Abstract
Bardet-Biedl syndrome (BBS) is primarily an autosomal recessive ciliopathy characterized by progressive retinal degeneration, obesity, cognitive impairment, polydactyly, and kidney anomalies. The disorder is genetically heterogeneous, with 11 BBS genes identified to date, which account for ∼70% of affected families. We have combined single-nucleotide–polymorphism array homozygosity mapping with in silico analysis to identify a new BBS gene, BBS12. Patients from two Gypsy families were homozygous and haploidentical in a 6-Mb region of chromosome 4q27. FLJ35630 was selected as a candidate gene, because it was predicted to encode a protein with similarity to members of the type II chaperonin superfamily, which includes BBS6 and BBS10. We found pathogenic mutations in both Gypsy families, as well as in 14 other families of various ethnic backgrounds, indicating that BBS12 accounts for ∼5% of all BBS cases. BBS12 is vertebrate specific and, together with BBS6 and BBS10, defines a novel branch of the type II chaperonin superfamily. These three genes are characterized by unusually rapid evolution and are likely to perform ciliary functions specific to vertebrates that are important in the pathophysiology of the syndrome, and together they account for about one-third of the total BBS mutational load. Consistent with this notion, suppression of each family member in zebrafish yielded gastrulation-movement defects characteristic of other BBS morphants, whereas simultaneous suppression of all three members resulted in severely affected embryos, possibly hinting at partial functional redundancy within this protein family.
Bardet-Biedl syndrome (BBS [MIM 209900]) is a clinically pleiotropic disorder transmitted primarily in an autosomal recessive fashion, and its hallmarks include progressive retinal degeneration, obesity, polydactyly, hypogenitalism, cognitive impairment, and kidney dysplasia.1,2 Its incidence has been estimated to be 1 in 150,000 individuals in European populations but is much higher in some populations with a high level of consanguinity (e.g., the Middle East and North Africa) or that are geographically isolated (e.g., Newfoundland).3–5 Since 1994,6 the use of homozygosity mapping in consanguineous families with BBS allowed the progressive recognition of a surprisingly high level of nonallelic genetic heterogeneity. Since the identification of the first gene in 2000 (BBS6, also called “MKKS” because it is mutated in the closely related McKusick-Kaufmann syndrome),7,8 mutations have been found in a total of 11 genes (BBS1–BBS11) in patients with BBS.9–19 Recent molecular evidence has revealed an unexpected connection between the BBS proteins and primary cilia, microtubule-based structures arising from the basal body that are notably involved as mechanosensors in kidney epithelium, in the organization of photoreceptor cells of the retina, and in the developmental phenomenon of planar cell polarity.14,20,21 This allowed the characterization of BBS as one of the ciliopathies, an expanding group of clinically distinct but overlapping disorders that includes autosomal dominant polycystic kidney disease, nephronophthisis, Alström syndrome, orofaciodigital syndrome type 1, and, most recently, Meckel and Joubert syndromes.21,22 The remarkable conservation of ciliary and basal-body proteins across phyla has catalyzed the identification of several BBS genes, on the basis of the assumption that these genes are conserved exclusively in ciliated organisms, such as Chlamydomonas, but are absent from nonciliated ones, such as Arabidopsis.12,16,17 However, this approach is insufficient to identify all genes mutated in this phenotype, as highlighted by the recent identification of the vertebrate-specific BBS10 locus.18 In fact, the first BBS gene identified (BBS6, or MKKS) and BBS10 are absent from all ciliary data sets, including the recently constructed, manually curated Ciliary Proteome Database.23 Interestingly, BBS6 and BBS10 are both atypical members of the superfamily of type II chaperonins, on the basis of a distant sequence homology.18,24
The extensive genetic heterogeneity of BBS has profound implications for diagnostic and genetics-counseling applications. Two major genes, BBS1 and BBS10, each account for ∼20% of the mutational load in families of European descent, whereas each of the other nine genes accounts for ⩽5%, and some of these were found mutated in only a few families or even a single family (the latter in the case of BBS11).18,19,25 Taken together, the 11 known BBS genes account for ∼70% of affected families,18 suggesting that additional BBS genes remain to be identified. A further complication is the finding that, in some cases, inheritance departs from classic autosomal recessive inheritance and involves three mutated alleles in two genes, defining oligogenic inheritance, and severity can be modulated by an allele of a modifier gene.25,26
Here, we report the identification of BBS12 on chromosome 4q27, starting with homozygosity mapping in two consanguineous Gypsy families with BBS. In silico studies pointed to a candidate gene, FLJ35630, which encodes a predicted chaperonin-like protein. We found mutations in both Gypsy families as well as in other BBS-affected families, suggesting that this novel locus accounts for some 5% of the overall mutational load of families with BBS.
BBS12, together with BBS6 and BBS10, defines a new vertebrate-specific and divergent branch of chaperonin-like proteins with particular sequence insertions, with respect to the typical group II chaperonins. Suppression of each member of the family in zebrafish recapitulated previously reported bbs morphant phenotypes,26 whereas combinatorial suppression of bbs6, bbs10, and bbs12 revealed strong genetic interaction, with the embryos exhibiting markedly more-severe phenotypes when all three chaperonin-like proteins were suppressed simultaneously. This branch of atypical chaperonins is of major importance for BBS, since the three genes together account for about one-third of the mutational load.
Patients and Methods
The patients analyzed in this study came from two cohorts, one recruited in France and one in the United States. The clinical inclusion criteria were defined elsewhere.1 Appropriate informed consent was obtained from all participants in the study, with respect to the tenets of the Declaration of Helsinki.
For the French cohort, >200 families with BBS have been collected during the past 3 years. Nine families that did not show any mutation in the BBS1–BBS10 genes were selected for a first screen by SNP array analysis because of consanguinity and/or because of the number of affected individuals. A second series of 106 patients with BBS, comprising mostly sporadic cases, was tested for mutations. We excluded from the testing any families that had two bona fide pathologic alleles in other BBS genes. For the US cohort, a series of 139 patients with BBS was tested, without preselection against probands with previously known BBS mutations.
SNP Homozygosity Mapping
Affected and unaffected siblings from consanguineous families were studied with the Affymetrix GeneChip Mapping 10K Array (Affymetrix). These arrays allow analysis of 10,000 SNPs with a mean genetic gap distance of 0.32 cM and an average heterozygosity of 0.37.
