Abstract
Prox1, an early specific marker for developing liver and pancreas in foregut endoderm has recently been shown to interact with α-fetoprotein transcription factor (FTF) and repress cholesterol 7α-hydroxylase (CYP7A1) gene transcription. Using yeast two-hybrid assay, we found that Prox1 strongly and specifically interacted with HNF4α, an important transactivator of the human CYP7A1 gene in bile acid synthesis and phosphoenolpyruvate carboxykinase (PEPCK) gene in gluconeogenesis. Real time PCR assay detected Prox1 mRNA expression in human primary hepatocytes and HepG2 cells. Reporter assay, GST pull-down, co-immunoprecipitation and yeast two-hybrid assays identified a specific interaction between the N-terminal LXXLL motif of Prox1 and the activation function 2 domain of HNF4α. Prox1 strongly inhibited HNF4α and peroxisome proliferators-activated receptor γ coactivator-1α (PGC-1α) co-activation of the CYP7A1 and PEPCK genes. Knock-down of the endogenous Prox1 by small interfering RNA (siRNA) resulted in significant increase of CYP7A1 and PEPCK mRNA expression and the rate of bile acid synthesis in HepG2 cells. These results suggest that Prox1 is a novel co-regulator of HNF4α that may play a key role in the regulation of bile acid synthesis and gluconeogenesis in the liver.
CYP7A1 catalyzes the first and rate-limiting step in the conversion of cholesterol to bile acids and plays an important role in maintaining whole body lipid homeostasis (1). Bile acids are physiological detergents that facilitate absorption, transport and distribution of sterols and lipid soluble vitamins, and disposal of toxic metabolites and xenobiotics. Bile acid synthesis and CYP7A1 gene transcription is feedback inhibited by bile acids returning to the liver via enterohepatic circulation of bile (1). Recent studies have identified farnesoid X receptor (FXR, NR1H4) as a bile acid-activated receptor that induces an atypical nuclear receptor small heterodimer partner (SHP, NR0B2), which interacts with FTF (NR5A2) and HNF4α (NR2A1) bound to an overlapping sequence located in the bile acid response element II (-144/-126) and represses CYP7A1 gene transcription (2). However, the molecular mechanism by which FTF and HNF4α regulate the CYP7A1 gene is not completely understood.
HNF4α is the most abundant nuclear receptor expressed in the liver and is involved in early liver development (3). Conditional knockout of the HNF4α gene in mouse liver caused accumulation of lipids in the liver, markedly reduced serum cholesterol and triglycerides and increased serum bile acids (4). CYP7A1, Na+taurocholate co-transport peptide, organic anion transporter 1, apolipoprotein B100, and scavenger receptor B-1 expression are reduced in these mice (4). It appears that HNF4α is a key regulator of bile acid and lipoprotein metabolism, and plays a central role in lipid homeostasis (5). HNF4α is involved in diabetes; mutation of the HNF4α gene causes maturity onset diabetes of the young type 1 (MODY1) (6). HNF4α regulates the HNF1α gene, a MODY 3 gene (7).
The transcriptional activities of nuclear receptors are largely dependent on ligand-binding and activation. Nuclear receptors interact with co-regulators and regulate their target genes in a tissue and gene-specific manner (8). Upon ligand binding, the helix 12 of nuclear receptor is exposed and binds to the co-activators and activates nuclear receptor activity. Recently, PGC-1α has been identified as a co-activator of HNF4α (9). PGC-1α is highly induced during starvation by glucocorticoids and glucagon to induce PEPCK, a rate-limiting enzyme in gluconeogenesis (10). It has been reported that PGC-1α co-activates HNF4α and induces CYP7A1 gene transcription during starvation in mice (11). It has been suggested that bile acid synthesis and gluconeogenesis may be coordinately regulated in fasted -to-fed cycle (12). Our recent study shows that glucagon and cAMP inhibit CYP7A1 by inducing phosphorylation of HNF4α (13).
Prox1 has recently been identified as a co-repressor of FTF/LRH-1 by yeast two-hybrid screening (14,15). Prox1 was originally cloned by homology to the Drosophila melanogaster gene prospero (16). Prox1 is expressed in lens, heart, liver, kidney, skeletal muscle, pancreas, and central nervous system (16,17). Earlier studies have linked Prox1 function to lens and lymphatic system development (18,19). More recent studies indicate that Prox1 is required for hepatocyte migration in developing liver and pancreas in the mammalian foregut endoderm (20,21). Prox1 interacts with the NR5 subfamily of nuclear receptors including Ff1b (NR5A4), a zebra fish homologue of nuclear receptor, steroidogenic factor 1 (SF-1, NR5A1) (22), and FTF (14,15), and represses their transactivation activity. We hypothesized that Prox1 may interact with HNF4α and suppressed CYP7A1 gene transcription. To test this hypothesis, we used yeast two-hybrid assay to study the interaction between Prox1 and HNF4α and studied the effect of Prox1 on the HNF4α transactivation of the human CYP7A1 gene. Our findings provide a novel molecular mechanism for Prox1 inhibition of bile acid synthesis and gluconeogenesis.
Experimental procedures
Cell culture - Primary human hepatocytes were isolated from human donors (HH1201, 69 yr male; HH1205, 45 yr male; HH1209 50 yr female; HH1234, 56 yr male; HH1247, 3 yr male; HH1248, 42 yr female) and were obtained from the Liver Tissue Procurement and Distribution System of National Institute of Health (S. Strom, University of Pittsburgh, PA). The HepG2 cells were obtained from the American Type Culture Collection (Manassas, VA). Cells were maintained as described previously (13).
Plasmids - The mammalian expression plasmids for HNF4α, Nur77, PGC-1α, Flag-Prox1, Gal4-HNF4α, pHNF4α-tk-Luc reporter, NurRE-Luc, 5X UAS-Luc and human CYP7A1 promoter luciferase reporters were as described previously (14,23,24). Human PEPCK promoter luciferase reporter was generously provided by Dr. Richard Hanson (Case Western Reserve University, Cleveland, OH). The various deletion constructs of Prox1 [Prox1-NT-WT, amino acid (a. a.) 1-312; Prox1-NT-MT, a.a. 1-312, alanine mutations were introduced to convert LRKLL (a.a. 70-74) to ARKAL; Prox1-Homeo (a.a. 313-736)] and HNF4α (HNF4α-NT, a.a. 1-128; HNF4α-LBD, a.a. 129-455; HNF4α-△AF2, a.a. 1-352) were made by PCR with suitable restriction endonucleases and inserted into the pcDNA3-HA or the yeast LexA or B42 expression vector (CLONTECH Laboratories, Inc., Palo Alto, CA). For bacterial expression, GST-fused Prox1 was constructed by inserting EcoRI-XhoI fragments of Prox1 from B42-Prox1 into pGEX4T-1 vector (Amersham Bioscience, Inc). All the clones were confirmed by sequencing analysis.
GST-pull down assay - [35S] Methionine-labeled proteins were prepared by in vitro translation using the TNT-coupled transcriptional translation system with conditions as described by the manufacturer (Promega, Madison, WI). GST fused Prox1 (GSTProx1) was expressed in the E. coli BL21 (DE3) strain and purified using glutathione-Sepharose 4B beads (Pharmacia, Piscataway, NJ). In vitro protein-protein interaction assays were carried out as described (23).
