Abstract
Maternal smoking is a recognized risk factor for orofacial clefts. Maternal or fetal pharmacogenetic variants are plausible modulators of this risk. In this work, we studied 5,427 DNA samples, including 1,244 from subjects in Denmark and Iowa with facial clefting and 4,183 from parents, siblings, or unrelated population controls. We examined 25 single-nucleotide polymorphisms in 16 genes in pathways for detoxification of components of cigarette smoke, to look for evidence of gene-environment interactions. For genes identified as related to oral clefting, we studied gene-expression profiles in fetal development in the relevant tissues and time intervals. Maternal smoking was a significant risk factor for clefting and showed dosage effects, in both the Danish and Iowan data. Suggestive effects of variants in the fetal NAT2 and CYP1A1 genes were observed in both the Iowan and the Danish participants. In an expanded case set, NAT2 continued to show significant overtransmission of an allele to the fetus, with a final P value of .00003. There was an interaction between maternal smoking and fetal inheritance of a GSTT1-null deletion, seen in both the Danish (P=.03) and Iowan (P=.002) studies, with a Fisher’s combined P value of <.001, which remained significant after correction for multiple comparisons. Gene-expression analysis demonstrated expression of GSTT1 in human embryonic craniofacial tissues during the relevant developmental interval. This study benefited from two large samples, involving independent populations, that provided substantial power and a framework for future studies that could identify a susceptible population for preventive health care.
Clefts of the upper lip and palate are common birth defects whose etiology includes genes, environment, and their interaction effects. Because they appear to have separate etiologies, affected subjects are categorized as having either cleft lip with or without cleft palate (CL/P) or as having cleft palate only (CPO).1 The prevalence (at birth) of orofacial clefts (OCs) is related to environmental factors, geographic origin, and socioeconomic status.2,3 Among the environmental factors, maternal cigarette smoking is one of the most widely studied,4–9 with some disparities in results that may be explained by population differences, sampling variation, and the variations in inherited pharmacogenetic susceptibilities to effects of cigarette smoking. Nevertheless, a recent meta-analysis—of 24 case-control and cohort studies—of the association between maternal smoking during pregnancy and offspring OC identified statistically significant associations between maternal smoking and CL/P (relative risk [RR]=1.34; 95% CI 1.25–1.44) and between maternal smoking and CPO (RR=1.22; 95% CI 1.10–1.35).10 Investigations of genetic modifiers remain preliminary, with genes involved in the phases I (CYP1A1 [MIM 108330] and EPHX1 [MIM 132810]) and II (GSTM1 [MIM 138350], GSTT1 [MIM 600436], and NAT2 [MIM 243400]) detoxification pathways of special interest.11–13 To investigate genes involved in the detoxification or the aryl hydrocarbon receptor–aryl hydrocarbon receptor nuclear translocator (AHR-ARNT [MIM 600253 and 126110]) pathway as cofactors with maternal smoking, we studied 25 allelic variants in 16 genes, using two large population-based OC-affected groups (Iowa and Denmark). Because effects of maternal cigarette smoking on embryonic development are complex, as is the role of fetal and maternal genes involved in detoxification, we performed statistical tests of several scenarios: (1) detoxification of the risk-relevant component in cigarette smoke is influenced by fetal genotype, (2) a favorable intrauterine environment is determined by the genotype of a smoking mother, and (3) the susceptibility detoxification alleles, either of the fetus or of the mother, have phenotypic expression only in the presence of the environmental factors (maternal smoking) or other susceptibility alleles (gene-environment or gene-gene interaction effects). We corroborated positive results with gene-expression data available for human embryonic craniofacial tissues.
Material and Methods
Study Populations
DNA samples used in this study came from Iowa and Denmark, as shown in table 1. The study was approved by the institutional review boards in Iowa and Denmark, and written informed consent was obtained from each individual (or the parents of minors) included in the study.
Table 1. .
Study Samples Used[Note]
| No. of Subjects with |
Total |
|||||||||
| Population | Study Design | No. of Families | No. of Studied Parents | No. of Unaffected Controls or Siblings |
CPO | CLCP | CLO | Anomalya | Casesb | Samples |
| DBS | Case-control | 755 | 0 | 485c | 68 | 0 | 270 | 755 | ||
| DCS | Case-parent | 517 | 978 | 664d | 101 | 201 | 156 | 70 | 529e | 2,171 |
| Iowa | Case-control triad | 857 |
1,637 |
419c |
110 |
157 |
103 |
42 |
445 |
2,501 |
| Total | 2,129 | 2,615 | 1,568 | 279 | 358 | 259 | 112 | 1,244 | 5,427 | |
Note.— There were 202 subjects in the DBS who had either CLCP or CLO; the total number of subjects with CLCP and CLO was 819.
OCs that occur together with other anomalies, such as heart disease, extremity anomalies, or anomalies associated with syndromic OC.
Cleft status is not known for some cases, so the numbers of cleft subphenotype will not add to the total number of cases.
Unaffected control.
Unaffected sibling.
A small proportion of families have multiple affected subjects. We randomly selected one affected child for the analysis.
Danish samples
The Danish samples are derived from two independent studies. One set, the Danish case-control (DBS) was obtained through a 3-year (December 1991 to August 1994) case-control study of CL/P and CPO in Denmark. Newborn-screening blood spots were used to extract DNA. The 1st-trimester maternal exposure information, including cigarette smoking, vitamin intake, alcohol use, and medication, was obtained via interview within a few weeks after birth and from birth records. More detail is provided in the work of Christensen et al.6 Because of the limitation of sample quantity, only a subset of the markers was tested on this set of samples. A second set of Danish samples—triads of an affected individual and parents (DCS), for whom DNA was extracted from cheek swabs—consisted of affected individuals (born 1981–1990), their parents, and, in some families, the unaffected sibling(s). These samples were collected between 1995 and 2003, with no overlap with the DBS samples. Epidemiologic exposure data on smoking and alcohol and vitamin consumption were available for the DCS samples.
Iowan population
Patients with clefts who were born between 1987 and 2001 in Iowa were identified through the Iowa Registry for Congenital and Inherited Disorders. Control children born without major anomalies and matched by birth month, year, and sex were also available; for details, see the work of Romitti et al.14 Most of the families provided complete triads, formed by the index case or control and both biologic birth parents (as determined by parent report and consistency with Mendelian transmission, by use of the 25 genotyped SNPs). Data on maternal cigarette-smoking status were collected by questionnaire, mostly within 12 mo of the proband’s birth, and were collected for the 3-mo period before conception and for each pregnancy trimester. Data on paternal smoking were available only for a subset of Iowa data, and preliminary analysis did not identify a paternal smoking effect, so it was not included in the analysis. Data on a number of other epidemiologic variables—such as alcohol and vitamin intake, medication during pregnancy, race/ethnicity, and education level—were also recorded. Selected demographic characteristics and maternal perinatal exposures are summarized in table 2.
Table 2. .
Selected Maternal Characteristics[Note]
| Iowa |
DBS |
DCS |
||||
| Characteristics | Case (n=445) |
Control (n=421) |
Case (n=270) |
Control (n=485) |
Case (n=663) |
Controla (n=529) |
| Age (years): | ||||||
| Data missing (N) | 69 | 7 | 0 | 0 | 17 | 9 |
| <21 (%) | 10.9 | 9.9 | 4.8 | 4.3 | 5.6 | 2.9 |
| 21–30 (%) | 62.4 | 60.1 | 63.7 | 67 | 69.6 | 67.6 |
| >30 (%) | 26.7 | 30 | 31.5 | 28.7 | 26.8 | 27.5 |
| Ethnicity (% non-Hispanic white) | 96.8 | 97.3 | 100 | 100 | 100 | 100 |
| Education: | ||||||
| Data missing (N) | 72 | 8 | ||||
| High school degree or less (%) | 73.2 | 52.5 | ||||
| Some college (%) | 19 | 25.7 | ||||
| Bachelor’s degree or higher (%) | 7.8 | 21.8 | ||||
| Smoking during pregnancy: | ||||||
| Data missing (N) | 66 | 5 | 0 | 0 | 13 | 11 |
| Yes (%) | 27.2 | 20 | 39.6 | 31.8 | 34.7 | 30.8 |
| No (%) | 72.8 | 80 | 60.4 | 68.2 | 65.3 | 69.2 |
| Multivitamin intake during pregnancy: | ||||||
| Data missing (N) | 118 | 24 | 4 | 2 | 87 | 104 |
| Yes (%) | 47.1 | 39.5 | 76.7 | 79.7 | 76.7 | 79.6 |
| No (%) | 52.9 | 60.5 | 23.3 | 20.3 | 23.3 | 20.4 |
| Alcohol intake during pregnancy: | ||||||
| Data missing (N) | 142 | 52 | 3 | 13 | 27 | 39 |
| Yes (%) | 74.3 | 62.3 | 62.2 | 68 | 36.7 | 37.2 |
| No (%) | 25.7 | 37.7 | 37.8 | 32 | 63.3 | 62.8 |
Note.— Percentages are based on the total nonmissing data.
Unaffected sibling.
Additional sets of samples were used to further investigate the apparent overtransmission at the NAT2 590 locus. The samples include 679 Philippine case triads (with children affected with OC), >3,000 additional Danish samples from 600 families (consisting of subjects with OC, unaffected sibling, the parents, and, for some families, grandparents), 375 CEPH samples (phenotype unknown),15 and 921 control triads from Iowa.
Genotyping Method
The studied candidate genes and alleles were selected on the basis of enzymatic activities, expression data during fetal development, and sufficient rare-allele frequencies (>5%). Samples were genotyped either by kinetic PCR16 or by Taqman assays (Applied Biosystems).17 For kinetic PCR, each DNA sample was amplified in two separate reactions, with allele-specific primers in each reaction, and PCR was done in duplicate. Primer sequences are available on request.
Taqman SNP genotyping assays were obtained through either the Assay-on-Demand or the Assay-by-Design service from Applied Biosystems. Reactions were performed in duplicate, with use of conditions set forth by the manufacturer. The sequences of primers and probes from the Assay-by-Design service are available on request.
We investigated 25 polymorphisms in 16 genes; information on genes and markers is summarized in table 3, with complete data listed in table 4. All genotyped families were checked for consistency with Mendelian inheritance, and each gene-SNP combination was checked for Hardy-Weinberg equilibrium. Allele frequencies were calculated separately for mothers, fathers, and children and were stratified by population and OC status (see table 5 for details).
Table 3. .
