Abstract
Nemaline myopathy (NM) is a congenital myopathy characterized by muscle weakness and nemaline bodies in affected myofibers. Five NM genes, all encoding components of the sarcomeric thin filament, are known. We report identification of a sixth gene, CFL2, encoding the actin-binding protein muscle cofilin-2, which is mutated in two siblings with congenital myopathy. The proband’s muscle contained characteristic nemaline bodies, as well as occasional fibers with minicores, concentric laminated bodies, and areas of F-actin accumulation. Her affected sister’s muscle was reported to exhibit nonspecific myopathic changes. Cofilin-2 levels were significantly lower in the proband’s muscle, and the mutant protein was less soluble when expressed in Escherichia coli, suggesting that deficiency of cofilin-2 may result in reduced depolymerization of actin filaments, causing their accumulation in nemaline bodies, minicores, and, possibly, concentric laminated bodies.
Nemaline myopathy (NM forms 1–6 [MIM 609284, 256030, 161800, 609285, 605355, and 609273]), the most common form of congenital myopathy, is a diagnosis applied to a clinically and genetically heterogeneous collection of patients characterized by weakness and the presence of rodlike structures called “nemaline bodies” in affected muscles.1–4 Ultrastructurally, nemaline bodies appear to originate from sarcomeric Z-disks involving the adjacent thin filaments where the primary abnormalities lie. Mutations of the genes encoding the thin-filament proteins nebulin (NEB) and skeletal muscle α-actin (ACTA1) account for a majority of NM cases,5–7 whereas mutations of the genes encoding troponin T1 (TNNT1),8 β-tropomyosin (TPM2),9 and α-tropomyosin (TPM3)10,11 are quite rare, each having been reported in only one or several independent families with NM. Additionally, NM in two families with autosomal dominant inheritance is in linkage with an unknown gene on chromosome 15q.12 Despite extensive searches, the genetic basis for NM, in a significant number of cases, remains unknown.
Cofilin-2, encoded by CFL2 on chromosome 14q12 (MIM 601443), is a member of the AC group of proteins that also includes cofilin-1 (encoded by CFL1) and destrin (encoded by DSTN), all of which regulate actin-filament dynamics.13,14 CFL2 encodes a skeletal muscle–specific isoform15 localized to the thin filaments where it exerts its effect on actin, in part through interactions with tropomyosins.16 Despite an earlier study that found no CFL2 mutations in 50 patients with NM,17 we considered that this remained an excellent candidate gene because of its critical role in regulation of sarcomeric actin filaments.
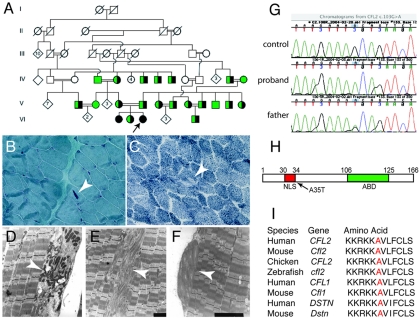
Using genomic PCR and DNA sequencing,17 we screened the CFL2 gene in 113 unrelated patients with NM and 58 patients with clinicopathological diagnoses of other congenital myopathies (i.e., 10 myotubular myopathy, 6 centronuclear myopathy, 9 multiminicore disease, 6 congenital muscular dystrophy, 2 spheroid body myopathy, 2 Walker-Warburg syndrome, and 23 with nonspecific congenital myopathies). All study subjects were enrolled, after appropriate informed consent, under the supervision of the Children’s Hospital Boston institutional review board. None of the patients had known mutations in previously identified genes. A homozygous missense mutation of CFL2 was identified in two affected siblings in a large family of Middle Eastern origin (fig. 1A). Both patients had similar clinical presentations, with hypotonia noted at birth, delayed early motor milestones, frequent falls, and the inability to run. The elder sister, now age 16 years, can walk short distances but uses a wheelchair outside the house. She was given a diagnosis of nonspecific congenital myopathy at age 4 years, when her muscle biopsy sample showed marked fiber-size variability, type I fiber predominance, and a few fibers on oxidative stains that exhibited focal loss of integrity consistent with minicores. The younger sibling (proband), now age 9 years, is ambulant and was given a diagnosis of NM at age 2 years, after a muscle biopsy sample showed nemaline bodies as well as occasional minicores and concentric laminated bodies (fig. 1B–1F). The course of the disease in both siblings has been like that of the “typical” form of NM, but the distribution of weakness is distinct, since no significant facial weakness or foot drop was evident.
Figure 1. .