Coimmunoprecipitation assay - The cell extracts of human primary hepatocytes were pre-cleared with whole rabbit serum adsorbed on Protein A-Sepharose beads (Amersham Biosciences, Piscataway, NJ) and subsequently subjected to immunoprecipitation with 10 μg of anti-HNF4α or non-immune serum (IgG) for overnight at 4 °C. The beads were washed several times with ice-cold modified radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.4, 1% Nonidet P-40, 0.25% Na+-deoxycholate, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 1 mM ethylendiamine tetraacetic acid, 1 mM sodium orthovanadate, 1 mM NaF), and the immune-complexes were subjected to electrophoresis on a 10% SDS-polyacrylamide gel and then transferred to a nitrocellulose membrane (Amersham Biosciences). Enhanced chemiluminescence Western blotting (Amersham Biosciences) was performed according to the manufacturer’s instructions. Prox1 and HNF4α proteins were detected by incubation of blots with an anti-Prox1 antibody (1:2000 dilution; Upstate, Lake Placid, NY) and anti-HNF4α antibody (1:2,000 dilution; Santa Cruz Biotechnology, Santa Cruz, CA), respectively.
Yeast two-hybrid assay - For the yeast two-hybrid system, LexA and B42 fusion plasmids for nuclear receptors or co-activators were co-transformed into Saccharomyces cerevisiae EGY48 strain containing the LacZ reporter plasmid (p80p-lac Z) under the control of the LexA binding site. β-galactosidase activity expressed on the plates was assayed as described previously (23). For assay of thyroid hormone receptor (TR), glucocorticoid receptor (GR), retinoid X receptor (RXR) and retinoic acid receptor (RAR), 100 μl of 1 μM stock solution of appropriate ligands (T3, dexamethasone, all-trans retinoic acid and 9-cis retinoid) was added before plating to test the effect of ligand activation on interaction. Assays were repeated at least three times.
Transient transfection and Luciferase reporter assay - For luciferase reporter assay, HepG2 cells were plated in 24-well plates 24 h before transfection with reporter or expression plasmids using LipofectAMINE 2000 reagent (Life Technologies, Inc., Gaithersburg, MD) according to the manufacturer’s instructions. Total DNA used in each transfection was adjusted by adding the appropriate amount of pcDNA3 vector. Luciferase activities are expressed as relative luciferase unit (RLU)/β-galactosidase activity as described previously (23).
Small Interfering RNA (siRNA) Experiments The SMART pool siRNAs for human Prox1 were purchased from Dharmacon Research (Lafayette, Co) and transfected into HepG2 cells using LipofectAMINE 2000 reagent (Life Technologies, Inc., Gaithersburg, MD) according to the manufacturer’s instructions. Forty-eight hours after transfection, cells were extracted and analyzed. The SMART pool is a mixture of four sequences located at different regions of mRNA. Two of them were tested to be effective in knockdown Prox1 in HepG2 cells. The siRNA sequences are: siRNA#1, nt 1009-1027; GGGCCAAACTCCTTACAAC, siRNA#2 nt 2096-2114; GCAAAGATGTTGATCCTTC. The control siRNA probe is a scrambled siRNA that was designed to have the same G-C content as the siRNA#2 but did not display sequence identity with Prox1: GATCGTGTGTAGTTCATAACC.
RNA isolation and Real-time Quantitative PCR (Q-PCR) - HepG2 cells was transfected with synthesized siRNA of human Prox1 and total RNA was isolated using Tri-reagent (Sigma, St. Louis, MO) according to the manufacturer’s instruction. Reverse-transcription and Q-PCR were performed to detect Prox1, CYP7A1, PEPCK, HNF4α, and cyclophilin B mRNAs as described previously (13).
Chromatin immunoprecipitation assay (ChIP) ChIP assays were performed using a ChIP Assay kit (Upstate Cell Signaling Solutions, Lake Placid, NY) according to the manufacturer’s instructions. HepG2 were transfected with pcDNA3 empty vector or Flag-Prox1 and chromatin was cross-linked in 1% formaldehyde and sonicated as reported previously (24). Cell lysate solution (5%) in ChIP dilution buffer was kept aside as “input”. Ten μg HNF4α antibody (Santa Cruz Biotechnology) or anti-Flag antibody (Sigma) was added to precipitate DNA-protein complexes and non-immune IgG was used as a control. A 391-bp DNA fragment (-432 to -41) containing the BARE-I and BARE-II of the CYP7A1 promoter was PCR amplified for 30 cycles using 5 μl of the DNA as template and analyzed on a 1.5% agarose gel. PCR primers for amplification were as follows: 5′-ATCACCGTCTCTCTGGCAAAGCAC-3′; reverse primer: 5′-CCATTAACTTGAGCTTGGTTGACAAAG-3′.
Bile Acid analysis - The siRNA for human Prox1 and control siRNA were transfected into HepG2 cells using LipofectAMINE 2000 reagent (Life Technologies, Inc., Gaithersburg, MD) according to the manufacturer’s instructions. Forty-eight hours after transfection, cells were washed and then incubated with serum-free media for a period of time from 3 to 24 h. The medium was collected at indicated time points and frozen at -80 °C for later analysis of bile acids. At the end of the incubation, the cells were harvested and stored at -80 °C until use. A Sep-Pak C18 reversed phase cartridge (Waters Associates, Inc., Milford, MA) was used for bile acid extraction from media as described previously (25). Total bile acid concentration was analyzed by enzymatic 3α-hydroxysteroid dehydrogenase method using total bile acid assay kit (Bio-quant Inc., San Diego, CA) according to the manufacturer’s instruction.
Results
Prox1 interacts with HNF4α in vivo and in vitro
The nuclear receptors that have been identified to interact with Prox1 all belong to the NR5A subfamily (14,15,22). To identify other potential interaction partners of Prox1, we performed yeast two-hybrid protein interaction assays using Lex A and B42 constructs available in our laboratory. Table 1 shows that Prox1 interacted with HNF4α. This interaction is stronger than Prox1 interaction with SF-1. On the other hand, Prox1 did not interact with TRα, TRβ, RXR, RAR, GR, SHP, dosage-sensitive sex reversal, AHC critical region on the X chromosome, gene 1 (DAX-1), and Nur77 regardless of the presence or absence of their respective ligands (Table 1). Transcriptional repressors N-CoR and SMRT did not interact with Prox1. These results showed that Prox1 interacts with HNF4α and NR5A family nuclear receptors.
TABLE 1.
Interaction of Prox1 with nuclear receptors in yeast two-hybrid interaction assay
| LexA | B42 | Interaction1 |
|---|---|---|
| - | - | - |
| Prox1 | - | - |
| - | Prox1 | - |
| TRα | Prox1 | - |
| TRβ | Prox1 | - |
| GR | Prox1 | - |
| RAR | Prox1 | - |
| RXR | Prox1 | - |
| SHP | Prox1 | - |
| DAX-1 | Prox1 | - |
| N-CoR | Prox1 | - |
| SMRT | Prox1 | - |
| Prox1 | Nur77 | - |
| Prox1 | SF-1 | +++ |
| Prox1 | HNF4α | ++++ |
| SHP | HNF4α | +++ |
“-” Indicates no interaction; number of “+” indicates strength of interaction.