Sample processing and labeling were performed in accordance with the manufacturer's instructions (Affymetrix Mapping 10K 2.0 Assay Manual, version 1.0). The arrays were hybridized on a GeneChip Hybridization Oven 640, were washed with a GeneChip Fluidics Station 450, and were scanned with a GeneChip Scanner 3000. Data were processed by the GeneChip DNA Analysis Software version 3.0.2, to generate SNP allele calls. An average call rate >99% was obtained. Homozygosity regions were identified as regions of homozygosity comprising >25 adjacent SNPs. One of us (F.P.) has developed a graphic interface (fig. 1A) that allows an easy comparison of homozygous regions between patients or unaffected siblings from different families, including a majority of small families that have limited informativeness for linkage and that are each consistent with BBS linkage to 2–5 chromosomal regions.
Figure 1. .
Mapping of a candidate BBS region to 4q27. A, Analysis of homozygosity in chromosome 4 in three consanguineous families. The areas of homozygosity with >25 SNPs are black, whereas homozygosity regions defined by 15–25 consecutive SNPs are gray. B, Detailed SNP homozygosity mapping and microsatellite segregation in the region of interest on chromosome 4 in the two Gypsy families. Light- and dark-gray regions represent homozygous SNPs (AA and BB, respectively), whereas white regions indicate heterozygous alleles (AB).
DNA Analysis with Microsatellite Markers
Genotyping of additional fluorescent microsatellite markers was performed on a CEQ8800 genetic analysis system (Beckman Coulter). Experimental conditions are available on request. Microsatellite sequences were obtained from the UCSC Genome Browser.
DNA Sequencing and Mutation Screening
PCR amplifications of exon 2 of FLJ35630 were performed with 50 ng of genomic DNA template per amplicon (see table 1 for oligonucleotide primers). The noncoding exon 1 was screened for all the US families and for one family in the French cohort (family III.10). Bidirectional sequencing of the purified PCR products was performed as described elsewhere.18
Table 1. .
Primer Sequences for BBS12
| Primer Sequence(5′→3′) |
|||
| Exon and Amplicon |
Forward | Reverse | Size (bp) |
| Exon 1 | CGCAGCCCGGCGTTAAGCGG | CCCTGGCCCGAAGAGGACAG | 508 |
| Exon 2: | |||
| Amplicon 1 | GCAGGGAGCATATCTTGTTT | CTACCTCTTCACTACAAAAG | 482 |
| Amplicon 2 | GGAGCAGTGCAGTTGAAGAA | GAGTAGCTGTAACATGTTCTTG | 495 |
| Amplicon 3 | CCTGCTGCAGACAGTCAATA | GCCCACAGTTCTTCTGAGCT | 520 |
| Amplicon 4 | GAGGGTGACCTCACAGAGAA | GCCAGAGATGAAGCCAGCCA | 665 |
| Amplicon 5 | GGTCTTCCTTGGAGGTGGTG | GGCACCATGACTTGGCTATC | 616 |
Database Searches
Systematic exploration in UniProt was performed with BLASTP on each candidate gene in the interval of interest. For each gene, the sequences detected with expect value <10−3 were aligned with the PipeAlign program.
For BBS12, a PSI-BLAST in UniProt and RefSeq was performed using a protein sequence from UniProt, accession number Q6ZW61_HUMAN, as the query and was used to obtain a multiple alignment as described elsewhere.18 The multiple alignment was manually adjusted, with attention paid to secondary structures, leading to nearly 300 sequences corresponding to α, β, γ, δ, ɛ, ζ, η, and θ chaperonin-containing TCP1 (CCT) chaperonin subunits, archeal chaperonins, BBS6, BBS10, and BBS12 sequences. The multiple sequence alignment is available at Bardet Biedl Syndrome: Multiple Alignment Web site.
Phylogenetic Tree
A phylogenetic tree was built with Phylo_win by use of representative sequences from each different chaperonin subunit and from BBS12, BBS10, and BBS6. The parameters were set to global gap removal by use of the neighbor-joining method. Branches were tested by bootstrapping (500 replicates), and the tree was edited and displayed with TreeView. The multiple alignment used for phylogenetic analyses contains 211 positions between the selected sequences.
Phylogenetic Distribution of BBS12 in Complete Genomes
Phylogenetic distribution of BBS12 was examined in 42 eukaryotic organisms for which the genome sequences are available. The presence or absence of each BBS12 was cross-validated at both the proteomic and genomic levels.
The 42 eukaryotic genomes used were Homo sapiens, Mus musculus, Rattus norvegicus, Gallus gallus, Xenopus laevis, Tetraodon nigroviridis, Danio rerio, Takifugu rubripes, Ciona intestinalis,Strongylocentrotus purpuratus, Drosophila melanogaster, Anopheles gambiae, Apis mellifera, Caenorhabditis elegans, Caenorhabditis briggsae, Schizosaccharomyces pombe, Saccharomyces cerevisiae, Candida albicans, Candida glabrata, Kluyveromyces lactis, Ashbya (Eremothecium) gossypii, Debaryomyces hansenii, Yarrowia lipolytica, Aspergillus fumigatus, Neurospora crassa, Cryptococcus neoformans, Encephalitozoon cuniculi, Dictyostelium discoideum, Entamoeba histolytica, Thalassiosira pseudonana, Trypanosoma cruzi, Leishmania major, Plasmodium falciparum, Cryptosporidium parvum, Cryptosporidium hominis, Theileria parva, Tetrahymena thermophila, Giardia lamblia, Arabidopsis thaliana, Oryza sativa, and Cyanidioschyzon merolae.
When available, the genomic sequences were queried by TBLASTN within the corresponding nucleotide-sequence database from the National Center for Biotechnology Information (NCBI) (including GenBank and RefSeq), UCSC Genome Browser, or Ensembl Genome Browser. We also used dedicated Web sites to retrieve the latest sequence version for C. briggsae (WormBase), T. pseudonana (JGI: T. pseudonana), D. discoideum (dictyBase), E. histolytica and L. major (The Wellcome Trust Sanger Institute), O. sativa (Rice Information System), and T. thermophila (The Institute for Genomic Research) and other sites for additional BLAST searches for cryptosporidia (CryptoDB) and C. merolae (Cyanidioschyzon merolae Genome Project). The putative BBS12 sequences were created for the following six vertebrate genomes: Bos taurus, Canis familiaris, X. laevis, D. rerio, T. rubripes, and T. nigroviridis.