Summary of Marker Information[Note]
| Gene Name and SNP or Deletion | Gene/SNP Designation | Allele Frequencya (%) |
Functional Effects of Variantsb |
| Aryl hydrocarbon receptor: | |||
| G1721A | AHR_snp1 | 7 | Higher inducibility for CYP1A1 |
| A→G | AHR_snp2 | 31 | Unknown |
| Cytochrome P450 1A1: | |||
| T3801C | CYP1A1c | 7 | Stabler mRNA, higher activity |
| Cytochrome P450 1A2: | |||
| −164C→A in intron 1 | CYP1A2c | 32 | Lower activity |
| Cytochrome P450 1B1: | |||
| Val432Leu | CYP1B1_snp1 | 49 | Lower activity |
| Asn453Ser | CYP1B1_snp2 | 15 | Unknown |
| Cytochrome P450 2E1: | |||
| G→A | CYP2E1 | 21 | Unknown |
| Microsomal epoxide hydrolase: | |||
| Y113H | EPHX1_snp1c | 32 | Higher activity |
| His139Arg | EPHX1_snp2 | 22 | Higher activity |
| Glutathione transferases alpha-4: | |||
| G→C | GSTA4_snp1 | 41 | Unknown |
| T→C | GSTA4_snp2 | 27 | Unknown |
| Glutathione transferases mu-1: | |||
| Null deletion | GSTM1_nullc | 56 | Loss of function |
| C→G | GSTM1_snp1 | 37 | Unknown |
| Glutathione transferases mu-3: | |||
| G→A | GSTM3 | 37 | Unknown |
| Glutathione transferases pi-1: | |||
| A313G | GSTP1_snp1c | 38 | Lower activity |
| C341T | GSTP1_snp2 | 10 | Unknown |
| G→T | GSTP1_snp3 | 46 | Unknown |
| Glutathione transferases theta-1: | |||
| Null deletion | GSTT1_nullc | 18 | Loss of function |
| Hypoxia-induced factor-1 alpha subunit: | |||
| A→C | HIF1A | 25 | Unknown |
| N-acetyltransferases (NATs) 2: | |||
| C481T | NAT2_snp1c | 41 | Unknown |
| G590A | NAT2_snp2c | 33 | Lower activity |
| NAD(P)H: quinone oxidoreductase: | |||
| C609 T | NQO1c | 17 | Loss of function |
| Sulfotransferases 1A1: | |||
| R213H | SULT1A1 | 38 | Lower activity |
| UDP-glucuronosyltransferases 1A7: | |||
| T387G, C391A, and G392A | UGT1A7_snp1 | 63 | Unknown |
| T662C | UGT1A7_snp2 | 36 | Unknown |
Note.— The SNP designations are used in other tables as references to markers.
Allele frequencies were based on Iowan control samples.
Change in enzymatic activities of the variant allele (the second allele listed in SNP column).
Only these markers were genotyped on the DBS samples.
Table 4. .
Detailed Marker Information[Note]
| Gene Name, Location, and Gene/SNP Designation | SNP or Deletion | Wild Type | Mutant Type | Reference SNP | Allele Frequency(%) | Available Assaysa | Assay-on-demand Product Number | Functional Effects of Variants |
| Aryl hydrocarbon receptor: | ||||||||
| 7p15: | ||||||||
| AHR_snp1 | G1721A | G | A | rs2066853 | 7 | TM | C_11170747_10 | Higher inducibility for CYP1A1 |
| AHR_snp2 | A→G | A | G | rs2282885 | 31 | TM | C__2541460_1_ | |
| Cytochrome P450 1A1: | ||||||||
| 15q22-q24: | ||||||||
| CYP1A1b | T3801C | G | A | rs4646421 | 7 | K/TM | C__1840674_10 | Stabler mRNA, higher activity |
| Cytochrome P450 1A2: | ||||||||
| 15q22-qter: | ||||||||
| CYP1A2b | −164C→A in intron 1 | A | C | rs762551 | 32 | K/TM | C__8881221_10 | Lower activity |
| Cytochrome P450 1B1: | ||||||||
| 2p22-p21: | ||||||||
| CYP1B1_snp1 | Val432Leu | G | C | rs1056836 | 49 | TM | Lower activity | |
| CYP1B1_snp2 | Asn453Ser | A | G | rs1800440 | 15 | TM | ||
| Cytochrome P450 2E1: | ||||||||
| 10q24.3-qter: | ||||||||
| CYP2E1 | G→A | G | A | rs2249695 | 21 | TM | C__2431848_1_ | |
| Microsomal epoxide hydrolase: | ||||||||
| 1q42.1: | ||||||||
| EPHX1_snp1b | Y113H | T | C | rs1051740 | 32 | K/TM | C___14938_1_ | Lower activity |
| EPHX1_snp2 | His139Arg | A | G | rs2234922 | 22 | TM/TM | C_11638783_10 | Higher activity |
| Glutathione transferases alpha-4: | ||||||||
| 6p12: | ||||||||
| GSTA4_snp1 | G→C | G | C | rs316133 | 41 | TM | C__1205160_1_ | |
| GSTA4_snp2 | T→C | T | C | rs3756980 | 27 | TM | C__1205169_1_ | |
| Glutathione transferases mu-1: | ||||||||
| 1p13.3: | ||||||||
| GSTM1_nullb | Null deletion | 56 | K | Loss of function | ||||
| GSTM1_snp1 | C→G | C | G | rs1010167 | 37 | TM | C__2617802_10 | |
| Glutathione transferases mu-3: | ||||||||
| 1p13.3: | ||||||||
| GSTM3 | G→A | G | A | rs1571858 | 37 | TM | C__3184520_10 | |
| Glutathione transferases pi-1: | ||||||||
| 11q13: | ||||||||
| GSTP1_snp1b | A313G | T | C | rs947894 | 38 | K/TM | C__3237198_1_ | Lower activity |
| GSTP1_snp2 | C341T | C | T | rs1799811 | 10 | TM | ||
| GSTP1_snp3 | G→T | G | T | rs762803 | 46 | TM | C__3237197_10 | |
| Glutathione transferases theta-1: | ||||||||
| 22q11.2: | ||||||||
| GSTT1_nullb | Null deletion | 18 | K | Loss of function | ||||
| Hypoxia-induced factor-1 alpha subunit: | ||||||||
| 14q21-24: | ||||||||
| HIF1A | A→C | A | C | rs2301113 | 25 | TM | C_15755917_10 | |
| N-acetyltransferases (NATs) 2: | ||||||||
| 8p23.1-21.3: | ||||||||
| NAT2_snp1b | C481T | C | T | rs1799929 | 41 | TM/TM | C__1204092_10 | |
| NAT2_snp2b | G590A | G | A | rs1799930 | 33 | K/TM | C__1204091_10 | Lower activity |
| NAD(P)H: quinone oxidoreductase: | ||||||||
| 16q21.1: | ||||||||
| NQO1b | C609 T | C | T | rs1800566 | 17 | K/TM | Loss of function | |
| Sulfotransferases 1A1: | ||||||||
| 16p12.1-p11.2: | ||||||||
| SULT1A1 | R213H | rs1042028 | 38 | K | Lower activity | |||
| UDP-glucuronosyltransferases 1A7: | ||||||||
| 2p37: | ||||||||
| UGT1A7_snp1 | T387G, C391A, and G392A | T387, C391, and G392 | G387, A391, and A392 | rs17868323,rs17863778,and rs17868324 | 63 | T | ||
| UGT1A7_snp2 | T662C | T | C | rs11692021 | 36 | K |
Note.— The SNP designations are used in other tables as references to markers.
TM = Taqman assays; K = kinetic PCR assays.
These makers were also genotyped in DBS samples.
Table 5. .
Allele Frequencies
| No. of Subjects with Allele | |||||||||||||||||||||||||
| Sample, Family Member, and Genotype | AHR_snp1 | AHR_snp2 | CYP1A1 | CYP1A2 | CYP1B1_snp1 | CYP1B1_snp2 | CYP2E1 | EPHX1_snp1 | EPHX1_snp2 | GSTA4_snp1 | GSTA4_snp2 | GSTM1_null | GSTM1_snp1 | GSTM3 | GSTP1_snp1 | GSTP1_snp2 | GSTP1_snp3 | GSTT1_null | HIF1A | NAT1_snp1 | NAT2_snp2 | NQO1 | SULT1A1 | UGT1A7_snp1 | UGT1A7_snp2 |
| DBS: | |||||||||||||||||||||||||
| Mothera: | |||||||||||||||||||||||||
| 11 | 534 | 250 | 555 | 330 | 140 | 449 | 428 | 311 | 392 | 236 | 425 | 307 | 181 | 320 | 296 | 560 | 219 | 570 | 420 | 196 | 312 | 425 | 194 | 82 | 262 |
| 12 | 110 | 315 | 85 | 279 | 348 | 189 | 186 | 278 | 222 | 293 | 207 | 343 | 223 | 280 | 267 | 91 | 309 | 83 | 198 | 323 | 299 | 191 | 245 | 256 | 299 |
| 22 | 1 | 74 | 2 | 42 | 163 | 13 | 25 | 55 | 36 | 115 | 24 | 73 | 37 | 84 | 4 | 115 | 35 | 134 | 43 | 20 | 97 | 227 | 78 | ||
| Fatherb: | |||||||||||||||||||||||||
| 11 | 461 | 218 | 453 | 303 | 113 | 360 | 356 | 286 | 365 | 217 | 399 | 255 | 165 | 268 | 255 | 493 | 142 | 492 | 374 | 215 | 280 | 354 | 215 | 68 | 223 |
| 12 | 92 | 273 | 89 | 210 | 293 | 186 | 168 | 235 | 190 | 287 | 162 | 313 | 203 | 238 | 246 | 79 | 288 | 76 | 169 | 246 | 238 | 193 | 212 | 238 | 241 |
| 22 | 9 | 73 | 9 | 43 | 162 | 20 | 34 | 39 | 17 | 59 | 14 | 0 | 55 | 43 | 69 | 1 | 128 | 0 | 21 | 109 | 52 | 12 | 61 | 191 | 92 |
| Control childc: | |||||||||||||||||||||||||
| 11 | 1,312 | 598 | 1,323 | 806 | 336 | 1,062 | 1,046 | 787 | 1,004 | 590 | 1,056 | 738 | 450 | 762 | 680 | 1,365 | 477 | 1,379 | 1,032 | 508 | 833 | 1,021 | 541 | 189 | 633 |
| 12 | 266 | 758 | 248 | 664 | 816 | 502 | 459 | 668 | 530 | 750 | 509 | 866 | 562 | 705 | 732 | 250 | 795 | 228 | 502 | 795 | 655 | 506 | 574 | 665 | 715 |