Pathologic and genetic findings in a family with CFL2 mutation A35T. A, Partial pedigree of the family illustrates several consanguineous loops. The proband is indicated by an arrow. The two affected sisters (filled circles) are homozygous for A35T, whereas an unaffected sister, both parents, and several other members of the extended family (half-filled circles) are heterozygous for the change and for a shared haplotype spanning ∼4.6-Mb pairs around the CFL2 gene. Green symbols indicate tested individuals with WT sequence. Light microscopic findings in proband’s muscle include presence of nemaline bodies (B, arrow) on Gomori trichrome staining and occasional minicores (C, arrow) on nicotinamide adenine nucleotide dehydrogenase-tetrazolium reductase staining. Electron microscopy confirmed identity of nemaline bodies (D, arrow), unstructured minicores (E, arrow), and concentric laminated bodies (F, arrow). G, DNA sequence analysis of genomic PCR products illustrating three genotypes for CFL2 c.103G→A seen in the family. H, Schematic representation of cofilin-2. Residue 35 is located next to NLS (30–34 aa); ABD = actin-binding domain. I, Altered alanine residue (red) is evolutionarily conserved among AC proteins (i.e., cofilin-2, cofilin-1, and destrin) across all sequenced vertebrates.
The proband and her affected sibling are homozygous for CFL2 c.103G→A, predicted to result in an alanine-to-threonine substitution at residue 35 (A35T) (fig. 1G and 1H). A35 is located next to a nuclear localization signal (NLS) (fig. 1H) in a region that is highly conserved across vertebrates (fig. 1I). An unaffected sibling, both unaffected parents, and a number of other extended family members were heterozygous for this change. Extensive intermarriage, with multiple consanguineous loops, strongly suggested identity by descent for the two mutant alleles, a supposition that was supported by linkage studies that used flanking markers. Linkage analysis, assuming autosomal recessive inheritance of a disease gene with a frequency of 0.005, gave a LOD score of 1.9 with a recombination fraction of 0. The mutation was ruled out in 282 unaffected control individuals, including 91 originating from the same geographic region and ethnic group as the family.
Indirect immunofluorescence analysis of the proband’s muscle with the use of anti-α-actinin-2 antisera showed that, as expected, the nemaline bodies contained this protein (fig. 2A–2F). Interestingly, despite the fact that mutation of the α-actin gene, ACTA1, had been previously ruled out, this biopsy sample also exhibited features of an actinopathy,5 since phalloidin staining of the same sections revealed that 4% of myofibers contained distinct actin-filament accumulations (fig. 2E and 2H) that stained equally well with phalloidin (specific for F-actin) and anti-sarcomeric actin, which binds both G- and F-actin (fig. 2G–2I).
Figure 2. .
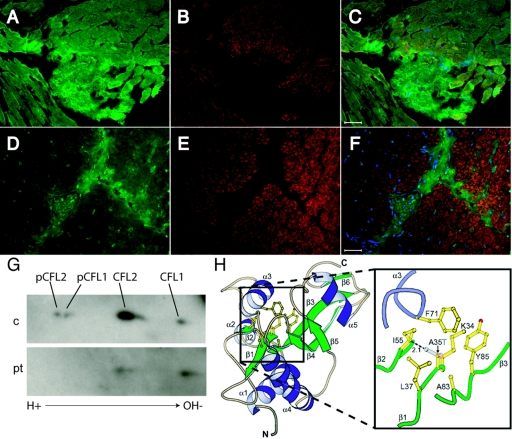
Fluorescence microscopic analysis revealing α-actinin-2–positive nemaline bodies and actin-filament accumulations. Unaffected control muscle (A–C) and the proband’s muscle (D–I) were immunostained with anti-α-actinin-218 (A and D), anti-skeletal actin (clone 5C5 anti-sarcomeric actin [Sigma A2172]) (G), and phalloidin Alexa Fluor 546 (Invitrogen) (B, E, and H). Merged images, including blue DAPI-stained nuclei, are shown in panels C, F, and I. Several α-actinin–positive nemaline bodies are indicated by arrows in panels D and F, whereas nonoverlapping F-actin accumulations are indicated by arrowheads in panels E–I. Scale bars equal 20 μm (C, F, and I).
To establish whether the A35T mutation was associated with altered cofilin-2 expression and/or localization, we performed immunofluorescence analysis of cofilins in the proband’s muscle biopsy sample and in several age-matched unaffected human skeletal muscle specimens. On longitudinal sections of control muscles, anti-cofilin antibodies stained in a sarcomeric pattern that colocalized with actin at I-bands, as expected for cofilin-2 (data not shown). Remarkably, sarcomeric cofilin-2 staining was significantly less intense in the patient muscle fibers relative to those of age-matched unaffected controls (fig. 3A–3F) and several other patients with NM (data not shown). Cross-reactivity of anti-cofilin-2 antibody with cofilin-1 led to interstitial tissue staining of both the patient and control muscle, providing an internal control for staining.