Comparative Sequence Analysis of All Known BBS Genes
When available, the BBS1–BBS11 protein sequences from H. sapiens to C. elegans were retrieved from protein databases and were aligned using the same protocol as for BBS12. Sequence identities and similarities were computed for each member of the BBS1–BBS11 family as the pairwise percent identity of each sequence against the human protein sequence. Positions in the alignment of these sequences corresponding to gaps were excluded from the calculation.
Embryo Collection and Morpholino (MO) Injection
Embryos were collected from natural matings of wild-type zebrafish immediately after fertilization. A translational MO against bbs12 was designed (Gene Tools) and was diluted to various concentrations for dose-response assays. For genetic interaction assays, 1 ng of bbs12 MO was injected, along with 4 ng of bbs6 MO and 6 ng of bbs10 MO. For all injections, 1 nl of solution was injected into one- to two-cell–stage embryos. Embryos were scored at midsomite stages.
Results
Identification of a Candidate Region on Chromosome 4q27 by Homozygosity Mapping
We analyzed probands from nine consanguineous and/or multiplex BBS-affected families with the 10K Affymetrix SNP array. A search for all common regions of homozygosity suggested potential linkage of three families (V.3, III.12, and III.10) to 4q27 (fig. 1A), included in the broader 4q24-q32.2 (58-Mb) region proposed recently as a candidate region for a novel BBS locus on the basis of a similar analysis with a different cohort.17 This region also contains the BBS7 gene described elsewhere.15 Analysis of unaffected sibs was consistent with the suggestive linkage, allowing us to restrict the candidate region to a 6-Mb region extending from 118.87 Mb to 124.77 Mb (fig. 1B). Interestingly, the three patients in families V.3 and III.12, known to be of Gypsy (Roms) origin, were haploidentical for a region of 47 Mb, including the region of interest, which suggests that they share a common mutation by descent (fig. 1B). The two families were not known to be related and are partially settled in two different regions of France.
We investigated this 6-Mb region for candidate genes. Forty genes were listed in Ensembl, of which 23 are well characterized (including BBS7 at position 122.97 Mb), whereas 17 have minimal function annotation. Sequencing of the 19 exons of BBS7 in the three families did not reveal any mutations.
Selection of Candidate Genes in the 4q26-q27 Region
After systematic bioinformatic analysis of genes in the candidate interval—encompassing database searches and multiple alignment analysis—we prioritized six genes for screening; three were listed in the cilia-related comparative genomic data sets23 (PRSS12, SEC24D, and SPATA5), whereas three others were selected because of an expression pattern or putative function compatible with BBS or with a ciliopathy. Of particular note was one transcript of unknown function, FLJ35630, annotated in the genome as “C4orf24,” that showed distant homology to group II chaperonins and to the previously identified BBS10 and BBS6 genes.
Sequencing of FLJ35630 in Two Gypsy Families
FLJ35630 consists of two exons, of which only the second is coding, for a predicted protein of 710 aa. Sequencing of exon 2 in the two Gypsy families revealed a homozygous C1062T change that induces a R355X nonsense mutation, which was present in all patients of both families (individuals V.3.C, III.12.C, and III.12.D). The parents were heterozygous, and none of the seven unaffected sibs were homozygous for this mutation, suggesting that mutations in FLJ35630 cause BBS, which thereby designates the 12th locus for the disorder, BBS12. No mutation was found in exons 1 and 2 in the third family (III.10), which showed putative linkage in the same region. Other potentially linked regions of homozygosity were detected for this family with low informativeness (maximum expected LOD score 1.32).
Sequencing of FLJ35630 in Other Families with BBS
To confirm the causality of BBS12 and to assess its contribution to the total BBS mutational load, we sequenced an additional 245 unrelated probands with BBS from the French and US cohorts. The French series included 30 BBS-affected families with no mutation identified in BBS1–BBS8 and BBS10 (BBS9 and BBS11 have not been tested yet) and 76 families tested partially for BBS genes, in which either no mutation or only a single one have yet been identified. We found homozygous or compound heterozygous likely pathogenic mutations in 10 of these 106 families (table 2). For the US cohort, we sequenced 139 families without preselection against families with mutations in another BBS gene. Three of these families were compound heterozygotes, each carrying one mutation causing a frameshift and premature termination and another mutation affecting the protein sequence. The proband of a fourth family had a single missense mutant allele (G540V) that is probably deleterious (see below).
Table 2. .
Mutations Detected in BBS12 in Families with BBS[Note]
| Amino Acid Change |
Nucleotide Change |
|||||
| Family | Allele 1 | Allele 2 | Allele 1 | Allele 2 | Ethnic Origin | Other Mutated BBS Genes |
| V.3; III.12 | R355X | R355X | 1063C→T | 1063C→T | Gypsy | NDD |
| VII.7 | V113del | V113del | 335delTAG | 335delTAG | Chinese | NDD |
| III.17 | F90fsX112 | T501M | 268delT | 1502C→T | White | NDD |
| IV.15 | N506K/P507del | G630fsX638 | 1518delCCC | 1890delCT | White | NDD |
| I.1 | E495fsX498 | E495fsX498 | 1483delGA | 1483delGA | Kurdish | NDD |
| I.11; VII.16 | F372fsX373 | F372fsX373 | 1114delTT | 1114delTT | White | NDD |
| I.7 | T257fsX266 | T257fsX266 | 769insA | 769insA | Algerian | BBS3a (R122Q/N) |
| III.30 | A289P | A289P | 865G→C | 865G→C | White | NDD |
| V.6 | G594fsX605 | G630fsX638 | 1779insA | 1890delCT | White | NDD |
| I.14 | P159L | I346T | 476C→T | 1037T→C | White | NDD |
| AR 198-04 | F372fsX373 | G540V | 1114delTT | 1619G→T | White | NDD |
| AR 273-04 | F372fsX373 | Q511-Q513 del | 1114delTT | 1529delACAGATGCA | White | NDD |
| AR 316-03 | E365fsX382 | X711Y-X728 | 1092delA | 2133G→C | White | NDD |
| AR 14-04 | G540V | … | 1619G→T | … | White | NDD |
Note.— The family numbering is done according to the laboratories' coding. NDD = no other mutation detected to date.
Mutation of uncertain pathogenecity.
Overall, we identified 16 BBS-affected families with pathogenic mutations (table 2), suggesting that BBS12 mutations account for ∼5% of families with BBS. Patients with mutations in BBS12 did not present obvious phenotypic differences compared with patients with mutations in other known BBS genes.