| 22 | 9 | 207 | 9 | 117 | 455 | 48 | 71 | 134 | 66 | 246 | 52 | 0 | 154 | 102 | 185 | 6 | 303 | 0 | 72 | 302 | 125 | 43 | 194 | 552 | 216 |
| DCS case: | |||||||||||||||||||||||||
| Case childd: | |||||||||||||||||||||||||
| 11 | 426 | 203 | 455 | 249 | 104 | 347 | 341 | 258 | 316 | 195 | 365 | 251 | 135 | 258 | 222 | 453 | 148 | 453 | 322 | 147 | 281 | 332 | 173 | 73 | 194 |
| 12 | 85 | 232 | 66 | 226 | 270 | 165 | 153 | 222 | 181 | 242 | 150 | 277 | 182 | 221 | 251 | 79 | 264 | 77 | 176 | 276 | 219 | 156 | 198 | 200 | 235 |
| 22 | 4 | 82 | 3 | 43 | 155 | 18 | 20 | 49 | 28 | 81 | 20 | 0 | 61 | 39 | 56 | 1 | 97 | 0 | 28 | 102 | 31 | 26 | 53 | 195 | 76 |
| Iowan control: | |||||||||||||||||||||||||
| Mothere: | |||||||||||||||||||||||||
| 11 | 293 | 139 | 288 | 192 | 66 | 274 | 260 | 196 | 270 | 132 | 248 | 171 | 103 | 173 | 150 | 310 | 134 | 306 | 230 | 118 | 174 | 243 | 185 | 65 | 119 |
| 12 | 77 | 170 | 81 | 139 | 219 | 116 | 119 | 163 | 99 | 184 | 124 | 213 | 179 | 175 | 155 | 77 | 178 | 77 | 147 | 192 | 160 | 93 | 116 | 159 | 164 |
| 22 | 3 | 53 | 6 | 23 | 110 | 5 | 9 | 20 | 12 | 57 | 21 | 46 | 34 | 41 | 2 | 70 | 14 | 65 | 36 | 23 | 42 | 145 | 54 | ||
| Fatherf: | |||||||||||||||||||||||||
| 11 | 267 | 122 | 261 | 183 | 66 | 226 | 235 | 178 | 218 | 113 | 219 | 150 | 106 | 150 | 127 | 293 | 107 | 273 | 214 | 100 | 181 | 189 | 141 | 51 | 108 |
| 12 | 62 | 151 | 68 | 113 | 171 | 124 | 97 | 141 | 109 | 167 | 115 | 189 | 143 | 152 | 140 | 53 | 168 | 67 | 115 | 182 | 135 | 111 | 115 | 141 | 152 |
| 22 | 2 | 38 | 6 | 18 | 125 | 12 | 18 | 29 | 16 | 52 | 19 | 0 | 46 | 23 | 43 | 3 | 64 | 0 | 18 | 57 | 16 | 16 | 39 | 131 | 46 |
| Childg: | |||||||||||||||||||||||||
| 11 | 1,531 | 739 | 1,497 | 993 | 386 | 1,387 | 1,288 | 1,002 | 1,317 | 655 | 1,299 | 862 | 543 | 942 | 773 | 1,705 | 672 | 1,558 | 1,208 | 622 | 993 | 1,221 | 815 | 344 | 603 |
| 12 | 373 | 884 | 398 | 700 | 993 | 610 | 589 | 810 | 598 | 907 | 663 | 1,066 | 738 | 822 | 824 | 327 | 924 | 380 | 710 | 993 | 774 | 567 | 681 | 848 | 801 |
| 22 | 17 | 250 | 33 | 150 | 672 | 69 | 79 | 158 | 87 | 305 | 87 | 0 | 272 | 145 | 245 | 11 | 342 | 0 | 90 | 331 | 144 | 87 | 246 | 687 | 354 |
| Iowan case: | |||||||||||||||||||||||||
| Motherh: | |||||||||||||||||||||||||
| 11 | 407 | 208 | 392 | 254 | 112 | 360 | 322 | 253 | 361 | 171 | 357 | 227 | 142 | 255 | 203 | 465 | 174 | 406 | 312 | 182 | 252 | 327 | 204 | 100 | 146 |
| 12 | 104 | 237 | 104 | 191 | 248 | 162 | 164 | 214 | 152 | 226 | 171 | 266 | 167 | 202 | 231 | 83 | 246 | 93 | 192 | 247 | 210 | 157 | 192 | 237 | 231 |
| 22 | 7 | 68 | 12 | 52 | 181 | 21 | 24 | 42 | 26 | 82 | 21 | 77 | 46 | 77 | 3 | 91 | 30 | 84 | 44 | 21 | 81 | 173 | 98 | ||
| Fatheri: | |||||||||||||||||||||||||
| 11 | 341 | 156 | 327 | 208 | 84 | 299 | 270 | 209 | 271 | 132 | 277 | 178 | 113 | 215 | 185 | 387 | 158 | 340 | 274 | 129 | 230 | 283 | 169 | 77 | 125 |
| 12 | 77 | 203 | 83 | 163 | 210 | 125 | 130 | 167 | 147 | 191 | 152 | 235 | 137 | 171 | 182 | 57 | 195 | 78 | 143 | 216 | 152 | 119 | 156 | 193 | 155 |
| 22 | 3 | 52 | 5 | 40 | 145 | 18 | 17 | 41 | 22 | 73 | 16 | 0 | 55 | 21 | 50 | 1 | 59 | 0 | 17 | 81 | 26 | 13 | 50 | 138 | 104 |
| Childj: | |||||||||||||||||||||||||
| 11 | 337 | 162 | 321 | 189 | 103 | 312 | 275 | 221 | 290 | 128 | 299 | 199 | 106 | 213 | 165 | 407 | 143 | 357 | 247 | 140 | 230 | 263 | 142 | 75 | 106 |
| 12 | 69 | 192 | 73 | 182 | 186 | 114 | 127 | 177 | 140 | 190 | 151 | 222 | 146 | 168 | 176 | 65 | 222 | 76 | 151 | 203 | 148 | 131 | 155 | 190 | 169 |
| 22 | 9 | 40 | 8 | 26 | 159 | 15 | 13 | 30 | 19 | 67 | 17 | 0 | 47 | 27 | 57 | 3 | 44 | 0 | 34 | 79 | 28 | 17 | 51 | 118 | 96 |
Allele/frequency: AHR_snp1/.087, AHR_snp2/.362, CYP1A1/.279, CYP1A2/.279, CYP1B1_snp1/.518, CYP1B1_snp2/.165, CYP2E1/.185, EPHX1_snp1/.301, EPHX1_snp2/.226, GSTA4_snp1/.406, GSTA4_snp2/.194, GSTM1_null/.528, GSTM1_snp1/.387, GSTM3/.278, GSTP1_snp1/.336, GSTP1_snp2/.076, GSTP1_snp3/.419, GSTT1_null/.127, HIF1A/.205, NAT1_snp1/.453, NAT2_snp2/.294, NQO1/.182, SULT1A1/.41, UGT1A7_snp1/.628, and UGT1A7_snp2/.356.
Allele/frequency: AHR_snp1/.098, AHR_snp2/.371, CYP1A1/.097, CYP1A2/.266, CYP1B1_snp1/.543, CYP1B1_snp2/.2, CYP2E1/.211, EPHX1_snp1/.279, EPHX1_snp2/.196, GSTA4_snp1/.36, GSTA4_snp2/.165, GSTM1_null/.551, GSTM1_snp1/.37, GSTM3/.295, GSTP1_snp1/.337, GSTP1_snp2/.071, GSTP1_snp3/.487, GSTT1_null/.134, HIF1A/.187, NAT1_snp1/.407, NAT2_snp2/.3, NQO1/.194, SULT1A1/.342, UGT1A7_snp1/.624, and UGT1A7_snp2/.382.
Allele/frequency: AHR_snp1/.089, AHR_snp2/.375, CYP1A1/.084, CYP1A2/.283, CYP1B1_snp1/.537, CYP1B1_snp2/.185, CYP2E1/.191, EPHX1_snp1/.295, EPHX1_snp2/.207, GSTA4_snp1/.392, GSTA4_snp2/.19, GSTM1_null/.54, GSTM1_snp1/.373, GSTM3/.29, GSTP1_snp1/.345, GSTP1_snp2/.081, GSTP1_snp3/.445, GSTT1_null/.142, HIF1A/.201, NAT1_snp1/.436, NAT2_snp2/.281, NQO1/.189, SULT1A1/.367, UGT1A7_snp1/.629, and UGT1A7_snp2/.367.
Allele/frequency: AHR_snp1/.09, AHR_snp2/.383, CYP1A1/.069, CYP1A2/.301, CYP1B1_snp1/.548, CYP1B1_snp2/.19, CYP2E1/.188, EPHX1_snp1/.302, EPHX1_snp2/.226, GSTA4_snp1/.39, GSTA4_snp2/.178, GSTM1_null/.525, GSTM1_snp1/.402, GSTM3/.289, GSTP1_snp1/.343, GSTP1_snp2/.076, GSTP1_snp3/.45, GSTT1_null/.145, HIF1A/.221, NAT1_snp1/.457, NAT2_snp2/.265, NQO1/.202, SULT1A1/.358, UGT1A7_snp1/.63, and UGT1A7_snp2/.383.
Allele/frequency: AHR_snp1/.111, AHR_snp2/.381, CYP1A1/.124, CYP1A2/.261, CYP1B1_snp1/.556, CYP1B1_snp2/.159, CYP2E1/.177, EPHX1_snp1/.268, EPHX1_snp2/.161, GSTA4_snp1/.399, GSTA4_snp2/.211, GSTM1_null/.555, GSTM1_snp1/.413, GSTM3/.318, GSTP1_snp1/.342, GSTP1_snp2/.104, GSTP1_snp3/.416, GSTT1_null/.201, HIF1A/.224, NAT1_snp1/.429, NAT2_snp2/.314, NQO1/.194, SULT1A1/.292, UGT1A7_snp1/.608, and UGT1A7_snp2/.404.
Allele/frequency: AHR_snp1/.1, AHR_snp2/.365, CYP1A1/.119, CYP1A2/.237, CYP1B1_snp1/.581, CYP1B1_snp2/.204, CYP2E1/.19, EPHX1_snp1/.286, EPHX1_snp2/.206, GSTA4_snp1/.408, GSTA4_snp2/.217, GSTM1_null/.558, GSTM1_snp1/.398, GSTM3/.305, GSTP1_snp1/.365, GSTP1_snp2/.085, GSTP1_snp3/.437, GSTT1_null/.197, HIF1A/.218, NAT1_snp1/.437, NAT2_snp2/.252, NQO1/.226, SULT1A1/.327, UGT1A7_snp1/.624, and UGT1A7_snp2/.399.
Allele/frequency: AHR_snp1/.106, AHR_snp2/.369, CYP1A1/.12, CYP1A2/.271, CYP1B1_snp1/.57, CYP1B1_snp2/.181, CYP2E1/.191, EPHX1_snp1/.286, EPHX1_snp2/.193, GSTA4_snp1/.406, GSTA4_snp2/.204, GSTM1_null/.553, GSTM1_snp1/0.413, GSTM3/.291, GSTP1_snp1/.357, GSTP1_snp2/.085, GSTP1_snp3/.415, GSTT1_null/.196, HIF1A/.222, NAT1_snp1/.425, NAT2_snp2/.278, NQO1/.198, SULT1A1/.337, UGT1A7_snp1/.591, and UGT1A7_snp2/.429.
Allele/frequency: AHR_snp1/.114, AHR_snp2/.364, CYP1A1/.126, CYP1A2/.297, CYP1B1_snp1/.564, CYP1B1_snp2/.188, CYP2E1/.208, EPHX1_snp1/.293, EPHX1_snp2/.189, GSTA4_snp1/.407, GSTA4_snp2/.194, GSTM1_null/.54, GSTM1_snp1/.416, GSTM3/.292, GSTP1_snp1/.377, GSTP1_snp2/.081, GSTP1_snp3/.419, GSTT1_null/.186, HIF1A/.236, NAT1_snp1/.404, NAT2_snp2/.294, NQO1/.197, SULT1A1/.371, UGT1A7_snp1/.572, and UGT1A7_snp2/.449.
Allele/frequency: AHR_snp1/.099, AHR_snp2/.373, CYP1A1/.112, CYP1A2/.296, CYP1B1_snp1/.569, CYP1B1_snp2/.182, CYP2E1/.197, EPHX1_snp1/.299, EPHX1_snp2/.217, GSTA4_snp1/.426, GSTA4_snp2/.207, GSTM1_null/.569, GSTM1_snp1/.405, GSTM3/.262, GSTP1_snp1/.338, GSTP1_snp2/.066, GSTP1_snp3/.38, GSTT1_null/.187, HIF1A/.204, NAT1_snp1/.444, NAT2_snp2/.25, NQO1/.175, SULT1A1/.341, UGT1A7_snp1/.575, and UGT1A7_snp2/.473.