Figure 3. .
Effects of the A35T mutation on cofilin-2 structure and expression in vivo. A–F, Indirect immunofluorescence analysis of cofilins (green stain [A, C, D, and F]) and skeletal actin (clone 5C5 anti-sarcomeric actin [Sigma A2172]) (red stain [B, C, E, and F]) merged with DAPI for visualization of nuclei (blue stain [C and F]) in muscle from control (A–C) and proband (D–F). Scale bar equals 50 μm. The anti-cofilin-2 polyclonal rabbit antibody (US Biologicals C7506-50) recognizes both sarcomeric cofilin-2 and nonmuscle cofilin-1 (seen in actin-negative connective tissue). Cofilin-2 staining in myofibers is markedly less intense in the proband compared with that in the control, but cofilin-1 staining is preserved in connective tissue and capillaries. G, Two-dimensional gel and Western blot analysis of cofilins in control (c) and proband muscle (pt) performed as described elsewhere.17 On each gel, 200 μg total muscle lysate proteins were loaded. Equal protein concentrations of lysates were confirmed by immunoblotting parallel SDS-PAGE gels that were stained for glyceraldehyde-3-phosphate dehydrogenase. Isoelectric focusing was accomplished on a pH gradient of 3–10, and immunodetection used an antibody that detects both phosphorylated and unphosphorylated cofilin-1 and cofilin-2 (catalog number C8736 [Sigma]). Identities of the various spots were confirmed using additional isoform-specific and phosphorylation-specific antibodies (not shown). Although unphosphorylated cofilin-1 spots are similar in intensity, both spots for cofilin-2 are significantly smaller in the patient’s muscle. H, A35T mutation, modeled using O and Molscript, illustrating side-chain clash of T35 with I55. Residue 35 is in the middle of a β-sheet, with its backbone amide and carbonyl making hydrogen bonds to the backbone carbonyl and amide of I55. In this model, the T35 side-chain hydroxyl forms part of a narrow canyon wall on the molecular surface, which is probably filled with solvent, allowing hydrogen bonds between water molecules and the T35 hydroxyl.
To determine if the immunofluorescence data reflected presence of smaller quantities of cofilin-2 in the patient’s muscle, we performed two-dimensional SDS-PAGE and immunoblotting, to separate phosphorylated and unphosphorylated forms of cofilins 1 and 2. Relative to age-matched control muscles, unphosphorylated cofilin-2 in the proband’s muscle was significantly lower, and phosphorylated forms were not detectable (fig. 3G).
Quantitative RT-PCR of CFL2 mRNA was used to determine if the apparent reduction in cofilin-2 protein seen in the patient’s muscle was caused by lower transcription and/or mRNA stability. Instead, we found that the proband’s muscle contained between 4- and 20-fold more CFL2 mRNA, compared with three unaffected, age-matched control muscle specimens. Thus, the relative absence of cofilin-2 in the patient’s myopathic muscles was likely a consequence of reduced protein stability and/or some other posttranscriptional mechanism(s).
To better understand the effects of the A35T mutation on cofilin-2 structure, we modeled this change into a nuclear magnetic resonance structure of chicken cofilin-2 (Protein Data Bank identification number 1TVJ). Chicken cofilin-2 differs from human cofilin-2 by three amino acid differences that are unlikely to affect the conformation of the region around residue 35. Two of these differences, P26Q and K44R, are located on highly solvent exposed loops that are spatially remote from residue 35. The third, A70S, is adjacent to F71 on α-helix 3. The F71 aromatic ring contributes to the hydrophobic core that includes the A35 methyl group. However, as the alanine or serine 70 side chains are solvent exposed, the difference is unlikely to distort the helix. A35 is in the middle of a β-sheet, with its backbone amide and carbonyl hydrogen bonded to the backbone carbonyl and amide of I55. A35 and I55 are both highly conserved among vertebrate members of the AC family.19 Modeling T35 with use of the common rotamer that minimizes clashes with neighboring side chains revealed a distance between Cγ2 of T35 and Cβ of I55 of only 2.1 Å, closer than that allowed by Van der Waals interactions (fig. 3H). Consequently, some distortion of the central β-sheet would be required to accommodate the T35 side chain, although this distortion would not necessarily break any hydrogen bonds.