To further confirm the results of yeast two-hybrid assay, we performed in vitro GST pull-down assays to study Prox1 and HNF4α interaction. Consistent with yeast two-hybrid assay results, GST-Prox1 interacted with HNF4α, but not RXR and RAR (Fig. 1A). The physical interaction between GST-Prox1 and 35S-labeled FTF was used as a positive control (Fig. 1A). To further verify the interaction between Prox1 and HNF4α, we performed a co-immunoprecipitation assay using human primary hepatocyte extracts. Cell extracts were immunoprecipitated with an anti-HNF4α antibody. The immunoprecipitated complexes were then analyzed on an immunoblot with anti-Prox1 antibody. As shown in Fig. 1B, anti-Prox1 antibody detected Prox1 in the immunoprecipitates, while non-immune IgG did not. These results indicated that Prox1 interacted with HNF4α in primary human hepatocytes, consistent with the results from yeast two-hybrid assay and GST pull-down assay.
Fig. 1.
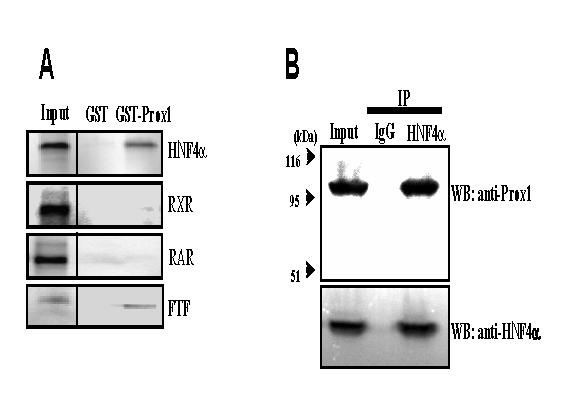
GST pull-down and co-immunoprecipitation assays of Prox1 interaction with HNF4α. A. GST pull-down assay. Purified GST alone (negative control) or GST-Prox1 bound to glutathione-Sepharose beads were incubated with 35S-labeled HNF4α, RXR, RAR, and FTF (positive control). The reactions were analyzed by SDS-polyacrylamide gel electrophoresis and bound proteins were visualized by autoradiography. The input represents 10% of the labeled proteins used for the pull-down assay. B. Co-immunoprecipitation assay. Protein extracts were prepared from human primary hepatocytes and immunoprecipitated with anti-HNF4α antibody or non-immune serum (IgG, as control). Immunoprecipitated proteins were resolved on SDS-polyacrylamide gel and analyzed by immunoblotting with anti-Prox1 and anti-HNF4α antibodies. Data represent one of three separate experiments.
Mapping of the interaction regions of Prox1 and HNF4α
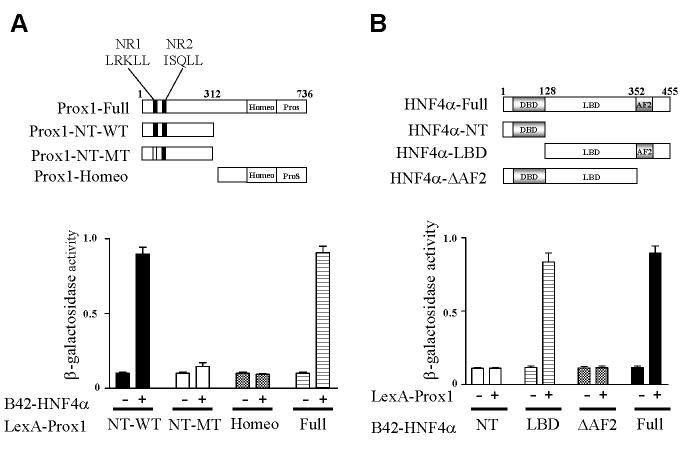
Co-regulators have conserved LXXLL motifs that are known to interact with the ligand binding domain (LBD) of nuclear receptors. Prox1 has two LXXLL motifs located in the N-terminus and another motif located in the C-terminus region. It has been reported that the N-terminal nuclear receptor box 1 (NR1, LRKLL) is critical for interaction with FTF/LRH-1, whereas NR2 (ISQLL) and NR3 are not essential for interaction (14,15,22). Yeast two-hybrid assay revealed that the full-length and N-terminal amino acid residues from 1 to 312 (Prox1-NT-WT) interacted with HNF4α (Fig. 2A). However, the C-terminal homeo and prospero domains (Prox1-Homeo) did not interact with HNF4α. When the LRKLL sequence (NR1) was mutated to ARKAL (Prox-NT-MT), this mutant did not interact with HNF4α. These results demonstrated that the N-terminal region of Prox1 interacted with HNF4α and the LRKLL motif is critical for Prox1 to interact with HNF4α.
Fig. 2.
Mapping of interaction domains between Prox1 and HNF4α. A. Prox1 N-terminal LRKLL motif (NR1) of Prox1 is required for HNF4α interaction. Various LexA-Prox1 constructs, as indicated in the upper panel, were co-transformed with B42 (-) or B42-HNF4α (+) into EGY48 yeast cells. B. The ligand binding domain of HNF4α interacts with Prox1. LexA (-) or LexA-Prox1 (+) and various B42-HNF4α constructs, as indicated upper panel were co-transformed into EGY48 yeast cells. Transformants were selected on plates with appropriate selection marker, and the β-galactosidase activity was measured. The results shown are the mean of β-galactosidase value from five independent transformant colonies. DBD, DNA-binding domain; LBD, ligand-binding domain; AF2, activation function 2.
We then investigated which region of HNF4α was required for interaction with Prox1. A series of deletion constructs of HNF4α were used to map the HNF4α interaction domain in yeast-two hybrid assay. Fig. 2B shows that the full-length HNF4α (HNF4α-full) and LBD which contains activation function 2 (AF2) domain (HNF4α-LBD) interacted with Prox1. However, the N-terminal DBD region and a construct without AF2 (HNF4α-△AF2) did not interact with Prox1. These results indicate that HNF4α interacts with Prox1 through the AF2 domain of HNF4α.
Prox1 is expressed in human hepatocytes
Although it has been reported that Prox1 is highly expressed in liver and pancreas (14), the expression of Prox1 in human hepatocytes has not been reported before. Using real time quantitative PCR, we were able to detect the mRNA expressions of CYP7A1, Prox1, PGC-1α and several nuclear receptors that are known to regulate CYP7A1 in five donor human primary hepatocytes and HepG2 cells. Table 2 shows Ct, the threshold cycle number, for each mRNA transcripts assayed in these hepatocytes. The mRNA expression levels were normalized to internal reference gene UBC and the △Ct values and standard deviations are shown in the Table 2. The expression patterns of these mRNA transcripts are similar in five donor hepatocytes and HepG2 cells. The Ct and △Ct values of CYP7A1 and PGC-1α are high reflecting low levels of mRNA expression. Those values for Prox1, HNF4α and SHP are low indicating relatively abundant expression of these three mRNA transcripts.
TABLE 2.