Overall, we found 17 different mutations, including 7 frameshifts, 1 nonsense mutation, 1 mutation that is predicted to extend the C-terminus of the protein, and 3 small in-frame deletions (fig. 2A). One of the frameshift mutations is recurrent and present in six alleles (four families). Five missense mutations were identified. One family was homozygous for an A289P mutation affecting a conserved residue. G540V and T501M were found in trans of frameshift mutations and correspond to nonconservative changes. G540V was also found in the heterozygous state in an additional patient for whom the second mutation was not detected (a large deletion cannot be excluded at this point). One patient was a compound heterozygote for P159L and I346T; both changes are nonconserved. The P159 residue is not conserved in evolution, whereas I346T is chemically conserved (hydrophobic residue or A replaces I in some vertebrate organisms). All missense changes were not observed in 288 control chromosomes and are not recorded in the dbSNP database. However, functional testing will be necessary to assess definitively their pathogenicity. We also observed four likely nonpathogenic variants (G15G, I39T, I170V, and V354V) that, although not previously reported in dbSNP, were found in our controls. All missense mutations are shown on the multiple sequence alignment (Bardet Biedl Syndrome: Multiple alignment Web site).
Figure 2. .
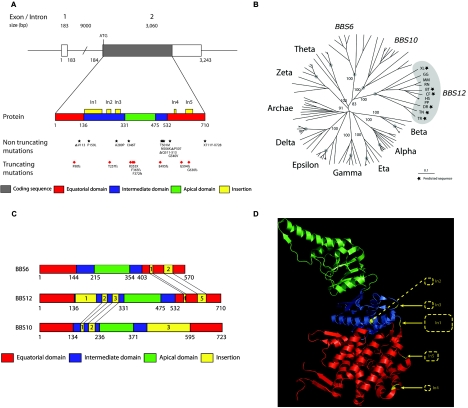
Schematic of the BBS12 locus and the position and/or nature of mutations. A, BBS12 genomic locus (top), corresponding to FLJ35630 (NCBI [RefSeq] accession number NM_152618). FLJ35630 encodes a predicted protein of 710 aa (UniProt accession number Q6ZW61_HUMAN). The bottom section is a schematic of the protein with the recognized domains in different colors (see key). The insertions in the intermediate and equatorial domains are shown as yellow boxes above the protein. All reported mutant alleles are shown underneath the protein. B, Phylogenetic tree of the BBS family. The tree contains BBS12, BBS10, and BBS6 sequences and representative sequences from all group II chaperonins. BBS12 genes are highlighted with organism abbreviations, and predicted sequences are marked with a black star. XL = X. laevis; GG = G. gallus; MM = M. musculus; RN = R. norvegicus; BT = B. taurus; CF = C. familiaris; HS = H. sapiens; PP = Pongo pygmaeus; DR = D. rerio; TN = T. nigroviridis; TR = T. rubripes. The roots of vertebrate branches are highlighted with gray dots. Bootstrap values are provided for significant nodes when they are >80%. C, Comparison of BBS chaperonin-like domain organization. BBS12, BBS10, and BBS6 are represented with the typical chaperonin group II organization in three domains (equatorial, intermediate, and apical). Specific insertions to the typical chaperonin group II are drawn in yellow and are numbered in order of appearance from N-ter to C-ter part. Points of insertions that appear common to BBS12 and BBS6 and to BBS12 and BBS10 are highlighted with black lines. D, Ribbon drawing of group II chaperonin α subunit (Protein Data Bank 1q2v). Domains are colored according to figure 3A. BBS12 insertions (In1-In5) relative to all group II chaperonin sequences are represented as yellow dashed rectangles.
The French cohort was not tested for oligogenism because families with two clearly pathogenic mutations in a single BBS gene were excluded from testing. Testing in US families was unbiased, since it included families with two mutated alleles. However, there is no conclusive evidence at present that BBS12 is significantly involved in oligogenic inheritance, since we observed only one family with two truncating mutations in BBS12 and a missense variant of uncertain significance in BBS3 (family I.7) or a few families with two mutations in other BBS genes and a missense variant of BBS12 also found in controls or that appears nonpathogenic (I170V) (not shown).
A Separate Branch of Type II Chaperonin Superfamily, Defined by BBS12, BBS6, and BBS10
Extensive database searches and sequence analyses revealed various BBS12 homologues and clearly relate BBS12 to the group II chaperonins and to a family of chaperonin-like sequences encompassing BBS10 and BBS6 (fig. 2B). As a result of the growing number of completely sequenced genomes available, the distribution of BBS12 across the eukaryotic phyla could be achieved by an in-depth analysis of the proteomes and genomes of 42 organisms. Like BBS10 and BBS6, BBS12 is only present in vertebrates, from fish to human (a distant BBS6 homologue, however, has been reported in the urochordate C. intestinalis).24 The analysis of the multiple alignments of complete representative sequences27 for all known chaperonin subunits and chaperonin-like proteins revealed the conservation of the classical chaperonin domain architecture (equatorial, intermediate, and apical domains)28 and the existence of specific insertions. Compared with the prototypic group II chaperonin, BBS12 contains five specific insertions in the intermediate and equatorial domain: one large insertion, In1 (82 aa at position 137); In2 (20 aa at position 241); In3 (25 aa at position 282); In4 (9 aa at position 557); and In5 (34 aa at position 612) (fig. 3C). In1 and In3 protrude from the same face of the molecule, suggesting that, like BBS10, they may cooperate to define an additional domain in BBS12 (fig. 2D). It is notable that the comparison of the positions of insertions with respect to the typical group II chaperonin sequence for BBS12, BBS10, and BBS6 reveals common insertion points, without detectable homology between any of the insertions. Indeed, BBS12 shares with BBS6 the same positions in the core chaperonin fold for In4 and In5 and with BBS10 for In2 and In3 (fig. 2C). In addition, the functional motif GDGTT[T/S]28 responsible for ATP hydrolysis in all typical group II chaperonins is not conserved in BBS12 and its orthologues, in that regard resembling BBS6.24
Figure 3. .