Allele/frequency: AHR_snp1/.105, AHR_snp2/.345, CYP1A1/.111, CYP1A2/.295, CYP1B1_snp1/.563, CYP1B1_snp2/.163, CYP2E1/.184, EPHX1_snp1/.277, EPHX1_snp2/.198, GSTA4_snp1/.421, GSTA4_snp2/.198, GSTM1_null/.527, GSTM1_snp1/.401, GSTM3/.272, GSTP1_snp1/.364, GSTP1_snp2/.075, GSTP1_snp3/.379, GSTT1_null/.176, HIF1A/.253, NAT1_snp1/.428, NAT2_snp2/.251, NQO1/.201, SULT1A1/.369, UGT1A7_snp1/.556, and UGT1A7_snp2/.487.
Embryonic Gene Expression
Embryonic-expression profiles of the detoxification genes used in this study were investigated to confirm that the genes were expressed in the relevant tissue during the relevant interval of embryonic development. One source of expression data is the COGENE (Craniofacial and Oral Gene Expression Network) Project, which investigates human gene-expression changes that occur during early stages of development, with a focus on craniofacial regions. Normal-appearing human embryos from RU-486–induced abortions—that were free of autosomal trisomies and for which no parental epidemiologic data were available—were used to generate RNA for micoarray analysis. Affymetrix microarray analysis was performed on microdissected, normal human craniofacial structures of 25 target tissues/stages to construct fetal gene-expression profiles.18
Statistical Analysis
Statistical analyses were performed separately on samples from Iowa and Denmark, and P values from these two sources were combined using Fisher’s method, as described in the “Multiple-Comparison Considerations” section. We also performed analysis on the basis of aggregated data, to improve estimation of RRs, if the estimates were compatible in the separate analysis. Since there were only a few families (n=12) with multiple affected offspring, one affected child was randomly chosen from such families to be included in the analysis. The data were analyzed under two statistical frameworks: a log-linear model (for case-parents triads) and logistic regression analysis (for case-control comparisons). Given that the outcome under study is rare, the RR parameters estimated under the former are equivalent to the odds ratio (OR) parameters estimated under the latter. When using the log-linear model in the analysis, we first used a 2-df test, which places no assumptions on the underlying genetic model (see the “Log-Linear Framework” section below for a detailed description). We then fitted the data in a log-additive model when a significant result was observed.
Multiple-Comparison Considerations
Interactions between genetic and environmental factors are statistically difficult to study; they are second-order effects, and many different combinations must be considered. Therefore, the present analysis faced a multiple-comparison issue. One can simply multiply the P values by the number of tests performed, in a full Bonferroni correction, but this method has rightly been described as “punitively conservative.”19(p505) Here is our approach. We consider that there are three levels of analysis performed here. Our primary interest is to identify interactions between genetic variants—in either the mother or the fetus—and the exposure: maternal smoking. For our 25 SNPs, this implies that we must include 50 separate tests in our formal hypothesis–testing framework. There are also two phenotype categories (CPO and CL/P) and two populations (Iowan and Danish). For the overall formal-testing framework, we combined the phenotypes and then also combined the P values for the two populations, using Fisher’s method.20 We considered an interaction to be significant within this framework if the P value is <.05/50—that is, 0.001. Another level of analysis has to do with the main effects for these factors, such as smoking, which are already known or strongly suspected to be related to risk or to the metabolism of xenobiotics. We provide an analysis of these, for improved estimation of their RRs. A third level should be considered exploratory, rather than providing formal-hypothesis tests, and that exploratory category includes gene-by-gene interactions, differences between the Danish and Iowan populations, and differences between the two subphenotypes. For the second and third sets of analyses, the P values should be taken as an index of strength of evidence rather than as providing formal-hypothesis tests.
Log-Linear Framework
Major gene effects
We used the log-linear approach to study the effects of fetal and maternal genotypes21,22 in family-based analyses. The expectation-maximization algorithm was applied, for full use of the families with a missing parental genotype.23 The log-linear approach tests fetal genetic effects by comparing the distribution of the case genotype after stratification by parental genotypes with the expected distribution under Mendelian inheritance. The unit of analysis is the triad, consisting of an affected offspring and the two parents. Tests of maternally mediated genetic effects—that is, effects on the fetus that are due to effects of the maternal genotype on the maternal phenotype during pregnancy—are based on the symmetry assumption of allele counts between the mothers and the fathers in the source population, as defined by Schaid and Sommer.24 The log-linear approach provides likelihood-ratio tests (LRTs) of the genetic effects as well as maximum-likelihood estimators of the genetic RRs for both offspring-mediated and mother-mediated genetic effects.21 This approach has the advantage of allowing for different RRs that correspond to carrying one and carrying two copies of a susceptibility-related allele, relative to no copies. Simulation studies have shown the log-linear approach to be more powerful under a dominant or a recessive disease model than with another transmission/disequilibrium–based test.25 A limitation of the triad approach is that, unlike the case-control design, it does not provide a way to detect effects of exposures on risk, although one can assess multiplicative interactions between exposure and genotypes.26
Effects of gene and maternal-smoking interaction
For the Iowan samples, smoking status during the critical period between 3 mo before and 3 mo after conception served as the indicator variable for smoking in testing gene–maternal-smoking interaction effects. For the Danish sample, for which the smoking exposure before conception was not ascertained directly, smoking status in the 1st trimester was used. For the triad-based analyses, we applied an extension of the log-linear approach26 in analyzing interaction effects between maternal genotypes and maternal cigarette smoking. We applied the polytomous logistic-regression approach,27 originally developed for testing linkage and association in relation to quantitative traits (QTs), in analysis of fetal-gene and maternal-smoking interaction effects, by substituting the maternal-smoking status for the QT. This approach compares the allele transmission to affected offspring for triads with smoking versus triads with nonsmoking mothers. A multiplicative interaction would be evidenced by a difference between the two transmission patterns.
Gene-gene interaction
The log-linear framework for testing gene-environment interaction was adapted for testing gene-gene interaction by treating one genetic factor as the environmental factor and applying the method described in the “Effects of gene and maternal-smoking interaction” section. To reduce the degrees of freedom of the test statistic by using a more parsimonious alternative model formulation, we assumed a dominant or a recessive effect of the genetic factor. The test does require that the “exposure” and the inherited genotype be independent, conditional on the parental genotypes and hence could be validly used only for unlinked loci. We included in this analysis two genes, IRF6 (MIM 607199)28 and MSX1 (MIM 142983),29 that have elsewhere shown association with OC and on which genotype data were available for the samples used in the present study.
Heterogeneity in genetic RR
When heterogeneity in genetic RRs exists between different populations, it is not appropriate to combine the data for analysis. We used the log-linear framework for testing gene-exposure interaction to test for the heterogeneity in genetic RRs across studies by treating population group as the exposure variable. When there was no evidence of heterogeneity, we tested fetal or maternal genetic effects, using the combined data.
Logistic-Regression Framework
GSTT1- and GSTM1-null deletion polymorphisms present a challenge, in that the genotyping method we applied can detect only the presence or absence of the normal allele, not the exact copy number (1 vs. 2 alleles). Logistic-regression analyses were applied in estimation of genetic, maternal, and gene-smoking interaction effects of these two polymorphisms for all the markers in case-control comparisons. For triad samples, we adjusted for parental genotypes, if available, in testing for child genetic effects or child gene–maternal smoking interaction.
In analysis of the combined data from different studies, we included a variable indicating the specific substudy, together with interactions between that indicator and the other covariates, whereby the parameters of the tested effects were constrained to be the same across substudies and all other parameters were free to vary. We fit logistic models both with and without adjustment of other epidemiologic factors, such as vitamin intake and alcohol use.30 Data collected from the DBS samples were analyzed using logistic regression in similar ways.
We applied the Cochran-Armitage trend test30 to assess the dosage effect of cigarette smoking (using smoking amounts as consecutive integers). We also calculated the attributable fraction for the interaction between GSTT1 and maternal smoking. Population attributable fraction for interaction on a multiplicative scale provides an estimate of the reduction in OC occurrence that would result from eliminating the interaction.31
Results
Smoking Effects
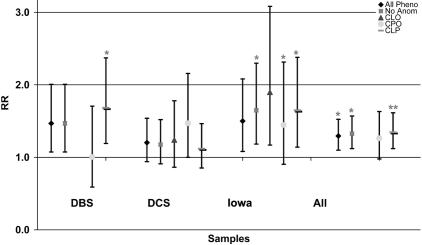
We observed an increased risk of CL/P in the offspring of smoking mothers in both the Danish (OR=1.53; 95% CI 1.08–2.16) and Iowan (OR=1.62; 95% CI 1.06–2.50) case-control substudies, after adjusting for maternal perinatal multivitamin and alcohol use (fig. 1). To quantify tobacco use, we divided maternal smoking into four groups for analysis (in the Danish samples: group 1, 0/d; group 2, 1–9/d; group 3, 10–19/d; group 4, ⩾ 20/d; in the Iowan samples: group 1, 0/d; group 2, 1–4/d; group 3, 5–15/d; group 4, >15/d). Mothers who smoked 10–19 cigarettes/d in the DBS samples and those who smoked >5 cigarettes/d in the Iowan samples had the highest risk of OC (table 6). We saw ORs of 1.33 (95% CI 1.09–1.62) for CL/P and 1.29 (95% CI 0.97–1.73) for CPO in the smoking mothers in the unified data set (Iowa and Denmark), after adjustment for sample origins and maternal perinatal multivitamin use and alcohol intake (fig. 1). It should be noted that, although the point estimate of OR is higher for the 10–19/d group than for the >20/d group in the Danish samples, this most likely results from the small sample size of the >20/d group, as indicated by the observation that the 95% CI of OR for the 10–19/d groups is contained within that for the >20/d group. Using the Cochran-Armitage trend test, we also identified dosage effect of maternal smoking for both populations (data not shown). Maternal perinatal multivitamin or alcohol use did not show significant association with OC, after adjustment for maternal smoking (data not shown).
Figure 1. .
ORs of maternal cigarette smoking in different population and different phenotypic subgroups. All Pheno = all subjects with OC (with or without other anomalies); No Anom = subjects with OC and without other associated anomalies.
Table 6. .
Effects of Smoking by Cigarettes per Day
| Phenotype, Population, and No. of Daily Cigarettes |
OR | 95% CI |
| All OCs: | ||
| DBS: | ||
| 1–9 | 1.12 | .65–1.92 |
| 10–19 | 1.55 | 1.06–2.25 |
| 20+ | .93 | .42–2.06 |
| Iowa: | ||
| 1–4 | .84 | .42–1.70 |
| 5–14 | 2.17 | 1.17–4.03 |
| 15+ | 2.17 | 1.15–4.07 |
| CL/P: | ||
| Denmark: | ||
| 1–9 | 1.14 | .62–2.08 |
| DBS: | ||
| 10–19 | 1.71 | 1.14–2.56 |
| 20+ | 1.16 | .51–2.64 |
| Iowa: | ||
| 1–4 | .93 | .42–2.07 |
| 5–14 | 2.24 | 1.11–4.50 |
| 15+ | 2.78 | 1.42–5.44 |
| CPO: | ||
| Denmark: | ||
| 1–9 | 1.03 | .42–2.57 |
| DBS: | ||
| 10–19 | 1.17 | .61–2.23 |
| 20+ | .35 | .05–2.67 |
| Iowa: | ||
| 1–4 | .91 | .30–2.75 |
| 5–14 | 2.52 | 1.09–5.81 |
| 15+ | 1.58 | .59–4.21 |
Genetic Effects
We tested a variety of genetic effects, including fetal genotype and maternal genotype, maternal smoking by gene (fetal or maternal gene) interaction, and gene-by-gene interaction effects. Tables A1–A7 represent a subset of these results that are at least suggestive of a possible effect (P⩽.03). The P values listed in tables 7 and A1–A7 have not been adjusted for multiple comparisons. In table A8, we summarized the markers with P values <.05 for individual tests in the three studied sample sets and for different effect types (fetal genotype, maternal genotype, fetal genotype and smoking interaction, and maternal genotype and smoking interaction).