Quantitative real time PCR analysis of mRNA expression levels of CYP7A1, nuclear receptors and co-regulators in human primary hepatocytes and HepG2 cells.
| HH1201 | HH1205 | HH1209 | HH1247 | HH1248 | HepG2 | ||
|---|---|---|---|---|---|---|---|
| UBC | Ct±SDCt | 20.6±0.3 | 19.8±0.3 | 19.9±0.05 | 22.7±0.03 | 21.6±0.08 | 20.9±0.07 |
| CYP7A1 | Ct±SDC | 30.4±0.09 | 25.0±0.19 | 29.2±0.05 | 35.9±0.49 | 35.5±0.31 | 31.2±0.03 |
| ΔCt±SDΔC | 9.7±0.31 | 4.9±0.36 | 9.3±0.07 | 13.1±0.49 | 13.4±0.32 | 10.2±0.076 | |
| t | |||||||
| PGC-1α | Ct±SDCt | 27.7±0.09 | 24.4±0.01 | 27.4±0.02 | 28.2±0.02 | 28.3±0.03 | 27.8±0.09 |
| ΔCt±SDΔC | 7.0±0.31 | 4.3±0.3 | 7.5±0.05 | 5.5±0.04 | 6.7±0.09 | 6.8±0.11 | |
| t | |||||||
| FXR: | Ct±SDCt | 26.1±0.4 | 22.8±0.4 | 23.3±0.07 | 26.1±0.05 | 24.8±0.04 | 27.8±0.3 |
| ΔCt±SDΔC | 5.5±0.5 | 2.7±0.5 | 3.4±0.07 | 3.4±0.06 | 3.2±0.09 | 6.7±0.3 | |
| t | |||||||
| FTF | Ct±SDCt | 24.4±0.03 | 24.2±0.5 | 24.2±0.03 | 27.2±0.03 | 06.3±0.08 | 25.8±0.08 |
| ΔCt±SDΔC | 3.7±0.3 | 4.1±0.58 | 4.3±0.06 | 4.5±0.04 | 4.7±0.11 | 4.9±0.11 | |
| t | |||||||
| PROX1 | Ct±SDCt | 24.8±0.23 | 23.9±0.2 | 24.2±0.08 | 26.1±0.08 | 25.3±0.08 | 24.1±0.03 |
| ΔCt±SDΔC | 4.2±0.38 | 3.8±0.36 | 4.3±0.09 | 3.4±0.09 | 3.7±0.11 | 3.1±0.08 | |
| t | |||||||
| HNF4α | Ct±SDCt | 24.6±0.01 | 21.4±0.08 | 21.9±0.01 | 25.2±0.02 | 23.4±0.04 | 23.8±0.05 |
| ΔCt±SDΔC | 3.9±0.3 | 1.3±0.31 | 2.0±0.05 | 2.5±0.04 | 1.8±0.09 | 2.8±0.08 | |
| t | |||||||
| SHP | Ct±SDCt | 24.4±0.03 | 21.7±0.4 | 23.4±0.1 | 25.7±0.07 | 23.9±0.14 | 21.8±0.07 |
| ΔCt±SDΔC | 3.7±0.3 | 1.6±0.5 | 3.5±0.11 | 3.0±0.08 | 2.3±0.16 | 0.9±0.10 | |
| t |
Five donor liver hepatocytes (HH1201, HH1205, HH1209, HH1247, HH1248) and HepG2 cells were analyzed for mRNA expression levels as described under material and methods. ΔCt was calculated by subtracting Ct of ubiquintin C (UBC) from Ct of target genes. ΔCt: Ct of target - Ct of UBC; SDΔCt : Square Root [(SD of Ct of UBC)2 +(SD of target)2]
Prox1 is a transcriptional repressor of HNF4α
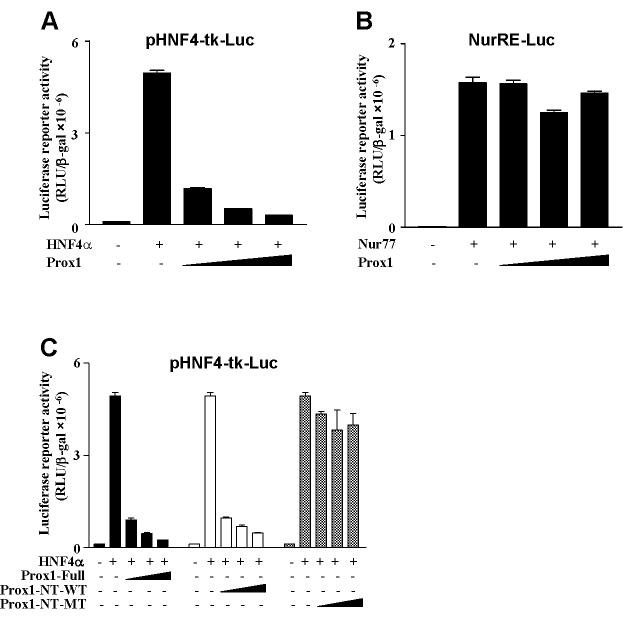
We then studied the transcriptional activity of Prox1 in reporter assays in HepG2 cells. As shown in Fig. 3A, ectopic expression of HNF4α increased a heterologous HNF4α-tk-luciferase reporter activity. Addition of Prox1 substantially repressed HNF4α transactivation activity in a dose-dependent manner. As a negative control, a reporter construct containing 3 copies of Nur77 response element (NurRE-Luc) was not affected by Prox1 (Fig. 3B). Since the N-terminal domain that contains an NR1 motif is important for Prox1 to interact with HNF4α, we performed transfection assays to test the effect of wild type and mutant Prox1 constructs on HNF4α reporter activity. Fig. 3C shows that wild-type Prox1-Full and Prox1-NT-WT represses HNF4α-mediated transactivation but Prox-1-NT-MT failed to repress the activity suggesting that the N-terminal region of the LXXLL motif of Prox1 is involved in the interaction and repression of HNF4α transactivation activity.
Fig. 3.
Prox1 abrogates HNF4α-mediated transactivation. A. HepG2 cells were co-transfected with the pHNF4α-tk-Luc reporter (200 ng) along with the HNF4α expression plasmid (200 ng) and increasing amount of Prox1 expression plasmid (10, 50 and 100 ng). B. HepG2 cells were cotransfected with the Nur77 response element-Luc reporter (NurRE-Luc reporter, 200 ng) along with the Nur77 plasmid (100 ng) and increasing amount of Prox1 plasmid (10, 50 and 100 ng). C. HepG2 cells were co-transfected with the pHNF4α-tk-Luc reporter (200 ng) along with the HNF4α plasmid (200 ng) and increasing amount of Prox1-Full, Prox1-NT-WT (aa 1-312), Prox1-NT-MT (aa 1-312, mutations that convert LRKLL (aa70-74) to ARKAL) expression plasmids (10, 50 and 100 ng). Luciferase activity was normalized to β-galactosidase activity. All experiments were done in duplicates and data represent the mean ± SD of three individual experiments.
Prox1 suppresses HNF4α transactivation of the human CYP7A1 gene
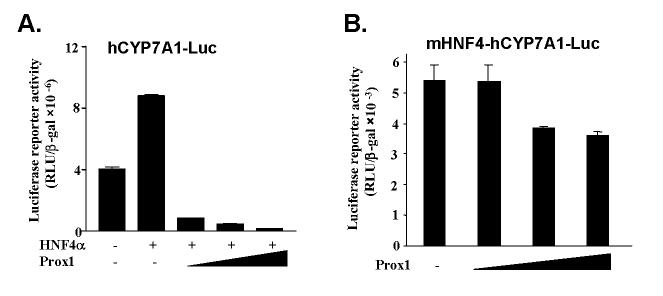
Recent studies have provided substantial evidences that HNF4α is an important transcription factor that regulates liver-specific expression of CYP7A1 (4). HNF4α is the only nuclear receptor that activates the human CYP7A1 gene in transfection assays in HepG2 cells (24,26). We thus investigated the effect of Prox1 on CYP7A1 transcription. As shown in Fig. 4A, ectopically expressed HNF4α modestly induced CYP7A1 reporter activity in HepG2 cells. This may be due to inhibition of HNF4α transactivation of CYP7A1 activity by endogenous Prox1. Co-expression of increasing amounts of Prox1 expression vectors caused marked repression of human CYP7A1 promoter reporter activity to a level below the basal activity. This suggests that exogenous Prox1 could suppress CYP7A1 basal reporter activity induced by endogenous HNF4α.