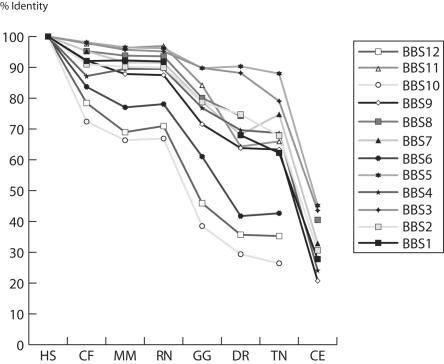
Sequence conservation of BBS proteins in vertebrates. The percent identities are given from human to the nematode by use of the human sequence as a reference. HS = H. sapiens; CF = C. familiaris; MM = M. musculus; RN = R. norvegicus; GG = G. gallus; DR = D. rerio; TN = T. nigroviridis; CE = C. elegans. No BBS6, BBS10, and BBS12 orthologues exist in the C. elegans genome.
Examination of a phylogenetic tree of the 12 subfamilies of the type II chaperonin superfamily also indicates clearly that the three BBS chaperonin-like molecules constitute a separate branch from the eight eukaryotic CCT chaperonins (α to ζ) or the archeobacteria subfamily (thermosome TF55).29 The branch lengths from vertebrate separation (indicated by a gray dot in fig. 2B) are much longer for the three BBS subfamilies than for the eight eukaryotic CCT subfamilies, indicating that the former have evolved faster in vertebrates.
Conservation of the BBS12 protein in vertebrates was also compared with that of the other known BBS proteins (fig. 3). The three chaperonin-like proteins (BBS6, BBS10, and BBS12) share a similar pattern of rapid sequence change that is strikingly different from the pattern of much greater conservation exhibited by all other known BBS proteins.
Suppression of BBS12 and Other BBS Chaperonins and Affect on Zebrafish Development
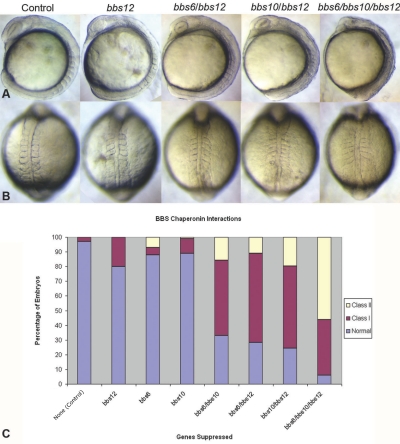
To assess the function of BBS12 in vivo, we investigated the effects of suppression of bbs12 in zebrafish, taking advantage of the BBS-related phenotypes in this model that we reported elsewhere.20,26 Consistent with most other known ciliary and all BBS genes, we found the zebrafish genome to contain a single copy of the likely BBS12 orthologue, to which we designed an oligonucleotide MO to block protein translation. Injection of the MO into wild-type embryos at progressively increasing concentrations produced dose-dependent effects, which could be rescued by coinjection of a capped bbs12 mRNA that escaped MO suppression. We observed phenotypes consistent with convergence and extension (CE) defects, including shortened body axis, broadened somites, kinked notochord, and dorsal thinning that increased with severity in a dose-dependent manner (fig. 4). To quantify the penetrance and variability of these phenotypes, we categorized embryos on the basis of the severity and extent of phenotypes: either poorly defined and irregularly shaped somites and slightly widened notochord in mild-to-moderately affected embryos (class I) or severe misshaping and lack of definition of somites, misshapen notochord, and significantly shorter body axes in severely affected embryos (class II). Because these phenotypes were consistent with those observed in morphants of other bbs genes, including those expressing the atypical chaperonin bbs6 and bbs10,18,20,26 we explored the possibility that there may be genetic interaction or functional redundancy between these genes. Elsewhere, we reported that coinjection of bbs6 and bbs10 MOs resulted in a modest but clear increase in the proportion of affected embryos.18 We found that coinjection of bbs6 and bbs12 or bbs10 and bbs12 MOs resulted in a substantial increase in the proportion of affected embryos (fig. 4). To explore the possibility that these genes may exhibit redundancy in function, we tested for suppression of all bbs chaperonin proteins. Coinjection of bbs6, bbs10, and bbs12 at subeffective concentrations, which by themselves led to phenotypes in <20% of embryos (n>100), resulted in the most severe increase in the proportion of affected embryos (fig. 4). For instance, coinjection of MOs against bbs6 and bbs12 resulted in 75% of embryos with CE-like phenotypes, whereas a bbs10 and bbs12 combination injection yielded ∼80% affected embryos. Strikingly, simultaneous suppression of all three members of this chaperonin subgroup gave rise to the most-severe phenotypes, both in terms of penetrance, with >90% of embryos exhibiting CE defects accompanied by other phenotypes that include cell detachment along the embryonic axis and poor head definition, and in terms of expressivity, with the severe (class II) fraction of affected embryos expanding from <20% in any single- or double-suppression combination to some 60% (fig. 4). These findings suggest that the three bbs chaperonins may share redundant functions necessary for proper embryo development.
Figure 4. .
CE defects and genetic interaction between the BBS chaperonins in zebrafish. A and B, Side (A) and dorsal (B) views of live embryos injected either with a control MO, or various translational blocking MOs against the chaperonin-like subclass of bbs genes. C, Distribution of phenotypes resulting from suppressing various combinations of bbs6, bbs10, and bbs12. Note the synthetic effect of double suppression and the pronounced effect of the combinatorial suppression of all three genes, with the expansion of the severe class of morphants (class II) being particularly prominent.
Discussion
Identification of BBS genes has been based largely on either traditional positional cloning approaches7–11 or, more recently, comparative genomic approaches subtracting likely ciliary genes from large regions of linkage that would otherwise be experimentally intractable.12,16,17 However, both approaches have significant limitations. Positional cloning requires large families for initial linkage, which, for BBS, are scarce. The alternative, evolutionarily based approaches, also fail to provide complete coverage, as evident by the fact that BBS6, BBS10, and now BBS12 are only present in the vertebrate lineage.
Despite our very large sample of families with BBS (combining cohorts from France and the United States), there are no more large families available to us that would allow singly the mapping of new BBS loci. We are thus using a strategy of homozygosity mapping in smaller consanguineous families, looking for homozygous regions shared by affected members of several families (each family showing several potentially linked regions). The SNP Affymetrix arrays allow much more efficient detection of homozygous-by-descent regions than do standard whole-genome microsatellite linkage scans (the 10K SNP array represents an approximately eightfold gain in informativeness). A similar approach has been reported recently by Nishimura et al., leading to the identification of BBS9, as well as 19 additional possible candidate chromosomal regions that do not map to any known BBS genes.17 Our analysis of nine consanguineous families indeed pointed to a region of potential linkage around 4q27 in three families (one of which has no mutation detected to date). The most informative one elicits a maximum LOD score of 2.05 (i.e., it is insufficient to prove linkage).