Since the modeling of T35 suggested a conformational change in the β-sheet, we next expressed the wild-type (WT) and A35T cofilin-2 proteins in eukaryotic and prokaryotic systems. We constructed matched expression vectors containing the WT and c.103G→A mutant sequences in the full-length human CFL2 cDNA cloned into mammalian expression vectors pcDNA-DEST-47 (C-terminal green fluorescent protein [GFP] fusion) and pcDNA-3.2-DEST (C-terminal V5 tag) and bacterial expression vectors pDEST-42 (C-terminal V5-6x His fusions) and pDEST-14 (no fusion) (all Invitrogen Life Technologies). No differences were observed between C2C12 myoblasts transfected with either GFP or V5-tagged WT and A35T cofilin-2 clones; both proteins were uniformly distributed throughout the cytoplasm.20 After several days of high-level expression, both transduced cofilins organized into cytoplasmic and nuclear rod structures, and continued high-level expression eventually resulted in loss of cofilin-expressing cells, presumably because of cell-cycle arrest21 (data not shown).
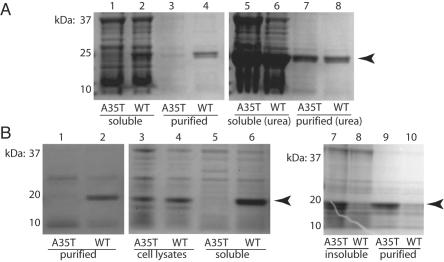
In contrast, A35T and WT cofilin-2 proteins behaved quite differently when expressed in bacteria. Both proteins were expressed as His fusions in Escherichia coli and were purified using His-affinity chromatography on Ni-NTA agarose columns (Qiagen). Although similar amounts of WT and A35T protein were found in the initial whole-cell lysates, the purified fractions repeatedly demonstrated selective loss of the mutant but not the WT proteins (fig. 4A). Cell lysis and purification in the presence of 6 M urea resulted in similar yields for both versions of the protein, suggesting that the loss of A35T mutant cofilin-2 was a result of decreased stability and/or solubility. To rule out the possibility that the His/V5 tag itself might affect the solubility, each protein was expressed without any fusion tags, and the expressed proteins were purified using Vivapure D anion exchange columns (Vivascience). As before, similar amounts of both proteins were present in total cell lysates, but purification resulted in complete loss of the A35T samples (fig. 4B). The first step of the purification procedure involved centrifugation to remove insoluble material from the initial lysates. When these pellets were treated with 6 M urea, we found that the mutant A35T protein was preferentially recovered, demonstrating that the mutation likely causes protein misfolding in this prokaryotic expression system, leading to precipitation and loss in the insoluble fractions during purification (fig. 4B).
Figure 4. .
Differential solubility of WT and mutant cofilin-2. A, SDS-PAGE analysis of cofilin-2 with His/V5 tag expressed in E. coli. The amount of mutant cofilin-2 (arrowhead) in the soluble and purified fractions (lanes 1 and 3) is markedly lower than in the WT (lanes 2 and 4), even though equal amounts of both proteins were present in whole bacterial cell lysates (not shown). In contrast, when 6 M urea is added to the lysis buffer, the amounts of mutant and WT cofilin-2 are similar in both supernatant and purified fractions (lanes 5–8). B, SDS-PAGE analysis of the native cofilin-2 proteins expressed in E. coli without epitope tags. Identity of cofilin-2 was confirmed by western blotting (not shown). As above, A35T protein failed to purify by standard methods (lanes 1 and 2). Analysis of whole bacterial-cell lysates showed roughly equal amounts of A35T and WT protein produced (lanes 3 and 4), but only WT protein was soluble and present in centrifuged supernatants (lanes 5 and 6). Treatment of the insoluble fractions with 6 M urea resulted in recovery and purification of the A35T proteins (lanes 7–10).
We hypothesize that misfolding or destabilization of the protein’s tertiary structure leads to premature degradation of the protein in vivo, so the mechanism of disease is likely related to reduction in the amount of cofilin-2. Determining whether or not reduced cofilin-2 is a useful diagnostic marker for CFL2 mutations will require identification of additional patients with different mutations, but it is noteworthy that only some pathogenic Caenorhabditis elegans UNC-60B (cofilin-2) mutations resulted in reduced levels of this protein.22
The pathology of affected muscles in our proband was remarkable for simultaneous presence of nemaline bodies, minicores, and concentric laminated bodies. Whereas nemaline bodies are associated with abnormalities of thin-filament proteins, concentric laminated bodies are postulated to be organized aggregates of excessive actin filaments,23 and both these and the minicores24 may in fact represent the ultrastructural correlates for the F-actin accumulations seen by immunofluorescence and phalloidin staining (fig. 2). Similar patterns of staining for both total sarcomeric actin and phalloidin-stained F-actin support a hypothesis that the lower amounts of cofilin-2 result in reduced depolymerization of actin filaments, causing their accumulation in concentric laminated bodies, nemaline bodies, and minicore regions.