Fig. 4.
Prox1 represses CYP7A1 promoter activity through the HNF4α response element. A. HepG2 cells were co-transfected with the human CYP7A1 promoter reporter (hCYP7A1-Luc, 200 ng) along with the HNF4α plasmid (200 ng) and increasing amounts of Prox1 plasmid (10, 50 and 100 ng). B. HepG2 cells were transfected with a human CYP7A1 promoter reporter with HNF4α-binding site mutation (mHNF4-hCYP7A1-Luc, 200 ng) and increasing amounts of Prox1 plasmid (10, 50 and 100 ng). Luciferase activity was normalized to β-galactosidase activity. All experiments were done in duplicates and data represent the mean ± SD of three individual experiments.
To further confirm the involvement of HNF4α in transactivation of CYP7A1 and its inhibition by Prox1, we studied the effect of Prox1 on a reporter with a mutation in the HNF4α binding site (mHNF4-hCYP7A1-Luc). This reporter has a basal activity that is about 1,000-fold lower than the wild-type reporter. Prox1 only slightly inhibited the HNF4α site mutant reporter (Fig. 4B). Taken together, these results suggest that Prox1 interacts with HNF4α and represses HNF4α-mediated human CYP7A1 gene expression.
Prox1 blocks HNF4α recruitment of PGC-1α to CYP7A1 chromatin
To investigate the molecular mechanism of Prox1 inhibition of HNF4α transactivation of CYP7A1, we first performed an in vivo ChIP assay to study the effect of Prox1 on the HNF4α binding to CYP7A1 chromatin. HepG2 cells were transfected with pcDNA3 or Flag-Prox1 plasmid and DNA/protein complex were immunoprecipitated with an anti-Flag or anti-HNF4α antibody as indicated for PCR amplification of the CYP7A1 promoter sequence from -432 to -41, which contains an HNF4α binding site. Fig. 5A shows that over-expressing Prox1 in HepG2 cells does not affect HNF4α binding to CYP7A1 chromatin (lane 2 vs Lane 3). CYP7A1 chromatin was also detected by PCR when anti-Flag antibody was used to immunoprecipitaed Flag-Prox1 (lane4). Since Prox1 does not bind to DNA, Prox1 must be recruited to CYP7A1 chromatin by HNF4α. This data is consistent with the results in Fig. 1. No promoter sequence was amplified in control experiments using none-immune rabbit IgG (lane 1). The negative control sequence from +860 to +1160 of hCYP7A1 was not amplified by immunoprecipitation with anti-HNF4α antibody (data not shown). These results suggested that Prox1 was associated with hCYP7A1 promoter via interaction with HNF4α and that Prox1 did not affect HNF4α binding to the CYP7A1 chromatin.
Fig. 5.
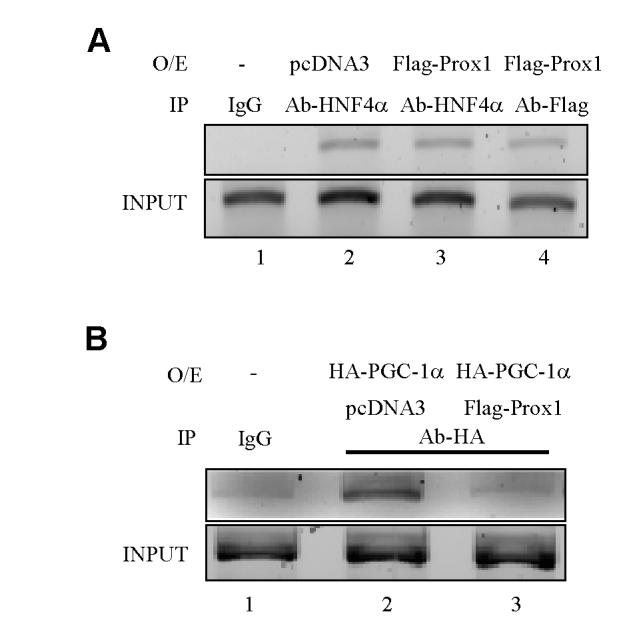
Chromatin immunoprecipitation assays of Prox1, HNF4α and PGC-1α binding to the human CYP7A1 gene. A. HepG2 cells were transfected with pcDNA3 empty vector (negative control) or Flag-Prox1 expression plasmid and subjected to formaldehyde cross-linking. Chromatin fragments were prepared by sonication and immunoprecipitated (IP) with anti-HNF4α or anti-Flag antibody, and promoter sequence containing HNF4α binding element was analyzed by PCR using primer sets specific for the CYP7A1 promoter. Cell lysate solution (5%) in ChIP dilution buffer was kept aside as “Input”. B. HepG2 cells were transfected with HA-PGC-1α alone or HA-PGC-1α together with Flag-Prox1 expression plasmid. Chromatin was immunoprecipitated with anti-HA antibody and DNA sequence containing HNF4α binding element was analyzed by PCR. Data represent one of three separate experiments.
Previously we have reported that HNF4α recruits PGC-1α to CYP7A1 chromatin (24). We thus studied the effect of Prox1 on HNF4α recruitment of PGC-1α to CYP7A1 chromatin. HepG2 cells were co-transfected with HA-PGC-1α and Flag-Prox1 or pcDNA3 empty vector (negative control) for ChIP assay using an antibody against HA. Fig. 5B shows that HAPGC-1α was recruited to CYP7A1 chromatin (land 2) via interaction with HNF4α because PGC-1α does not bind to DNA. Co-expression of Flag-Prox1 together with HA-PGC-1α markedly reduced the DNA fragment immunoprecipitated with anti-HA antibody (lane 3) indicating that Flag-Prox1 blocked HNF4α recruitment of HAPGC-1α to CYP7A1 chromatin. These data suggested that Prox-1 and PGC-1α might compete for binding to HNF4α in CYP7A1 chromatin.