Table 7. .
Transmission/Disequilibrium Tests of NAT2 G590A Marker
| No. of Subjects with Transmission |
|||||
| Sample | G | A | No. of Triads (1,1,1)a | χ2 | P |
| Case set 1b | 278 | 204 | 58 | 9.2 | .002 |
| Control set 1b | 313 | 248 | 62 | 6.2 | .013 |
| Case set 2c | 357 | 275 | 78 | 8.5 | .003 |
| Control set 2d | 450 | 434 | 111 | .2 | .630 |
| All casese | 635 | 479 | 136 | 17.6 | 3×10-5 |
| All controls | 763 | 682 | 173 | 3.7 | .056 |
Triad (1,1,1) represents a triad in which mother, father, and child are all heterozygous (GA).
Set 1 consists of the Iowa and DCS samples that are used in testing other markers.
Case set 2 consists of the Philippine triad samples and an additional set of Danish familial samples.
Control set 2 consists of the CEPH samples, affected siblings in the second set of Danish familial samples, and white control triad samples.
“All cases” and “All controls” represent the combined data set of set 1 and set 2.
Maternal effects
Tests on maternal effects, either maternal genotype effects or interaction effects between maternal genotype and maternal smoking, identified certain genes that may play a role through maternal genotypes, including EPHX1, GSTP1 (MIM 134660), NAT2, and UGT1A7 (MIM 606432) (tables A1 and A2).
Fetal effects
Tests specific for genetic effects of the offspring genotype identified suggestive effects of variants in the CYP1A1, CYP1A2 (MIM 124060), CYP1B1 (MIM 601771), GSTA4 (MIM 605450), GSTP1, and NAT2 genes (table A3). Whereas we saw most of the apparent effects in either the Iowan or the Danish samples but not in both, for variants in CYP1A1 and NAT2, the overall results suggest an effect in both populations. The risk of OC was estimated to decrease by 20% (40%) and 30% (50%) respectively, when a child carries 1 copy (2 copies) of the variant allele of CYP1A1 or NAT2.
Fetal effects: GSTT1
Several genes—including EPHX1, GSTA4, GSTP1, UGT1A7 (table A4), and GSTT1 (tables 8, A5, and A6)—showed significant interaction effects between the fetal genotype and maternal smoking, with the effect of GSTT1 remaining significant after correction for multiple comparisons. Similar to results for fetal genetic effects, the apparent interaction effects were usually observable in only one population. However, interaction effects between the GSTT1-null genotype and maternal smoking were detected in the Iowan (P=.003) and DBS samples (P=.03) (table A5), with a Fisher’s combined P value of .0009, which remained significant after Bonferroni correction for 50 tests. An estimated increase in risk of CL/P was observed when the child carried the null genotype and the mother smoked. The interaction effects were stronger in the CPO subgroup in both populations, and the combined data showed even more-significant results (P=.001) (table A5). ORs of maternal smoking for OC were most elevated in the GSTT1-null fetuses whose mothers smoked 10–19 cigarettes/d for the Danish samples (OR=4.2; 95% CI 1.4–12.4) and in the GSTT1-null genotyped fetuses whose mothers smoked >15 cigarettes/d for the Iowan samples (OR=17.1; 95% CI 2.1–141.4) (table 8). The Cochran-Armitage trend tests detected significant dosage effects in the GSTT1-null genotyped subjects with OC in both the Danish samples and the Iowan samples (table 8). Table A6 shows the 2×4 table for studying gene-environment interaction effects suggested by Botto et al.32
Table 8. .
ORs by Fetal GSTT1 Genotype and Maternal Smoking Categories
| Population, GSTT1 Genotype, and No. of Daily Cigarettes |
Case | Control | OR | 95% CI |
| DBS: | ||||
| +/+ or +/−a: | ||||
| 0 | 113 | 223 | Reference | |
| 1–9 | 34 | 61 | 1.10 | .68–1.77 |
| 10–19 | 54 | 82 | 1.30 | .86–1.96 |
| 20+ | 8 | 18 | .88 | .37–2.08 |
| −/−b: | ||||
| 0 | 18 | 44 | Reference | |
| 1–9 | 8 | 9 | 2.17 | .72–6.52 |
| 10–19 | 12 | 7 | 4.19 | 1.42–12.36 |
| 20+ | 3 | 3 | 2.44 | .45–13.27 |
| Iowa: | ||||
| +/+ or +/−c: | ||||
| 0 | 172 | 180 | Reference | |
| 1–4 | 8 | 15 | 0.56 | .23–1.35 |
| 5–14 | 22 | 14 | 1.64 | .82–3.32 |
| 15+ | 24 | 14 | 1.79 | .90–3.58 |
| −/−d: | ||||
| 0 | 30 | 57 | Reference | |
| 1–4 | 3 | 3 | 1.90 | .36–10 |
| 5–14 | 6 | 2 | 5.70 | 1.83–29.99 |
| 15+ | 9 | 1 | 17.10 | 2.07–141.4 |
Cochran-Armitage trend test Z=.83; P=.409.
Cochran-Armitage trend test Z=2.56; P=.011.
Cochran-Armitage trend test Z=1.85; P=.064.
Cochran-Armitage trend test Z=3.87; P=.0001.
With the assumption of a smoking prevalence of 25% in pregnant women and a GSTT1-deletion prevalence of 15%, the attributable fraction of the interaction on a multiplicative scale between GSTT1 deletion and maternal smoking is calculated to be 6%.31 The parameter estimates show that the GSTT1-null genotype confers a considerable risk when combined with smoking (double the risk) and no risk (maybe even a protective effect) if the mothers do not smoke.
Fetal effects: NAT2
Although fetal genetic effects were not part of the formal hypothesis–testing framework, the result of the NAT2 G590A marker is of special interest. The NAT2 G590A SNP showed association in both the Iowan (P=.04) and the Danish samples (P=.01). The test of heterogeneity in genetic RRs between the Iowan and Danish samples was not significant and supported the pooling of genotype data for analysis. The NAT2 G590A marker also demonstrated significant transmission distortion in the initial studied control samples (see the “Danish samples” and “Iowan population” sections) as well, although to a less extreme degree (P=.021) (last line of table A3). To further evaluate the apparent overtransmission, we investigated additional case and control samples. The overtransmission at the NAT2 590 locus continued to be observed in the case samples but was no longer observed in the control samples (table 7).
Gene-Gene Interaction Effects
Preliminary analyses also suggested gene-gene interactions between several pairs of genes (table A7).
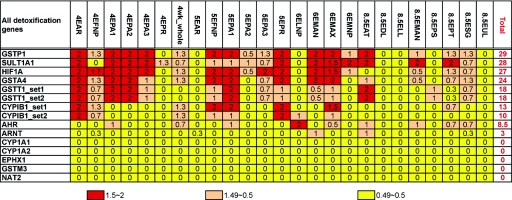
Expression Data
Expression profiles of the detoxification genes used in this study were investigated using COGENE expression data, as summarized in figure 2. Over 97% of human genes show expression below the level shown by GSTT1 in this study. GSTT1 demonstrated an elevated expression in the human craniofacial embryonic tissues, and its expression correlates with that of fibroblast growth factor receptors 1 and 2 (FGFR1 [MIM 136350] and FGFR2 [MIM 176943])33 (data not shown), genes that are known to be involved in craniofacial development. Several genes involved in detoxification of exogenous exposures are also thought to play a role in metabolism of endogenous morphogens in the fetus, to regulate development, so the role of these gene products may be different in the mother and the fetus.34
Figure 2. .
COGENE expression summary of detoxification genes. For evaluation of the expression level, the numerical values were assigned to the results of chip analysis with p (present)=2, m (marginal)=1, and a (absent)=0, and expression scores were calculated from the average of the replicate experiments. The column headings indicate the developmental stage and sample tissue.
Discussion
We report here a comprehensive study of effects of polymorphisms in genes involved in detoxification pathways and the interaction effects between these genetic variants and maternal cigarette smoking on a common birth defect: OC. The study benefited from large sample sizes in both Denmark and Iowa, which allowed comparisons and replications.
Maternal cigarette smoking adversely affects the health of both the mother and child, with increased risks of low birth weight, premature rupture of membranes, placenta previa, neonatal mortality, and stillbirth.35 It also causes increased health-care expenditures. The estimated smoking-attributable neonatal expenditures were $366 million, or $704 per maternal smoker, in the United States in 1996.36 In the present study, we identified an increased risk of OC in the offspring of smoking mothers in Denmark and Iowa. This finding lends further support for advocation of smoking-cessation programs.
Studies of gene-environment interaction effects have become increasingly important for complex traits such as clefting, whose etiology probably involves both genes and environmental factors. Gene-knockout mice that lack expression of AHR and certain CYPs (CYP1A1, CYP1A2, CYP1B1, CYP2E1 [MIM 124040]), EPHX1, GSTP1, GSTA4, and NQO1 [MIM 125860]) and are raised in a clean environment have no deleterious phenotypes, indicating that these genes have no direct roles in murine embryonic development. However, the knockout mice do have different response profiles when challenged with toxicants and carcinogens.37–39 This accentuates the importance of studying gene-environment interactions. Of the detoxification genes showing suggestive gene-environment interaction effects in this study, EPHX1, GSTA4, and GSTP1 were reported to be expressed in developing embryos, and the COGENE data showed elevated expression of GSTP1, GSTA4, GSTT1, and HIF1A (MIM 603348) at the critical regions and time during craniofacial development. In addition, an association was observed between the fetal genotype in GSTA4 and clefting (table A4), although it was seen in the Danish and not in the Iowan data. Such differences could suggest that the variant we studied is a marker in linkage disequilibrium with a clefting locus in the Danish but not in the Iowan population.