The identification of BBS12 provides further impetus in the effort to describe the complete genetic load in BBS, which is of significant clinical benefit. It is notable that we detected pathogenic mutations in a variety of ethnic groups, including Rom Gypsies, Europeans, Chinese, and Middle Eastern and/or Arabic populations. One recurrent mutation (F372fsX373) represents 20% of mutated alleles. Since, in the analysis of the French cohort, families with two bona fide mutations in another BBS gene were excluded from mutation screening, one can estimate that the contribution of BBS12 to the total mutation load in patients with BBS is ∼5%. This represents a significant improvement from the point of view of diagnostic and genetics counseling, especially because there is a single coding exon to screen for mutations, compared with the many exons present in most other BBS genes (with the exception of BBS6, BBS10, and the very rarely mutated BBS11).
With regard to oligogenic inheritance, only the US cohort (139 families) fully addressed this point, although the French cohort also included all families in which a single deleterious or probably deleterious allele had been observed elsewhere.18,30 The absence of families with unambiguously pathogenic mutations in two genes (table 2) suggests that BBS12 is rarely involved in oligogenic inheritance. However, it is possible that some of the missense polymorphisms that are rather frequent in BBS12 may affect the severity of the phenotype, as observed for a variant in the MGC1203 modifier gene, which affects the efficiency of splicing by modulating the relative strength of an exonic splice enhancer.26
The identification of BBS12 further highlights the important role of atypical type II chaperonins to the pathogenesis of this disorder, since the three BBS-causing chaperonin-like assemblies account for some 30% of the mutation load in BBS, and to the function of cilia and flagella in general. Our in silico analysis did not reveal other similar chaperonins encoded in the human genome that could indicate candidates for novel BBS genes. Both BBS6 and BBS10 localize to the pericentriolar region of basal bodies and centrosomes24 (N.K., unpublished data). We predict that BBS12 is also likely to localize to the same subcellular domain. It is notable that each of these molecules (BBS6, BBS10, and BBS12) arose in the vertebrate lineage (or perhaps in chordates for BBS6), despite the fact that both pericentriolar material components and the architecture and broad function of the basal body and its attached cilium have been conserved rigorously during evolution. Some recent data have hinted at the possibility that vertebrates have expanded the uses of cilia to transduce a wide variety of morphogenetic signals.31 We speculate that the acquisition of new ciliary functions might be coincident with the emergence of this branch of the type II subfamily. Consistent with this notion, at least one of these molecules, BBS6, was shown recently to be involved in the planar cell polarity pathway, a noncanonical Wnt pathway involved in the elaboration of structures in three-dimensional space.20 As such, it will be important to determine whether BBS6, BBS10, and BBS12 each play overlapping roles in Wnt signal transduction. It is striking, however, that despite the clear distinction in evolutionary patterns of these three BBS genes compared with the eight other known ones, there is no obvious clear-cut phenotypic difference linked to the nature of the mutated gene.
The identification of BBS12 allowed us to reanalyze the evolution of the type II chaperonin superfamily. Typical group II chaperonins are multisubunit assemblies of eight proteins (α to ζ) known to be mainly involved in protein folding. The three BBS genes are characterized by the conservation of the structural architecture called the “chaperonin fold,” but, in all three cases, with insertions of extraneous sequences that do not share sequence similarity with other chaperonins. Such insertions are not present in the nine other chaperonin subfamilies. They have orthologues only in vertebrates, and they present a rapid and parallel rate of evolution (fig. 3B). This distinguishes them from the eight CCT chaperonins that appeared very early in the evolution of eukaryotic organisms.29 Whereas prototypic type II chaperonins have an ATPase hydrolysis site that is important for their function, BBS12, like BBS6,24 appears to lack the key residues necessary for this activity. This feature and the presence of insertions that would prevent association with the canonical multimeric CCT complex24 suggest a different mode of function.
The striking similarity in rapid evolution rate of BBS6, BBS10, and BBS12 suggests that they have coevolved. Two alternative hypotheses might be proposed: either the three proteins interact in a common multisubunit complex or they coevolved with other vertebrate-specific protein targets, involved in basal body and cilia assembly. Such putative vertebrate-specific proteins might then be targets of BBS mutations. It is notable that suppression of each of these chaperonin-like molecules in zebrafish yielded highly overlapping phenotypes but, most intriguingly, that simultaneous suppression of the entire subfamily grossly exaggerated the penetrance and expressivity of these phenotypes. We speculate that this might underlie either some partial functional redundancy within the subfamily or might reflect the progressive loss of pericentriolar function, although the striking exacerbation of the zebrafish phenotype of triple morphants compared with any double morphant combination suggests the former is the more likely possibility.
In conclusion, the identification of BBS12, mutated in 5% of families with BBS, allowed the individualization of a novel branch of vertebrate-specific chaperonin-related proteins that account for one-third of the mutational load in patients with BBS. It will be of major importance for the understanding of the pathophysiology of this pleiotropic disease to analyze the cellular localization and function of the three chaperonin-related BBS proteins and the nature of their interactions.