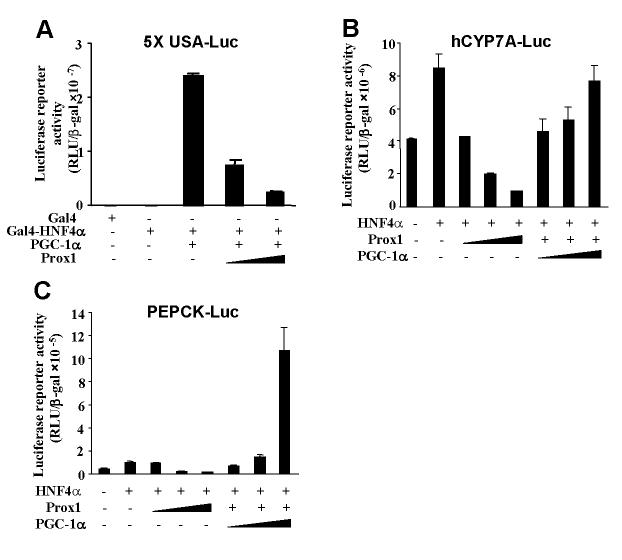
Prox1 and PGC-1α compete for interaction with HNF4α and regulate CYP7A1 and PEPCK genes
We then performed transfection assay to study the effect of Prox1 on HNF4α and PGC-1α co-activation of a heterologous reporter, 5xUASLuc and native human CYP7A1-luc and PEPCK-Luc reporters. As shown in Fig. 6A, Gal4-HNF4α alone slightly stimulates Gal4 reporter activity. Addition of PGC-1α drastically stimulated Gal4 reporter activity. Addition of Prox1 dose-dependently inhibited Gal4 reporter activity. These results suggest that Prox1 strongly inhibited HNF4α and PGC-1α transactivation activity. Fig. 6B shows that the Prox1 inhibition of HNF4α-mediated hCYP7A1-Luc reporter activity could be reversed by expression of PGC-1α in a dose-dependent manner. The PEPCK gene is known to be induced by HNF4α and PGC-1α (9). We thus performed similar experiments to study effect of Prox1 on PEPCK-Luc reporter activity. Fig. 6C shows that HNF4α stimulation of PEPCK-Luc reporter activity was repressed by Prox1 and increasing amounts of PGC-1α reversed the repressive effect of Prox1. These results further support that direct interaction between Prox1 and HNF4α prevented the recruitment of co-activator, PGC-1α to stimulate HNF4α transactivation of the CYP7A1 and PEPCK genes.
Fig. 6.
Effects of Prox1 on HNF4α and PGC-1α co-activation of gene transcription. A. pGAL4-HNF4α (200 ng) was co-transfected with PGC-1α (100 ng) and increasing amounts of Prox1 plasmid (50 and 100 ng) into HepG2 cells as indicated. B. HepG2 cells were co-transfected with a human CYP7A1 promoter reporter (hCYP7A1-Luc) and increasing amounts of Prox1 plasmid (10, 50 and 100 ng; +, 50 ng) or PGC-1α plasmid (50, 100, and 200 ng) as indicated. C. HepG2 cells were transfected with a human PEPCK promoter reporter (PEPCK-Luc) (200 ng) along with the HNF4α plasmid (200 ng) and increasing amounts of Prox1 plasmid (10, 50 and 100 ng; +, 50 ng) or PGC-1α (50, 100 and 200 ng) as indicated. Luciferase activity was normalized to β-galactosidase activity. All experiments were done in duplicates and data represent the mean ± SD of three individual experiments.
Knock-down of Prox1 increase CYP7A1 and PEPCK expression in HepG2 cells
To further confirm the role of endogenous Prox1 in HNF4α function, we examined the mRNA expression of HNF4α target genes (CYP7A1 and PEPCK) in HepG2 cells upon knock-down of Prox1 using Prox1 siRNA. We transfected Prox1 oligonucleotide siRNA in HepG2 cells and analyzed protein and mRNA expression of the Prox1, and the effect on CYP7A1 and PEPCK mRNA expression levels by real-time PCR. Among four different regions of human Prox1 siRNA we have tested, siRNA against the N-terminal region (nt 1009-1027; GGGCCAAACTCCTTACAAC) and the C-terminal region (nt 2096-2114;GCAAAGATGTTGATCCTTC) were found to efficiently knock-down Prox1 protein expression in HepG2 cells as shown by Western blot analysis (Fig. 7A). A control siRNA, which has the same G-C content but different sequence from the siRNA probe #2 did not affect Prox1 protein expression. Real time PCR assay shows that the siRNA #1 and #2 decreased Prox1 mRNA expression by about 50%, but increased CYP7A1 expression by 2.53- and 2.59-fold, and PEPCK expression by 1.75- and 2.28-fold, respectively (Fig. 7B). Prox1 siRNA had no effect on HNF4α and cyclophilin B mRNA expression levels. These data support that Prox1 siRNA specifically knockdown Prox1 expression and results in induction of the HNF4α target genes CYP7A1 and PEPCK in HepG2 cells.
Fig. 7.
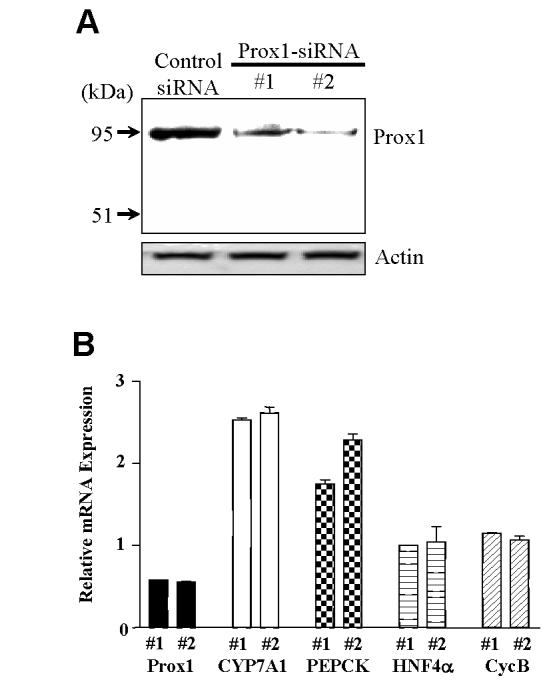
The siRNA knockdown of Prox1 increases the expression of CYP7A1 and PEPCK mRNA in HepG2 cells. A. HepG cells were transfected with the Prox1 siRNA #1, #2 and control siRNA. The effects of siRNAs on the Prox1 expression were measured by Western blot analysis. Actin expression was used as a loading control. Data represent one of three separate experiments, varies from 35 to 88% reduction of protein expression. B. HepG cells were transfected with the Prox1 siRNA #1, #2 and control siRNA. Total RNA was isolated for real-time quantitative PCR analysis of Prox1, CYP7A1, PEPCK, HNF4α and Cyclophilin B (CycB) mRNA levels. Data show relative mRNA expression of Prox1 siRNA treated to the control siRNA treated samples. Data represent the mean ± SD of at least three individual experiments.
Knock-down of Prox1 increases the rate of bile acid synthesis
To confirm whether knock-down of Prox1 expression affects the rate of bile acid synthesis, we analyzed the total bile acid synthesized in HepG2 cells. Knock-down of Prox1 mRNA expression levels resulted in increasing total bile acid synthesis by about 50% in Prox1 knock-down cells compared to control cells (Fig. 8), indicating that Prox1 negatively regulated bile acid synthesis in hepatocytes.
Fig. 8.
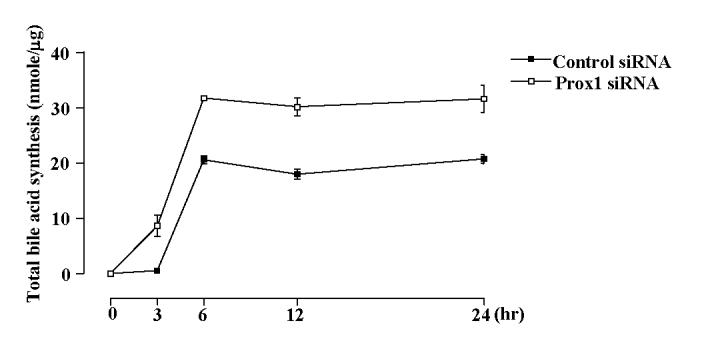
Knock-down of Prox1 increases the rate of bile acid synthesis. HepG2 cells were transfected with Prox1 siRNA and control siRNA and total bile acid were extracted from the media at the indicated times. The amount of total bile acid was determined by enzymatic 3α-hydroxysteroid dehydrogenase method and expressed as nmol/μg protein. The data represent one of three independent experiments.