Preferential transmission of the common allele of NAT2 G590A was observed in samples from the Iowan and Danish populations (table 7). However, this preferential transmission was also observed, albeit with less transmission distortion, in unaffected controls. Accordingly, a supplemental study was performed using additional control samples (see the “Danish samples” and “Iowan population” sections). No preferential transmission was seen in the second (larger) set of controls (P=.63), and the apparent transmission distortion was of only borderline significance for all controls combined (P=.056). Several possible explanations can be proposed, including sampling variation, systematic genotyping mistakes, actual effects of the variant allele on fetal survival, and segregation distortion. Consistency in genotypes of >300 samples, through use of kinetic PCR and Taqman assays designed for this marker and DNA sequencing of 90 samples, argues against genotyping errors. Zöllner et al. reported evidence of extensive transmission ratio distortion in several regions in the human genome, one of which was located on chromosome 8, coinciding with the chromosomal location of NAT2.40 In the final, complete data set for the NAT2 G590A marker, we observed overtransmission in the OC samples but not in the controls, which argues for an etiologic role of this variant. One other study9 has investigated the association between variants in the fetal NAT2 gene—including G590A—and clefting, and the researchers did not detect a significant association with the NAT2 G590A marker. That study, however, was performed using a case-control study design, which could be subject to bias due to genetic population stratification. Further study of the NAT2 G590A locus is required to fully understand the observed overtransmission. NAT2 is a member of the N-acetyltransferases family, which comprises two protein-encoding genes (NAT1 and NAT2) and a pseudogene. NAT2 catalyzes the N-acetylation of carcinogens and other xenobiotics such as arylamines, hydrazines, and hydrazides.41 NAT2 G590A has reduced catalytic activity and protein stability in vitro,42 and, whereas the COGENE data did not identify craniofacial embryonic expression for NAT2, the homolog, NAT1, is expressed in mouse fetal liver.43
GSTT1 is widely studied in the investigations of environmental effects, and the null genotype has been associated with increased chromosome aberrations and certain cancers.44 Using a case-control design, Van Rooij et al. reported increased risk of OC when mothers carry the GSTT1-null genotype and smoke cigarettes or when both mothers and infants carry the null genotype and mothers smoke.12 We saw increases in risk of OC associated with the GSTT1-null genotypes in the fetus in the Iowan and DBS components of our study and similar estimated ORs. In a population-based case-control study, Lammer et al. identified a doubling of risk of cleft lip only (CLO) for fetuses who had both a null genotype for GSTT1 and a mother who smoked, and there was a nearly sixfold increased risk of CL for fetuses with a combination of a mother who smoked and an absence of GSTM1 and GSTT1.45 The COGENE data show that GSTT1 is highly expressed in developing fetal craniofacial structures, which suggests that its absence in null-genotype individuals could contribute to abnormalities. One possibility is that GSTT1 is present in the embryo to metabolize endogenous morphogens, as has been suggested for the cytochrome P450 pathway genes and retinoic acid.34 Absence of GSTT1 in the presence of xenobiotics present in cigarette smoke might result in disruptions of normal signaling. These results are consistent with our finding that fetal GSTT1-null genotype combined with maternal smoking increases the risk of OC. In the present study, the estimated RR for the GSTT1-null mothers who smoked during pregnancy was 2.38, whereas that for the GSTT1-null mothers who did not smoke was 0.67.
We used a Bonferroni correction to correct our gene-environment interaction analysis for multiple-testing, and we observed significant association for interactive effects between fetal GSTT1-null genotype and maternal smoking, after the correction. Nevertheless, it is worth noting that, for such complex diseases as OC, multiple genetic and environmental factors are involved in the etiology, each contributing, at best, a moderate effect. It is likely that true-positive signals will be missed with strict adherence to the multiple-testing correction rule. Rather, the significant results identified in the present study provide preliminary evidence for their involvement, and future confirmation studies are required.
The specific SNPs selected for analysis may not be functionally important variants but may serve as markers in linkage disequilibrium with a functionally relevant haplotype. As markers, some SNPs may be more informative in some populations than in others, and this could explain some of the discrepancies in our analyses between results for the Iowan and the Danish populations. One should note that, even with a power of 0.8 at the causal SNP, the chance of replication of association is 0.64 in two independent samples and 0.51 in three independent samples.
In summary, our investigations of the effects of detoxification genes and maternal smoking on the etiology of clefting are the most comprehensive to date, in terms of the number of genes studied and the number of samples included. Results supporting a role for genetic, maternal, gene-smoking interactive, and gene-gene interaction effects were identified, which provide a valuable resource for future investigations. The demonstration of an interaction between maternal smoking and GSTT1 variants may make it possible for risk counselors to identify couples for whom behavior modification may help substantially reduce OC risk.
Acknowledgments
This study was supported by National Institutes of Health (NIH) grants R01 DE11948, DE08559, and P50 DE016215; by Centers for Disease Control and Prevention grant U50/CCU 71328; and by institutional support from the University of Iowa. This work was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences. We particularly acknowledge the generous participation of the patients and their families and their contribution of DNA resources. We thank Emily Kistner, for sharing the SAS macros with us, and Allen Wilcox and David Umbach, for their helpful comments on the manuscript. We also thank Diana Caprau, Ryan Grady, Susanne Knudsen, Sarah O’Brien, Sandy Daack-Hirsch, and Cathy Dragan for organizing and processing samples. The CEPH has generously provided DNA from families for use in this work. The administrative support of Nancy Davin, Susie McConnell, and Dan Benton is greatly appreciated.
Appendix A
Table A1. .
Maternal Effects[Note]
| Denmark |
Iowa |
Combined Data |
|||||||||||||
| General-Effect Model |
Log-Additive Model |
General-Effect Model |
Log-Additive Model |
General-Effect Model |
Log-Additive Model |
||||||||||
| Gene and Phenotype |
RR1 | RR2 | LRT (P) |
R_A | LRT (P) |
RR1 | RR2 | LRT (P) |
R_A | LRT (P) |
RR1 | RR2 | LRT (P) |
R_A | LRT (P) |
| EPHX1_snp1: | |||||||||||||||
| CL/P | .65 | 1.43 | .003 | .91 | .43 | 1.07 | .78 | .65 | .95 | .70 | .78 | 1.09 | .09 | .93 | .40 |
| CPO | 2.82 | .86 | <.001 | 1.34 | .10 | .96 | .64 | .65 | .86 | .47 | 1.69 | .76 | .01 | 1.11 | .44 |
| GSTP1_snp3: | |||||||||||||||
| CPO | .81 | .32 | .002 | .57 | .001 | 1.18 | 1.88 | .25 | 1.34 | .11 | THRRa | ||||
| GSTP1_snp1: | |||||||||||||||
| CPO | 1.18 | .42 | .02 | .75 | .08 | 1.33 | 2.13 | .27 | 1.42 | .11 | 1.24 | .77 | .25 | .95 | .69 |
| NAT2_snp1: | |||||||||||||||
| Allb | 1.37 | 1.40 | .06 | 1.20 | .03 | .78 | .77 | .28 | .85 | .16 | 1.06 | 1.12 | .69 | 1.06 | .39 |
| No anomaliesc | 1.26 | 1.51 | .08 | 1.23 | .02 | .77 | .79 | .27 | .85 | .18 | 1.01 | 1.18 | .47 | 1.07 | .32 |
| CLO | 1.28 | 1.22 | .61 | 1.12 | .443 | .41 | .33 | .03 | .51 | .02 | THRRa | ||||
| CPO | 1.95 | 2.56 | .01 | 1.64 | .004 | 1.16 | .71 | .46 | .92 | .67 | 1.52 | 1.53 | .09 | 1.28 | .05 |
| UGT1A7_snp2: | |||||||||||||||
| Allb | .84 | .68 | .16 | .83 | .051 | 1.14 | .78 | .193 | .89 | .306 | .96 | .7 | .047 | .86 | .034 |
| CPO | .82 | .42 | .08 | .69 | .037 | 1.61 | .84 | .13 | .91 | .665 | 1.04 | .55 | .048 | .79 | .076 |
Note.— RR1 is the genetic RR when the mother carries one copy of the variant allele; RR2 is that when the mother carries two copies of the variant alleles; R_A is the RR when the mother carries one copy of the variant allele, assumed to be a log additive model, a model in which the RR of carrying two copies of the variant alleles is the square of that of carrying one copy.
For test of heterogeneity in RR (THRR), P<.05.
Includes data from all subjects with nonsyndromic OC.
Includes data from subjects without other minor anomalies.
Table A2. .
Maternal Genotype and Maternal Smoking Interactive Effects[Note]
| Denmark |
Iowa |
||||||
| Gene and Phenotype |
I1 | I2 | LRT (P) |
I1 | I2 | LRT (P) |
Pa |
| NAT2_snp2: | |||||||
| Allb | 2.03 | .87 | .03 | .84 | .63 | .80 | .110 |
| No Anomaliesc | 2.48 | .79 | .006 | .79 | .54 | .68 | |
| UGT1A7_snp1: | |||||||
| Allb | 1.73 | .68 | .01 | 2.58 | 5.58 | .01 | .001e |
| No anomaliesc | 1.71 | .71 | .04 | 2.37 | 5.37 | .01 | |
| CLO | .71 | .36 | .29 | 10.5 | 29.8 | .024 | |
| CPO | 7.76 | 1.51 | .03 | … | … | … | |
Note.— I1 is the estimated interaction effect of one maternal copy of the variant allele together with maternal smoking; I2 is the estimated interaction effect of two maternal copies of the variant allele together with maternal smoking.
For formal-hypothesis testing, we used Fisher’s method to combine P values that were based on data from all OC cases in the Danish and Iowan populations.
Includes data from all subjects with nonsyndromic OC.
Includes data from subjects without other minor anomalies.
Estimates for interaction effects were different in the Danish and Iowan populations.
Table A3. .
Effects of Offspring Genotypes[Note]
| Denmark |
Iowa |
Combined |
|||||||||||||
| Gene/SNP and Phenotype |
RR1 | RR2 | LRT with 2 df (P) |
R_A | Additive Model LRT with 1 df (P) |
RR1 | RR2 | LRT with 2 df (P) |
R_A | Additive Model LRT with 1 df (P) |
RR1 | RR2 | LRT with 2 df (P) |
R_A | Additive Model LRT with 1 df (P) |
| CYP1B1_snp1: | |||||||||||||||
| Alla | 1.2 | 1.2 | .53 | 1.1 | .45 | .7 | .8 | .03 | 1.0 | .71 | 1.0 | 1.0 | .54 | 1.0 | .71 |
| CL/P | 1.2 | 1.2 | .60 | 1.1 | .42 | .6 | .7 | .05 | .8 | .22 | .9 | 1.0 | .72 | 1.0 | 1.00 |
| CPO | 1.0 | .9 | .93 | .9 | .70 | 1.3 | 2.5 | .04 | 1.7 | .02 | 1.1 | 1.4 | .37 | 1.2 | .23 |
| CYP1A1: | |||||||||||||||
| Alla | .6 | .8 | .03 | .7 | .01 | .7 | 1.1 | .20 | .8 | .17 | .7 | 1.0 | .006 | .7 | .007 |
| No anomaliesb | .7 | 1.0 | .11 | .7 | .068 | .6 | .8 | .09 | .7 | .05 | .7 | .9 | .01 | .7 | .008 |
| CYP1A2: | |||||||||||||||
| CPO | 2.1 | .8 | .005 | 1.5 | .05 | 1.4 | 1.2 | .58 | 1.3 | .41 | 1.7 | 1.3 | .01 | 1.4 | .03 |
| GSTA4_snp2: | |||||||||||||||
| No anomaliesb | .7 | .8 | .007 | .7 | .01 | 1.0 | 1.4 | .61 | 1.1 | .61 | .8 | 1.0 | .04 | .9 | .11 |
| CLO | .6 | 1.7 | .005 | .9 | .40 | .6 | .5 | .12 | .6 | .06 | .6 | 1.2 | .001 | .8 | .07 |
| GSTM1_snp1: | |||||||||||||||
| CPO | 1.3 | 3.5 | .02 | 1.7 | .01 | 1.1 | 1.1 | .98 | 1.1 | .82 | THRRc | ||||
| GSTP1_snp3: | |||||||||||||||
| Alla | 1.1 | 1.0 | .47 | 1.0 | .86 | 1.2 | .6 | .01 | .9 | .19 | THRRc | ||||
| CL/P | 1.1 | 1.1 | .60 | 1.0 | .70 | 1.1 | .5 | .01 | .8 | .16 | THRRc | ||||
| NAT2_snp2: | |||||||||||||||
| Alla | .8 | .6 | .04 | .8 | .01 | .8 | .6 | .12 | .8 | .04 | .8 | .6 | .006 | .8 | .001 |
| No anomaliesb | .8 | .6 | .07 | .8 | .02 | .8 | .6 | .20 | .8 | .07 | .8 | .6 | .01 | .8 | .003 |
| CL/P | .8 | .4 | .01 | .7 | .01 | .9 | .6 | .43 | .8 | .24 | .8 | .5 | .007 | .7 | .002 |
| Control | .7 | .8 | .01 | .8 | .02 | 1.0 | .8 | .67 | .9 | .43 | .8 | .8 | .03 | .8 | .02 |
Note.— RR1 is the genetic RR when the child carries one copy of the variant allele; RR2 is that when the child carries two copies of the variant alleles; R_A is the RR when the child carries one copy of the variant allele, assumed to be a log additive model, a model in which the RR of carrying two copies of the variant alleles is the square of that of carrying one copy.
