Yamamoto N, Han H, Tsukada J, Inoue H, Kubo M, Honjo T. Immunity. 2004;20:611–622. doi: 10.1016/s1074-7613(04)00109-8. [DOI] [PubMed] [Google Scholar]
- 35.Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, Flavell RA. Cell. 2004;117:515–526. doi: 10.1016/s0092-8674(04)00451-9. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka S, Tsukada J, Suzuki W, Hayashi K, Tanigaki K, Tsuji M, Inoue H, Honjo T, Kubo M. Immunity. 2006;24:689–701. doi: 10.1016/j.immuni.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 37.Felli MP, Maroder M, Mitsiadis TA, Campese AF, Bellavia D, Vacca A, Mann RS, Frati L, Lendahl U, Gulino A, Screpanti I. Int Immunol. 1999;11:1017–1025. doi: 10.1093/intimm/11.7.1017. [DOI] [PubMed] [Google Scholar]
- 38.Yokota T, Huang J, Tavian M, Nagai Y, Hirose J, Zuniga-Pflucker JC, Peault B, Kincade PW. Development (Cambridge, UK) 2006;133:2041–2051. doi: 10.1242/dev.02349. [DOI] [PubMed] [Google Scholar]
- 39.Masuda K, Itoi M, Amagai T, Minato N, Katsura Y, Kawamoto H. J Immunol. 2005;174:2525–2532. doi: 10.4049/jimmunol.174.5.2525. [DOI] [PubMed] [Google Scholar]
- 40.Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, et al. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 41.Schmitt TM, Zuniga-Pflucker JC. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 42.Miyazaki M, Kawamoto H, Kato Y, Itoi M, Miyazaki K, Masuda K, Tashiro S, Ishihara H, Igarashi K, Amagai T, et al. J Immunol. 2005;174:2507–2516. doi: 10.4049/jimmunol.174.5.2507. [DOI] [PubMed] [Google Scholar]
- 43.Masuda K, Kubagawa H, Ikawa T, Chen CC, Kakugawa K, Hattori M, Kageyama R, Cooper MD, Minato N, Katsura Y, Kawamoto H. EMBO J. 2005;24:4052–4060. doi: 10.1038/sj.emboj.7600878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yun TJ, Bevan MJ. J Immunol. 2003;170:5834–5841. doi: 10.4049/jimmunol.170.12.5834. [DOI] [PubMed] [Google Scholar]
- 45.Pirot P, van Grunsven LA, Marine JC, Huylebroeck D, Bellefroid EJ. Biochem Biophys Res Commun. 2004;322:526–534. doi: 10.1016/j.bbrc.2004.07.157. [DOI] [PubMed] [Google Scholar]
- 46.Bain G, Engel I, Robanus Maandag EC, te Riele HP, Voland JR, Sharp LL, Chun J, Huey B, Pinkel D, Murre C. Mol Cell Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heemskerk MH, Blom B, Nolan G, Stegmann AP, Bakker AQ, Weijer K, Res PC, Spits H. J Exp Med. 1997;186:1597–1602. doi: 10.1084/jem.186.9.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nie L, Xu M, Vladimirova A, Sun XH. EMBO J. 2003;22:5780–5792. doi: 10.1093/emboj/cdg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitamoto T, Takahashi K, Takimoto H, Tomizuka K, Hayasaka M, Tabira T, Hanaoka K. Biochem Biophys Res Commun. 2005;331:1154–1162. doi: 10.1016/j.bbrc.2005.03.241. [DOI] [PubMed] [Google Scholar]
- 50.Wilson A, Ferrero I, MacDonald HR, Radtke F. J Immunol. 2000;165:5397–5400. doi: 10.4049/jimmunol.165.10.5397. [DOI] [PubMed] [Google Scholar]
- 51.Sakamoto M, Hirata H, Ohtsuka T, Bessho Y, Kageyama R. J Biol Chem. 2003;278:44808–44815. doi: 10.1074/jbc.M300448200. [DOI] [PubMed] [Google Scholar]
- 52.Yabe D, Komuro R, Liang G, Goldstein JL, Brown MS. Proc Natl Acad Sci USA. 2003;100:3155–3160. doi: 10.1073/pnas.0130116100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.