Discussion
Bile acid synthesis is tightly regulated under physiological conditions to protect the liver from accumulation of highly toxic bile acids. Under normal physiological conditions, expression of CYP7A1 must be suppressed by various factors including insulin, glucagon, bile acids and cholesterol in the human (13). Nuclear receptors and co-regulators apparently play important roles in regulation of CYP7A1 gene transcription (27). Prox1 is constitutively expressed at high levels in adult livers and may be the major repressor of CYP7A1 gene transcription in human hepatocytes.
This study revealed a strong and specific interaction between Prox1 and HNF4α by yeast two-hybrid assay, in vivo coimmunoprecipitation assay using human primary hepatocytes, and in vitro GST pull-down assay. Our results indicate that Prox1 directly interacts with HNF4α via the N-terminal LXXLL motif of Prox1 and C-terminal AF2 domain of HNF4α. Co-regulators are known to interact with nuclear receptors via interaction of the LXXLL motif of co-regulators and the AF2 domain of nuclear receptor (8,28). However, the interaction of Prox1 with FTF also requires helices 2 and 10 of the LBD of FTF (15). Thus, it is likely that the interaction domain of Prox1 might be quite diverse compared with other nuclear receptors to allow interaction with various co-regulators such as SHP (29,30). It is noted that Prox1 may inhibit HNF4α and FTF by somewhat different mechanisms. Prox1 impairs the FTF binding to DNA (15) while our data show that Prox1 does not affect HNF4α binding to CYP7A1 chromatin as demonstrated by ChIP assays (Fig. 5). Our results suggest that Prox1 competes with PGC-1α for interaction with HNF4α and thus counteracts PGC-1α co-activating activity. This is because both Prox1 and PGC-1α interacts with the AF2 domain of HNF4α. Competition for binding and squelching of the limited co-activators could be a common mechanism for the negative regulation of gene transcription by nuclear receptors. In accordance with this scenario, increasing the amounts of PGC-1α could counteract Prox1-mediated repression of HNF4α activity. Conversely, increasing the amounts of Prox1 may interfere with PGC-1α co-activation of HNF4α activity. Thus, our finding that Prox1 interferes with the co-activator recruitment of HNF4α provides a novel mechanism for Prox1 repression of CYP7A1 gene transcription. Moreover, knockdown of Prox1 expression in a hepatocyte cell line increased CYP7A1 mRNA expression and a corresponding increase in bile acid synthesis (Fig.7 and 8). This study reveals a new biological function of Prox1, which has previously been shown to play an essential role for lymphatic system (19,31) and lens (18) development, in hepatocytes.
It is believed that FTF is an activator of gene transcription. More recent studies support that FTF is a negative transcription factor in vivo because ablation of one FTF allele strongly induced CYP7A1 and CYP8B1 mRNA expression in mouse liver (32). Reporter assays in HepG2 cells also suggest that FTF is a repressor of the human CYP7A1 gene (26). The repressor function of FTF can now be explained by the presence of high levels of Prox1 and low levels of PGC-1α in hepatocytes. It is noteworthy that the tissue expression patterns of Prox1 (14), FTF (14) and HNF4α (5) are similar. They all express very early in embryogenesis and expression in liver and pancreas is conserved throughout the vertebrates suggesting the spatial and temporal correlation of Prox1, FTF and HNF4α in regulation of development and function of the liver and pancreas. Prox1, PGC-1α, FTF and HNF4α play central roles in regulation of the CYP7A1 and CYP8B1 in bile acid synthesis, and PEPCK in gluconeogenesis. These factors play critical roles in regulating a variety of metabolic pathways that are involved in pathogenesis of diabetes (33). It is interesting that the putative endogenous ligands for FTF and HNF4α are phospholipids and fatty acids, respectively (34-36). The relative expression levels of these nuclear receptors as well as their co-regulators Prox1 and PGC-1α may regulate lipid homeostasis. A recent report suggests that Prox1 may play a role in adult-onset obesity (37). PGC-1α is greatly induced in the liver of streptozotocin-injected mice, a model of type 1 diabetes of insulin deficiency (9), and in ob/ob mice, a model of type 2 diabetes of insulin resistance (38). PGC-1α and HNF4α induce PEPCK, the rate-limiting enzyme in gluconeogenesis, to prevent hypoglycemia during starvation (9). On the other hand, Prox1 interaction with HNF4α may down regulate PEPCK gene expression to prevent hyperglycemia during the postprandial period. Thus, regulation of PEPCK gene expression by HNF4α and its co-activators and co-repressors may play a critical role in obesity and diabetes in humans.
In conclusion, here we provide direct experimental evidences that Prox1 acts as a novel co-repressor of nuclear receptor HNF4α. Prox1-mediated repression of HNF4α transactivation may play an important role in the regulation of HNF4α target genes. The intricate regulatory circuitry of Prox1, PGC-1α, HNF4α, SHP and FTF may maintain lipid and glucose homeostasis and prevent diabetes and obesity.
Abbreviations
- Prox1
prospero-related homeodomain protein
- CYP7A1
cholesterol 7α-hydroxylase
- CYP8B1
sterol 12α-hydroxylase
- PEPCK
phosphoenolpyruvate carboxykinase
- SHP
small heterodimer partner
- FTF
α-fetoprotein transcription factor
- LRH-1
liver related homologue
- FXR
farnesoid X receptor
- HNF4α
hepatocyte nuclear factor 4α
- DAX-1
dosage-sensitive sex reversal
- AHC critical region on the X chromosome
gene 1
- TR
thyroid hormone receptor
- GR
glucocorticoid receptor
- RXR
retinoid X receptor
- RAR
retinoic acid receptor
- Luc
luciferase
- PGC-1α
peroxisome proliferators-activated receptor γ co-activator 1α
- siRNA
small interfering RNA
- SMRT
silencing mediator of retinoid and thyroid receptors
- N-CoR
nuclear receptor co-repressor (N-CoR)
- MODY1
maturity onset diabetes of the young type 1
- SRC-1
steroid receptor co-activator 1
- ChIP
Chromatin immunoprecipitation
- Q-PCR
Real-time Quantitative PCR
- AF2
activation function 2
Footnotes
This study is supported by NIH grants DK58379 and DK44442.
Contributor Information
Kwang-Hoon Song, Department of Microbiology, Immunology and Biochemistry, Northeastern Ohio Universities College of Medicine, Rootstown, Ohio 44272.
Tiangang Li, Department of Microbiology, Immunology and Biochemistry, Northeastern Ohio Universities College of Medicine, Rootstown, Ohio 44272.