Acknowledgments
We thank the patients with BBS for their continued support and enthusiastic participation. We also thank the Centre National de Genotypage of Evry and the Affymetrix platform of Institut de Génétique et Biologie Moléculaire et Cellulaire/Genopole de Strasbourg. We acknowledge the financial support of Projet Hospitalier de Recherche Clinique national 2002, RETINA France, Lions Club du Kochersberg, Fédération des Maladies Orphelines, and the Programme National pour la Recherche sur la Vision INSERM program (all to H.D.). This study was also funded by grants from the College de France (to J.-L.M.); the National Institute of Child Health and Development (to N.K.); the National Institute of Diabetes, Digestive and Kidney Disorders (to N.K.); and the Polycystic Kidney Disease Foundation (to N.K.). J.M. was supported by grants from the National Research Fund and the Ministère de la Culture, de l'Enseignement Supérieur et de la Recherche of Luxembourg. We thank Dr. Mireille Cossée (Laboratoire de Diagnostic Génétique, Hôpitaux Universitaires de Strasbourg) for her contribution.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- Bardet Biedl Syndrome: Multiple Alignment, http://bips.u-strasbg.fr/BBS/ (for alignment of BBS12)
- Ciliary Proteome Database, http://www.ciliaproteome.org/
- CryptoDB, http://cryptodb.org/cryptodb/ (for cryptosporidia)
- Cyanidioschyzon merolae Genome Project, http://merolae.biol.s.u-tokyo.ac.jp/ (for C. merolae)
- dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP/
- dictyBase, http://dictybase.org/ (for D. discoideum)
- Ensembl Genome Browser, http://www.ensembl.org/index.html
- JGI: T. pseudonana,http://genome.jgi-psf.org/thaps1/thaps1.home.html
- NCBI, http://www.ncbi.nlm.nih.gov (for BBS12 [accession numbers NM_152618 and NP_689831])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for BBS) [PubMed]
- PipeAlign, http://bips.u-strasbg.fr/PipeAlign/
- Rice Information System, http://rise.genomics.org.cn/rice/index2.jsp (for O. sativa)
- The Institute for Genomic Research, http://www.tigr.org/ (for T. thermophila)
- The Wellcome Trust Sanger Institute, http://www.sanger.ac.uk/ (for E. histolytica and L. major)
- TreeView, http://taxonomy.zoology.gla.ac.uk/rod/treeview.html
- UCSC Genome Browser, http://genome.ucsc.edu/
- UniProt, http://www.expasy.uniprot.org/ (for BBS12 [accession numbers Q6ZW61_HUMAN, Q7Z482_HUMAN, and Q7Z342_HUMAN])
- WormBase, http://www.wormbase.org/ (for C. elegans)
References
- 1.Beales PL, Elcioglu N, Woolf AS, Parker D, Flinter FA (1999) New criteria for improved diagnosis of Bardet-Biedl syndrome: results of a population survey. J Med Genet 36:437–446 [PMC free article] [PubMed] [Google Scholar]
- 2.Green JS, Parfrey PS, Harnett JD, Farid NR, Cramer BC, Johnson G, Heath O, McManamon PJ, O‘Leary E, Pryse-Phillips W (1989) The cardinal manifestations of Bardet-Biedl syndrome, a form of Laurence-Moon-Biedl syndrome. N Engl J Med 321:1002–1009 [DOI] [PubMed] [Google Scholar]
- 3.Farag TI, Teebi AS (1988) Bardet-Biedl and Laurence-Moon syndromes in a mixed Arab population. Clin Genet 33:78–82 [DOI] [PubMed] [Google Scholar]
- 4.Farag TI, Teebi AS (1989) High incidence of Bardet-Biedl syndrome among the Bedouin. Clin Genet 36:463–464 [DOI] [PubMed] [Google Scholar]
- 5.Moore SJ, Green JS, Fan Y, Bhogal AK, Dicks E, Fernandez BA, Stefanelli M, Murphy C, Cramer BC, Dean JC, Beales PL, Katsanis N, Bassett AS, Davidson WS, Parfrey PS (2005) Clinical and genetic epidemiology of Bardet-Biedl syndrome in Newfoundland: a 22-year prospective, population-based, cohort study. Am J Med Genet A 132:352–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leppert M, Baird L, Anderson KL, Otterud B, Lupski JR, Lewis RA (1994) Bardet-Biedl syndrome is linked to DNA markers on chromosome 11q and is genetically heterogeneous. Nat Genet 7:108–112 10.1038/ng0594-108 [DOI] [PubMed] [Google Scholar]
- 7.Slavotinek AM, Stone EM, Mykytyn K, Heckenlively JR, Green JS, Heon E, Musarella MA, Parfrey PS, Sheffield VC, Biesecker LG (2000) Mutations in MKKS cause Bardet-Biedl syndrome. Nat Genet 26:15–16 10.1038/79116 [DOI] [PubMed] [Google Scholar]
- 8.Katsanis N, Beales PL, Woods MO, Lewis RA, Green JS, Parfrey PS, Ansley SJ, Davidson WS, Lupski JR (2000) Mutations in MKKS cause obesity, retinal dystrophy and renal malformations associated with Bardet-Biedl syndrome. Nat Genet 26:67–70 10.1038/79201 [DOI] [PubMed] [Google Scholar]
- 9.Mykytyn K, Braun T, Carmi R, Haider NB, Searby CC, Shastri M, Beck G, Wright AF, Iannaccone A, Elbedour K, Riise R, Baldi A, Raas-Rothschild A, Gorman SW, Duhl DM, Jacobson SG, Casavant T, Stone EM, Sheffield VC (2001) Identification of the gene that, when mutated, causes the human obesity syndrome BBS4. Nat Genet 28:188–191 10.1038/88925 [DOI] [PubMed] [Google Scholar]
- 10.Nishimura DY, Searby CC, Carmi R, Elbedour K, Van Maldergem L, Fulton AB, Lam BL, Powell BR, Swiderski RE, Bugge KE, Haider NB, Kwitek-Black AE, Ying L, Duhl DM, Gorman SW, Heon E, Iannaccone A, Bonneau D, Biesecker LG, Jacobson SG, Stone EM, Sheffield VC (2001) Positional cloning of a novel gene on chromosome 16q causing Bardet-Biedl syndrome (BBS2). Hum Mol Genet 10:865–874 10.1093/hmg/10.8.865 [DOI] [PubMed] [Google Scholar]
- 11.Mykytyn K, Nishimura DY, Searby CC, Shastri M, Yen HJ, Beck JS, Braun T, Streb LM, Cornier AS, Cox GF, Fulton AB, Carmi R, Luleci G, Chandrasekharappa SC, Collins FS, Jacobson SG, Heckenlively JR, Weleber RG, Stone EM, Sheffield VC (2002) Identification of the gene (BBS1) most commonly involved in Bardet-Biedl syndrome, a complex human obesity syndrome. Nat Genet 31:435–438 [DOI] [PubMed] [Google Scholar]