Includes data from all subjects with nonsyndromic OC.
Includes data from subjects without other minor anomalies.
For test of heterogeneity in RR (THRR), P<.05.
Table A4. .
Interactions between Fetal Genotype and Maternal Smoking[Note]
| Danish |
Iowa |
||||||
| Gene and Phenotype |
C×E 1 (I1) |
C×E 2 (I2) |
C×E LRT (P) |
CxE 1 (I1) |
CxE 2 (I2) |
C×E LRT (P) |
Pa |
| EPHX1_snp1: | |||||||
| Allb | .82 | .68 | .62 | .92 | 16.6 | .004 | .017 |
| No anomaliesc | .94 | .77 | .87 | .96 | 14.0 | .01 | |
| GSTA4_snp1: | |||||||
| CPO | .20 | 1.78 | .01 | 1.37 | 1.72 | .78 | |
| GSTA4_snp2: | |||||||
| CPO | .23 | 3.63 | .01 | .78 | 2.60 | .65 | |
| GSTP1_snp1: | |||||||
| CL/P | .50 | 1.94 | .02 | 1.26 | .86 | .89 | |
| HIF1A: | |||||||
| Allb | .82 | .37 | .20 | 3.02 | 1.13 | .02 | .027 |
| No anomaliesc | .82 | .39 | .28 | 2.89 | 1.19 | .04 | |
| UG1A7_snp1: | |||||||
| Allb | 2.30 | 1.44 | .08 | 2.96 | .53 | .02 | .014 |
| No anomaliesc | 2.14 | 1.63 | .12 | 3.34 | .53 | .01 | |
| CPO | 13.5 | 1.19 | .02 | 4.94 | .70 | .25 | |
Note.— I1 is the estimated interaction effect of one fetal copy of the variant allele together with maternal smoking; I2 is the estimated interaction effect of two fetal copies of the variant allele together with maternal smoking.
For formal hypothesis testing, we used Fisher’s method to combine P values that were based on data from all OC cases in the Danish and Iowan populations.
Includes data from all subjects with nonsyndromic OC.
Includes data from subjects without other minor anomalies.
Table A5. .
Interaction between Fetal GSTT1 Genotypes and Maternal Smoking
| Population and Phenotype |
RR (unexpa) |
95% CI of RR– |
RR (expb) |
95% CI of RR– |
RR (interactionc) |
95% CI of RR |
χ2 | P | Adjusted Pd |
| DBS: | |||||||||
| Alle | .84 | .48–1.46 | 2.34 | 1.12–4.91 | 2.79 | 1.11–7.01 | 4.9 | .03f | .03 |
| CPO | .87 | .35–2.19 | 4.22 | 1.39–12.85 | 4.83 | 1.14–20.43 | 4.5 | .03 | .03 |
| CL/P | .83 | .44–1.55 | 1.97 | .89–4.38 | 2.38 | .86–6.55 | 2.8 | .09 | .12 |
| Iowa triad: | |||||||||
| Alle | .65 | .37–1.15 | 4.54 | 1.20–17.15 | 7.03 | 1.67–28.98 | 8.7 | .003f | .02 |
| No anomaliesg | .60 | .33–1.10 | 4.61 | 1.20–17.76 | 7.72 | 1.80–32.83 | 9.2 | .002 | .03 |
| CLO | .48 | .15–1.50 | 6.21 | 1.20–31.64 | 12.9 | 1.90–88.01 | 7.3 | .007 | .01 |
| CPO | .63 | .26–1.55 | 6.95 | 1.48–31.76 | 11.1 | 1.95–60.70 | 8.3 | .004 | .04 |
| CL/P | .75 | .39–1.41 | 3.33 | .78–14.12 | 4.45 | .94–21.43 | 3.8 | .05 | .13 |
| DBS and Iowa triad: | |||||||||
| Alle | .70 | .47–.97 | 2.23 | 1.20–3.71 | 3.17 | 1.59–6.13 | 9.5 | .002 | <.001 |
| No anomaliesg | .69 | .45–.96 | 2.21 | 1.19–3.72 | 3.21 | 1.62–6.34 | 9.5 | .002 | <.001 |
| CPO | .60 | .37–1.19 | 4.25 | 1.51–7.33 | 7.13 | 1.88–13.41 | 11.4 | <.001 | <.001 |
| CL/P | .79 | .49–1.13 | 1.76 | .91–3.17 | 2.23 | 1.08–4.81 | 3.7 | .05 | .03 |
RR of GSTT1-null genotype in nonsmoking mothers.
RR of GSTT1-null genotype in smoking mothers.
RR of the interaction effect between GSTT1-null genotype and maternal cigarette smoking.
Adjusted for vitamin and alcohol use.
Includes data from all subjects with nonsyndromic OC.
The combined P value by use of Fisher’s method is .0009, which remains significant after correction for multiple tests (see the “Material and Methods” section).
Includes data from subjects without other minor anomalies.
Table A6. .
ORs of Fetal GSTT1 Deletion and Maternal Smoking Effects in OC[Note]
| No. of |
OR |
|||||
| Population, Fetal GSTT1 Genotypea, and Maternal Smokingb |
Cases | Controls | Total | Effectc | Value | 95% CI |
| Iowa: | ||||||
| −−: | ||||||
| Yes | 19 | 9 | 28 | ORge | 2.10 | .92–4.70 |
| No | 30 | 57 | 87 | ORg | .52 | .32–.85 |
| ++/+−: | ||||||
| Yes | 63 | 54 | 117 | ORe | 1.16 | .76–1.76 |
| No | 180 | 179 | 359 | Reference | ||
| DBS: | ||||||
| −−-: | ||||||
| Yes | 20 | 14 | 34 | ORge | 2.80 | 1.37–5.61 |
| No | 21 | 49 | 70 | ORg | .84 | .48–1.46 |
| ++/+−: | ||||||
| Yes | 78 | 128 | 206 | ORe | 1.20 | .84–1.70 |
| No | 131 | 257 | 388 | Reference | ||
| Iowa and DBS: | ||||||
| −−: | ||||||
| Yes | 39 | 23 | 62 | ORge | 2.38 | 1.39–4.01 |
| No | 51 | 106 | 157 | ORg | .67 | .47–.97 |
| ++/+−: | ||||||
| Yes | 141 | 182 | 323 | ORe | 1.09 | .83–1.41 |
| No | 311 | 436 | 747 | Reference | ||
Note.— Data from all subjects with OC are included.
−− Indicates GSTT1-null genotype; ++/+− indicates not-null GSTT1 genotype (either one or two copies of the wild alleles).
Yes or no.
ORge is the OR of the joint GSTT1 deletion and maternal smoking versus none; ORg is OR of the GSTT1 deletion alone versus none; ORe is the OR of maternal smoking alone versus none.
Table A7. .
Gene-Gene Interactive Effects[Note]
| Gene 1 | Gene 2 | χ2 | P |
| CYP1B1_snp2 | MSX1_snp5 | 12.213 | .002 |
| NAT2_snp1 | IRF6_snp3 | 11.02 | .004 |
| UGT1A7_snp2 | NAT2_snp1 | 10.7 | .005 |
| CYP1B1_snp2 | AHR_snp2 | 10.54 | .005 |
| GSTM1_snp1 | MSX1_snp1 | 10.536 | .005 |
| EPHX_snp1 | CYP1B1_snp2 | 10.41 | .005 |
| GSTP1_snp1 | EPHX1_snp1 | 9.67 | .008 |
| EPHX1_snp2 | AHR_snp2 | 9.52 | .009 |
| EPHX1_snp1 | GSTP1_snp1 | 9.45 | .009 |
| CYP1A2 | EPHX1_snp2 | 9.17 | .010 |
| GSTP4_snp1 | GSTP1_snp2 | 9.13 | .010 |
| HIF1A | EPHX1_snp2 | 9.04 | .011 |
| HIF1A | MSX1_snp6 | 9.033 | .011 |
| HIF1A | GSTM1_snp1 | 8.98 | .011 |
| CYP1B1_snp1 | MSX1_snp3 | 8.745 | .013 |
| GSTM3 | MSX1_snp1 | 7.989 | .018 |
| CYP1A2 | IRF6_snp50 | 7.94 | .019 |
| AHR_snp2 | UGT1A7_snp2 | 7.88 | .019 |
| EPHX1_snp1 | IRF6_snp32 | 7.34 | .025 |
| SULT1A1 | MSX1_snp3 | 7.128 | .028 |
| NAT2_snp2 | GSTM3 | 7.08 | .029 |
| UGT1A7_snp1 | GSTM_null | 7.83 | .020 |
Note.— Only results with χ2>7 are included.
Table A8. .
Summary of Markers with P<.05 in Tests for Different Effects in the Sample Sets Studied[Note]
| Genes |
Gene-Smoking Interaction |
|||
| Population | Offspring | Mother | Offspring | Mother |
| Iowa | AhR_snp2, CYP1B1_snp1, CYP1A1, CYP1A2, GSTP1_snp3, NAT2_snp2, and NQO1 | CYP1A1, GSTM3, and NAT2_snp1 | EPHX1_snp1, GSTP1_snp2, HIF1A, UGT1A7_snp1, and GSTT1_del | EPHX1_snp1 and UGT1A7_snp1 |
| DCS | CYP1A1, CYP1A2, GSTA4_SNP2, GSTM1_snp1, and NAT2_snp2 | EPHX1_snp1, GSTP1_snp3, NAT2_snp1, and UGT1A7_snp2 | CYP1B1_snp2, CYP1A1, EPHX1_snp2, GSTA4_snp1, GSTA4_snp2, GSTP1_snp3, GSTP1_snp1, and UGT1A7_snp1 | CYP2E1, GSTP1_snp3, HIF1A, NAT2_snp2, and UGT1A7_snp1 |
| DBSa | CYP1A1 | NA | GSTT1_del | NA |
Note.— SNPs showing statistical significance in more than one population are shown in bold.
No maternal samples were available in the DBS sample set, so tests of maternal genotype or maternal genotype and smoking interaction effects were not applicable (NA). Only 9 (shown in table 3) of the 25 markers were genotyped in Danish case control samples.