John Y.L. Chiang, Department of Microbiology, Immunology and Biochemistry, Northeastern Ohio Universities College of Medicine, Rootstown, Ohio 44272
References
- 1.Chiang JY. Front. Biosci. 1998;3:d176–193. doi: 10.2741/a273. [DOI] [PubMed] [Google Scholar]
- 2.Stroup D, Chiang JY. J. Lipid Res. 2000;41:1–11. [PubMed] [Google Scholar]
- 3.Li J, Ning G, Duncan SA. Genes Dev. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]
- 4.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Mol. Cell. Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sladek R, Giguere V. Adv Pharmacol. 2000;47:23–87. doi: 10.1016/s1054-3589(08)60109-x. [DOI] [PubMed] [Google Scholar]
- 6.Navas MA, Munoz-Elias EJ, Kim J, Shih D, Stoffel M. Diabetes. 1999;48:1459–1465. doi: 10.2337/diabetes.48.7.1459. [DOI] [PubMed] [Google Scholar]
- 7.Kuo CJ, Conley PB, Chen L, Sladek FM, Darnell JE, Jr., Crabtree GR. Nature. 1992;355:457–461. doi: 10.1038/355457a0. [DOI] [PubMed] [Google Scholar]
- 8.Glass CK, Rosenfeld MG. Genes & Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 9.Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, Newgard CB, Spiegelman BM. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 10.Chakravarty K, Cassuto H, Reshef L, Hanson RW. Crit. Rev. Biochem. Mol. Biol. 2005;40:129–154. doi: 10.1080/10409230590935479. [DOI] [PubMed] [Google Scholar]
- 11.Shin DJ, Campos JA, Gil G, Osborne TF. J. Biol. Chem. 2003;278:50047–50052. doi: 10.1074/jbc.M309736200. [DOI] [PubMed] [Google Scholar]
- 12.De Fabiani E, Mitro N, Gilardi F, Caruso D, Galli G, Crestani M. J. Biol. Chem. 2003;278:39124–39132. doi: 10.1074/jbc.M305079200. [DOI] [PubMed] [Google Scholar]
- 13.Song KH, Chiang JY. Hepatology. 2006;43:117–125. doi: 10.1002/hep.20919. [DOI] [PubMed] [Google Scholar]
- 14.Steffensen KR, Holter E, Bavner A, Nilsson M, Pelto-Huikko M, Tomarev S, Treuter E. EMBO Rep. 2004;5:613–619. doi: 10.1038/sj.embor.7400147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin J, Gao DM, Jiang QF, Zhou Q, Kong YY, Wang Y, Xie YH. Mol. Endocrinol. 2004;18:2424–2439. doi: 10.1210/me.2004-0009. [DOI] [PubMed] [Google Scholar]
- 16.Oliver G, Sosa-Pineda B, Geisendorf S, Spana EP, Doe CQ, Gruss P. Mech. Dev. 1993;44:3–16. doi: 10.1016/0925-4773(93)90012-m. [DOI] [PubMed] [Google Scholar]
- 17.Zinovieva RD, Duncan MK, Johnson TR, Torres R, Polymeropoulos MH, Tomarev SI. Genomics. 1996;35:517–522. doi: 10.1006/geno.1996.0392. [DOI] [PubMed] [Google Scholar]
- 18.Wigle JT, Chowdhury K, Gruss P, Oliver G. Nat Genet. 1999;21:318–322. doi: 10.1038/6844. [DOI] [PubMed] [Google Scholar]
- 19.Wigle JT, Oliver G. Cell. 1999;98:769–778. doi: 10.1016/s0092-8674(00)81511-1. [DOI] [PubMed] [Google Scholar]
- 20.Sosa-Pineda B, Wigle JT, Oliver G. Nat Genet. 2000;25:254–255. doi: 10.1038/76996. [DOI] [PubMed] [Google Scholar]
- 21.Burke Z, Oliver G. Mech. Dev. 2002;118:147–155. doi: 10.1016/s0925-4773(02)00240-x. [DOI] [PubMed] [Google Scholar]
- 22.Liu YW, Gao W, Teh HL, Tan JH, Chan WK. Mol. Cell. Biol. 2003;23:7243–7255. doi: 10.1128/MCB.23.20.7243-7255.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song KH, Park YY, Park KC, Hong CY, Park JH, Shong M, Lee K, Choi HS. Mol. Endocrinol. 2004;18:1929–1940. doi: 10.1210/me.2004-0043. [DOI] [PubMed] [Google Scholar]
- 24.Li T, Chiang JY. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;288:G74–84. doi: 10.1152/ajpgi.00258.2004. [DOI] [PubMed] [Google Scholar]
- 25.Feldmann D, Fenech C, Cuer JF. Clin. Chem. 1983;29:1694. [PubMed] [Google Scholar]
- 26.Chen W, Owsley E, Yang Y, Stroup D, Chiang JY. J. Lipid Res. 2001;42:1402–1412. [PubMed] [Google Scholar]
- 27.Chiang JY. Endocr. Rev. 2002;23:443–463. doi: 10.1210/er.2000-0035. [DOI] [PubMed] [Google Scholar]
- 28.McKenna NJ, O’Malley BW. Endocrinology. 2002;143:2461–2465. doi: 10.1210/endo.143.7.8892. [DOI] [PubMed] [Google Scholar]
- 29.Lee YK, Dell H, Dowhan DH, Hadzopoulou-Cladaras M, Moore DD. Mol. Cell. Biol. 2000;20:187–195. doi: 10.1128/mcb.20.1.187-195.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee YK, Moore DD. J. Biol. Chem. 2002;277:2463–2467. doi: 10.1074/jbc.M105161200. [DOI] [PubMed] [Google Scholar]
- 31.Petrova TV, Makinen T, Makela TP, Saarela J, Virtanen I, Ferrell RE, Finegold DN, Kerjaschki D, Yla-Herttuala S, Alitalo K. Embo. J. 2002;21:4593–4599. doi: 10.1093/emboj/cdf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.del Castillo-Olivares A, Campos JA, Pandak WM, Gil G. J. Biol. Chem. 2004;279:16813–16821. doi: 10.1074/jbc.M400646200. [DOI] [PubMed] [Google Scholar]
- 33.Silander K, Mohlke KL, Scott LJ, Peck EC, Hollstein P, Skol AD, Jackson AU, Deloukas P, Hunt S, Stavrides G, Chines PS, Erdos MR, Narisu N, Conneely KN, Li C, Fingerlin TE, Dhanjal SK, Valle TT, Bergman RN, Tuomilehto J, Watanabe RM, Boehnke M, Collins FS. Diabetes. 2004;53:1141–1149. doi: 10.2337/diabetes.53.4.1141. [DOI] [PubMed] [Google Scholar]
- 34.Krylova IN, Sablin EP, Moore J, Xu RX, Waitt GM, MacKay JA, Juzumiene D, Bynum JM, Madauss K, Montana V, Lebedeva L, Suzawa M, Williams JD, Williams SP, Guy RK, Thornton JW, Fletterick RJ, Willson TM, Ingraham HA. Cell. 2005;120:343–355. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 35.Ortlund EA, Lee Y, Solomon IH, Hager JM, Safi R, Choi Y, Guan Z, Tripathy A, Raetz CR, McDonnell DP, Moore DD, Redinbo MR. Nat. Struct. Mol. Biol. 2005;12:357–363. doi: 10.1038/nsmb910. [DOI] [PubMed] [Google Scholar]
- 36.Petrescu AD, Hertz R, Bar-Tana J, Schroeder F, Kier AB. J. Biol. Chem. 2002;277:23988–23999. doi: 10.1074/jbc.M201241200. [DOI] [PubMed] [Google Scholar]
- 37.Harvey NL, Srinivasan RS, Dillard ME, Johnson NC, Witte MH, Boyd K, Sleeman MW, Oliver G. Nat. Genet. 2005;37:1072–1081. doi: 10.1038/ng1642. [DOI] [PubMed] [Google Scholar]
- 38.Kakuma T, Wang ZW, Pan W, Unger RH, Zhou YT. Endocrinology. 2000;141:4576–4582. doi: 10.1210/endo.141.12.7804. [DOI] [PubMed] [Google Scholar]