- 12.Chiang AP, Nishimura D, Searby C, Elbedour K, Carmi R, Ferguson AL, Secrist J, Braun T, Casavant T, Stone EM, Sheffield VC (2004) Comparative genomic analysis identifies an ADP-ribosylation factor-like gene as the cause of Bardet-Biedl syndrome (BBS3). Am J Hum Genet 75:475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan Y, Esmail MA, Ansley SJ, Blacque OE, Boroevich K, Ross AJ, Moore SJ, Badano JL, May-Simera H, Compton DS, Green JS, Lewis RA, van Haelst MM, Parfrey PS, Baillie DL, Beales PL, Katsanis N, Davidson WS, Leroux MR (2004) Mutations in a member of the Ras superfamily of small GTP-binding proteins causes Bardet-Biedl syndrome. Nat Genet 36:989–993 10.1038/ng1414 [DOI] [PubMed] [Google Scholar]
- 14.Ansley SJ, Badano JL, Blacque OE, Hill J, Hoskins BE, Leitch CC, Kim JC, Ross AJ, Eichers ER, Teslovich TM, Mah AK, Johnsen RC, Cavender JC, Lewis RA, Leroux MR, Beales PL, Katsanis N (2003) Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature 425:628–633 10.1038/nature02030 [DOI] [PubMed] [Google Scholar]
- 15.Badano JL, Ansley SJ, Leitch CC, Lewis RA, Lupski JR, Katsanis N (2003) Identification of a novel Bardet-Biedl syndrome protein, BBS7, that shares structural features with BBS1 and BBS2. Am J Hum Genet 72:650–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, Lewis RA, Green JS, Parfrey PS, Leroux MR, Davidson WS, Beales PL, Guay-Woodford LM, Yoder BK, Stormo GD, Katsanis N, Dutcher SK (2004) Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117:541–552 10.1016/S0092-8674(04)00450-7 [DOI] [PubMed] [Google Scholar]
- 17.Nishimura DY, Swiderski RE, Searby CC, Berg EM, Ferguson AL, Hennekam R, Merin S, Weleber RG, Biesecker LG, Stone EM, Sheffield VC (2005) Comparative genomics and gene expression analysis identifies BBS9, a new Bardet-Biedl syndrome gene. Am J Hum Genet 77:1021–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoetzel C, Laurier V, Davis EE, Muller J, Rix S, Badano JL, Leitch CC, et al (2006) BBS10 encodes a vertebrate-specific chaperonin-like protein and is a major BBS locus. Nat Genet 38:521–524 10.1038/ng1771 [DOI] [PubMed] [Google Scholar]
- 19.Chiang AP, Beck JS, Yen HJ, Tayeh MK, Scheetz TE, Swiderski RE, Nishimura DY, Braun TA, Kim KY, Huang J, Elbedour K, Carmi R, Slusarski DC, Casavant TL, Stone EM, Sheffield VC (2006) Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11). Proc Natl Acad Sci USA 103:6287–6292 10.1073/pnas.0600158103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross AJ, May-Simera H, Eichers ER, Kai M, Hill J, Jagger DJ, Leitch CC, Chapple JP, Munro PM, Fisher S, Tan PL, Phillips HM, Leroux MR, Henderson DJ, Murdoch JN, Copp AJ, Eliot MM, Lupski JR, Kemp DT, Dollfus H, Tada M, Katsanis N, Forge A, Beales PL (2005) Disruption of Bardet-Biedl syndrome ciliary proteins perturbs planar cell polarity in vertebrates. Nat Genet 37:1135–1140 10.1038/ng1644 [DOI] [PubMed] [Google Scholar]
- 21.Badano JL, Mitsuma N, Beales PL, Katsanis N (2006) The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet 7:125–148 10.1146/annurev.genom.7.080505.115610 [DOI] [PubMed] [Google Scholar]
- 22.Sayer JA, Otto EA, O‘Toole JF, Nurnberg G, Kennedy MA, Becker C, Hennies HC, et al (2006) The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat Genet 38:674–681 10.1038/ng1786 [DOI] [PubMed] [Google Scholar]
- 23.Gherman A, Davis EE, Katsanis N (2006) The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nat Genet 38:961–962 10.1038/ng0906-961 [DOI] [PubMed] [Google Scholar]
- 24.Kim JC, Ou YY, Badano JL, Esmail MA, Leitch CC, Fiedrich E, Beales PL, Archibald JM, Katsanis N, Rattner JB, Leroux MR (2005) MKKS/BBS6, a divergent chaperonin-like protein linked to the obesity disorder Bardet-Biedl syndrome, is a novel centrosomal component required for cytokinesis. J Cell Sci 118:1007–1020 10.1242/jcs.01676 [DOI] [PubMed] [Google Scholar]
- 25.Katsanis N (2004) The oligogenic properties of Bardet-Biedl syndrome. Hum Mol Genet Spec 1 13:R65–R71 10.1093/hmg/ddh092 [DOI] [PubMed] [Google Scholar]
- 26.Badano JL, Leitch CC, Ansley SJ, May-Simera H, Lawson S, Lewis RA, Beales PL, Dietz HC, Fisher S, Katsanis N (2006) Dissection of epistasis in oligogenic Bardet-Biedl syndrome. Nature 439:326–330 10.1038/nature04370 [DOI] [PubMed] [Google Scholar]
- 27.Lecompte O, Thompson JD, Plewniak F, Thierry J, Poch O (2001) Multiple alignment of complete sequences (MACS) in the post-genomic era. Gene 270:17–30 10.1016/S0378-1119(01)00461-9 [DOI] [PubMed] [Google Scholar]
- 28.Ditzel L, Lowe J, Stock D, Stetter KO, Huber H, Huber R, Steinbacher S (1998) Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT. Cell 93:125–138 10.1016/S0092-8674(00)81152-6 [DOI] [PubMed] [Google Scholar]
- 29.Archibald JM, Logsdon JM Jr, Doolittle WF (2000) Origin and evolution of eukaryotic chaperonins: phylogenetic evidence for ancient duplications in CCT genes. Mol Biol Evol 17:1456–1466 [DOI] [PubMed] [Google Scholar]
- 30.Hichri H, Stoetzel C, Laurier V, Caron S, Sigaudy S, Sarda P, Hamel C, Martin-Coignard D, Gilles M, Leheup B, Holder M, Kaplan J, Bitoun P, Lacombe D, Verloes A, Bonneau D, Perrin-Schmitt F, Brandt C, Besancon AF, Mandel JL, Cossee M, Dollfus H (2005) Testing for triallelism: analysis of six BBS genes in a Bardet-Biedl syndrome family cohort. Eur J Hum Genet 13:607–616 10.1038/sj.ejhg.5201372 [DOI] [PubMed] [Google Scholar]
- 31.Davis EE, Brueckner M, Katsanis N (2006) The emerging complexity of the vertebrate cilium: new functional roles for an ancient organelle. Dev Cell 11:9–19 10.1016/j.devcel.2006.06.009 [DOI] [PubMed] [Google Scholar]