Web Resources
The URLs for data presented herein are as follows:
- COGENE Project, http://hg.wustl.edu/COGENE/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CYP1A1, EPHX1, GSTM1, GSTT1, NAT2, AHR-ARNT, IRF6, MSX1, GSTP1, UGT1A7, CYP1A2, CYP1B1, GSTA4, FGFR1, FGFR2, CYP2E1, NQO1, and HIF1A)
References
- 1.Harville EW, Wilcox AJ, Lie RT, Vindenes H, Abyholm F (2005) Cleft lip and palate versus cleft lip only: are they distinct defects? Am J Epidemiol 162:448–453 10.1093/aje/kwi214 [DOI] [PubMed] [Google Scholar]
- 2.Mossey P, Little J (2002) Epidemiology of oral clefts: an international perspective. In: Wyszynski DF (ed) Cleft lip and palate: from origin to treatment. Oxford University Press, New York, pp 127–158 [Google Scholar]
- 3.Murray JC (2002) Gene/environment causes of cleft lip and/or palate. Clin Genet 61:248–256 10.1034/j.1399-0004.2002.610402.x [DOI] [PubMed] [Google Scholar]
- 4.Hayes C, Werler MM, Willett WC, Mitchell AA (1996) Case-control study of periconceptional folic acid supplementation and oral clefts. Am J Epidemiol 143:1229–1234 [DOI] [PubMed] [Google Scholar]
- 5.Mitchell LE, Healey SC, Chenevix-Trench G (1995) Evidence for an association between nonsyndromic cleft lip with or without cleft palate and a gene located on the long arm of chromosome 4. Am J Hum Genet 57:1130–1136 [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen K, Olsen J, Norgaard-Pedersen B, Basso O, Stovring H, Milhollin-Johnson L, Murray JC (1999) Oral clefts, transforming growth factor alpha gene variants, and maternal smoking: a population-based case-control study in Denmark, 1991–1994. Am J Epidemiol 149:248–255 [DOI] [PubMed] [Google Scholar]
- 7.Lieff S, Olshan AF, Werler M, Strauss RP, Smith J, Mitchell A (1999) Maternal cigarette smoking during pregnancy and risk of oral clefts in newborns. Am J Epidemiol 150:683–694 [DOI] [PubMed] [Google Scholar]
- 8.Mitchell LE, Murray JC, O’Brien S, Christensen K (2001) Evaluation of two putative susceptibility loci for oral clefts in the Danish population. Am J Epidemiol 153:1007–1015 10.1093/aje/153.10.1007 [DOI] [PubMed] [Google Scholar]
- 9.Lammer EJ, Shaw GM, Iovannisci DM, Van Waes J, Finnell RH (2004) Maternal smoking and the risk of orofacial clefts: susceptibility with NAT1 and NAT2 polymorphisms. Epidemiology 15:150–156 10.1097/01.ede.0000112214.33432.cc [DOI] [PubMed] [Google Scholar]
- 10.Little J, Cardy A, Munger RG (2004) Tobacco smoking and oral clefts: a meta-analysis. Bull World Health Organ 82:213–218 [PMC free article] [PubMed] [Google Scholar]
- 11.Hartsfield JK Jr, Hickman TA, Everett ET, Shaw GM, Lammer EJ, Finnell RA (2001) Analysis of the EPHX1 113 polymorphism and GSTM1 homozygous null polymorphism and oral clefting associated with maternal smoking. Am J Med Genet 102:21–24 [DOI] [PubMed] [Google Scholar]
- 12.van Rooij IA, Wegerif MJ, Roelofs HM, Peters WH, Kuijpers-Jagtman AM, Zielhuis GA, Merkus HM, Steegers-Theunissen RP (2001) Smoking, genetic polymorphisms in biotransformation enzymes, and nonsyndromic oral clefting: a gene-environment interaction. Epidemiology 12:502–507 10.1097/00001648-200109000-00007 [DOI] [PubMed] [Google Scholar]
- 13.Shaw GM, Nelson V, Iovannisci DM, Finnell RH, Lammer EJ (2003) Maternal occupational chemical exposures and biotransformation genotypes as risk factors for selected congenital anomalies. Am J Epidemiol 157:475–484 10.1093/aje/kwg013 [DOI] [PubMed] [Google Scholar]
- 14.Romitti PA, Lidral AC, Munger RG, Daack-Hirsch S, Burns TL, Murray JC (1999) Candidate genes for nonsyndromic cleft lip and palate and maternal cigarette smoking and alcohol consumption: evaluation of genotype-environment interactions from a population-based case-control study of orofacial clefts. Teratology 59:39–50 [DOI] [PubMed] [Google Scholar]
- 15.Dausset J CH, Cohen D, Lathrop M, Lalouel J-M, White R (1990) Centre d’Etude du Polymorphisme Humain (CEPH): collaborative genetic mapping of the human genome. Genomics 6:575–577 10.1016/0888-7543(90)90491-C [DOI] [PubMed] [Google Scholar]
- 16.Shi M, Caprau D, Dagle J, Christiansen L, Christensen K, Murray JC (2004) Application of kinetic polymerase chain reaction and molecular beacon assays to pooled analyses and high-throughput genotyping for candidate genes. Birth Defects Res Part A Clin Mol Teratol 70:65–74 10.1002/bdra.10153 [DOI] [PubMed] [Google Scholar]
- 17.de Kok JB, Wiegerinck ET, Giesendorf BA, Swinkels DW (2002) Rapid genotyping of single nucleotide polymorphisms using novel minor groove binding DNA oligonucleotides (MGB probes). Hum Mutat 19:554–559 10.1002/humu.10076 [DOI] [PubMed] [Google Scholar]
- 18.Cai J, Ash D, Kotch LE, Jabs EW, Attie-Bitach T, Auge J, Mattei G, Etchevers H, Vekemans M, Korshunova Y, et al (2005) Gene expression in pharyngeal arch 1 during human embryonic development. Hum Mol Genet 14:903–912 10.1093/hmg/ddi083 [DOI] [PubMed] [Google Scholar]
- 19.Lin DY (2006) Evaluating statistical significance in two-stage genomewide association studies. Am J Hum Genet 78:505–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher R (1958) Statistical methods for research workers. Hafner, New York [Google Scholar]
- 21.Weinberg CR, Wilcox AJ, Lie RT (1998) A log-linear approach to case-parent–triad data: assessing effects of disease genes that act either directly or through maternal effects and that may be subject to parental imprinting. Am J Hum Genet 62:969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilcox AJ, Weinberg CR, Lie RT (1998) Distinguishing the effects of maternal and offspring genes through studies of “case-parent triads.” Am J Epidemiol 148:893–901 [DOI] [PubMed] [Google Scholar]
- 23.Weinberg CR (1999) Allowing for missing parents in genetic studies of case-parent triads. Am J Hum Genet 64:1186–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaid DJ, Sommer SS (1993) Genotype relative risks: methods for design and analysis of candidate-gene association studies. Am J Hum Genet 53:1114–1126 [PMC free article] [PubMed] [Google Scholar]
- 25.Starr JR, Hsu L, Schwartz SM (2005) Performance of the log-linear approach to case-parent triad data for assessing maternal genetic associations with offspring disease: type I error, power, and bias. Am J Epidemiol 161:196–204 10.1093/aje/kwi021 [DOI] [PubMed] [Google Scholar]
- 26.Umbach DM, Weinberg CR (2000) The use of case-parent triads to study joint effects of genotype and exposure. Am J Hum Genet 66:251–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kistner EO, Weinberg CR (2004) Method for using complete and incomplete trios to identify genes related to a quantitative trait. Genet Epidemiol 27:33–42 10.1002/gepi.20001 [DOI] [PubMed] [Google Scholar]
- 28.Zucchero TM, Cooper ME, Maher BS, Daack-Hirsch S, Nepomuceno B, Ribeiro L, Caprau D, Christensen K, Suzuki Y, Machida J, et al (2004) Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med 351:769–780 10.1056/NEJMoa032909 [DOI] [PubMed] [Google Scholar]
- 29.Jezewski PA, Vieira AR, Nishimura C, Ludwig B, Johnson M, O’Brien SE, Daack-Hirsch S, Schultz RE, Weber A, Nepomucena B, et al (2003) Complete sequencing shows a role for MSX1 in non-syndromic cleft lip and palate. J Med Genet 40:399–407 10.1136/jmg.40.6.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rothman K, Greenland S (1998) Modern Epidemiology. Lippincott Williams and Wilkins, Philadelphia [Google Scholar]
- 31.Yang Q, Khoury MJ, Friedman JM, Flanders WD (2003) On the use of population attributable fraction to determine sample size for case-control studies of gene-environment interaction. Epidemiology 14:161–167 10.1097/00001648-200303000-00009 [DOI] [PubMed] [Google Scholar]
- 32.Botto LD, Khoury MJ (2001) Commentary: facing the challenge of gene-environment interaction: the two-by-four table and beyond. Am J Epidemiol 153:1016–1020 10.1093/aje/153.10.1016 [DOI] [PubMed] [Google Scholar]
- 33.Wilkie AO, Morriss-Kay GM (2001) Genetics of craniofacial development and malformation. Nat Rev Genet 2:458–468 10.1038/35076601 [DOI] [PubMed] [Google Scholar]
- 34.Reijntjes S, Blentic A, Gale E, Maden M (2005) The control of morphogen signalling: regulation of the synthesis and catabolism of retinoic acid in the developing embryo. Dev Biol 285:224–237 10.1016/j.ydbio.2005.06.019 [DOI] [PubMed] [Google Scholar]
- 35.Center for Disease Control and Prevention (2004) Smoking during pregnancy—United States, 1990–2002. MMWR Morb Mortal Wkly Rep 53:911–915 [PubMed] [Google Scholar]
- 36.Center for Disease Control and Prevention (2004) State estimates of neonatal health-care costs associated with maternal smoking—United States, 1996. MMWR Morb Mortal Wkly Rep 53:915–917 [PubMed] [Google Scholar]
- 37.Gonzalez FJ (2001) The use of gene knockout mice to unravel the mechanisms of toxicity and chemical carcinogenesis. Toxicol Lett 120:199–208 10.1016/S0378-4274(01)00296-X [DOI] [PubMed] [Google Scholar]
- 38.Gonzalez FJ, Kimura S (2003) Study of P450 function using gene knockout and transgenic mice. Arch Biochem Biophys 409:153–158 10.1016/S0003-9861(02)00364-8 [DOI] [PubMed] [Google Scholar]
- 39.Engle MR, Singh SP, Czernik PJ, Gaddy D, Montague DC, Ceci JD, Yang Y, Awasthi S, Awasthi YC, Zimniak P (2004) Physiological role of mGSTA4-4, a glutathione S-transferase metabolizing 4-hydroxynonenal: generation and analysis of mGsta4 null mouse. Toxicol Appl Pharmacol 194:296–308 10.1016/j.taap.2003.10.001 [DOI] [PubMed] [Google Scholar]
- 40.Zöllner S, Wen X, Hanchard NA, Herbert MA, Ober C, Pritchard JK (2004) Evidence for extensive transmission distortion in the human genome. Am J Hum Genet 74:62–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hein DW (2002) Molecular genetics and function of NAT1 and NAT2: role in aromatic amine metabolism and carcinogenesis. Mutat Res 506–507:65–77 [DOI] [PubMed] [Google Scholar]
- 42.Fretland AJ, Leff MA, Doll MA, Hein DW (2001) Functional characterization of human N-acetyltransferase 2 (NAT2) single nucleotide polymorphisms. Pharmacogenetics 11:207–215 10.1097/00008571-200104000-00004 [DOI] [PubMed] [Google Scholar]
- 43.Mitchell MK, Futscher BW, McQueen CA (1999) Developmental expression of N-acetyltransferases in C57BI/6 mice. Drug Metab Dispos 27:261–264 [PubMed] [Google Scholar]
- 44.Landi S (2000) Mammalian class theta GST and differential susceptibility to carcinogens: a review. Mutat Res 463:247–283 10.1016/S1383-5742(00)00050-8 [DOI] [PubMed] [Google Scholar]
- 45.Lammer EJ, Shaw GM, Iovannisci DM, Finnell RH (2005) Maternal smoking, genetic variation of glutathione s-transferases, and risk for orofacial clefts. Epidemiology 16:698–701 10.1097/01.ede.0000172136.26733.4b [DOI] [PubMed] [Google Scholar]