In summary, we have identified mutation of a sixth thin-filament–related gene, CFL2, associated with an unusual form of NM. To our knowledge, this represents the first reported instance of mutation in any AC-gene-family member in humans. We estimate the frequency of CFL2 mutations in patients with NM at well below 0.6%. The rarity of CFL2 mutations is not surprising in the context that only a single instance of TNNT1 mutation has been found to date,8 whereas only two TPM2 mutations are known in patients with NM.9 The neuropathologist’s diagnosis of the proband's condition was unequivocally NM; however, occasional minicores and areas of filamentous-actin accumulation were also present in few fibers. Since both nemaline bodies and minicores may occasionally be nonspecific and the older sister presented with an undefined congenital myopathy, mutations of CFL2 should be considered possible in patients with any of these findings.
Acknowledgments
The authors gratefully acknowledge the participation of all the patients and family members, as well as many referring physicians, without whom this study would not have been possible. Thanks also to Dr. Christopher Pierson for assistance in interpreting pathological materials, to Elizabeth Taylor for patient recruitment and education, and to Molly O’Connell for assistance with cell-culture experiments. This work was supported by generous donations from the Joshua Frase Foundation and the Lee and Penny Anderson Family Foundation, by grants from the Muscular Dystrophy Association (USA), and by National Institutes of Health grant AR44345 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases. The Helsinki group (V.-M.L. and C.W.-P.) was supported by grants from the Sigrid Jusélius Foundation, the Finska Laekaresaellskapet, the Association Francaise contre les Myopathies, the Liv och Haelsa Foundation, and the Academy of Finland. N.G.L. is supported by the Australian National Health and Medical Research Council Fellowship grant 403904 and Project Grant 403941.
Web Resources
The URLs for data presented herein are as follows:
- MolScript, http://www.avatar.se/molscript/
- O, http://www.bioxray.dk/~mok/o-files.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for NM forms 1–6 and CFL2)
- RCSB Protein Data Bank, http://www.rcsb.org/pdb/ (for cofilin-2)
References
- 1.North KN, Laing NG, Wallgren-Pettersson C (1997) Nemaline myopathy: current concepts. The ENMC International Consortium and Nemaline Myopathy. J Med Genet 34:705–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryan MM, Schnell C, Strickland CD, Shield LK, Morgan G, Iannaccone ST, Laing NG, Beggs AH, North KN (2001) Nemaline myopathy: a clinical study of 143 cases. Ann Neurol 50:312–320 10.1002/ana.1080 [DOI] [PubMed] [Google Scholar]
- 3.Sanoudou D, Beggs AH (2001) Clinical and genetic heterogeneity in nemaline myopathy—a disease of skeletal muscle thin filaments. Trends Mol Med 7:362–368 10.1016/S1471-4914(01)02089-5 [DOI] [PubMed] [Google Scholar]
- 4.Ryan MM, Ilkovski B, Strickland CD, Schnell C, Sanoudou D, Midgett C, Houston R, Muirhead D, Dennett X, Shield LK, et al (2003) Clinical course correlates poorly with muscle pathology in nemaline myopathy. Neurology 60:665–673 [DOI] [PubMed] [Google Scholar]
- 5.Nowak KJ, Wattanasirichaigoon D, Goebel HH, Wilce M, Pelin K, Donner K, Jacob RL, Hubner C, Oexle K, Anderson JR, et al (1999) Mutations in the skeletal muscle α-actin gene in patients with actin myopathy and nemaline myopathy. Nat Genet 23:208–212 10.1038/13837 [DOI] [PubMed] [Google Scholar]
- 6.Pelin K, Hilpela P, Donner K, Sewry C, Akkari PA, Wilton SD, Wattanasirichaigoon D, Bang ML, Centner T, Hanefeld F, et al (1999) Mutations in the nebulin gene associated with autosomal recessive nemaline myopathy. Proc Natl Acad Sci USA 96:2305–2310 10.1073/pnas.96.5.2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wallgren-Pettersson C, Pelin K, Nowak KJ, Muntoni F, Romero NB, Goebel HH, North KN, Beggs AH, Laing NG (2004) Genotype-phenotype correlations in nemaline myopathy caused by mutations in the genes for nebulin and skeletal muscle α-actin. Neuromuscul Disord 14:461–470 10.1016/j.nmd.2004.03.006 [DOI] [PubMed] [Google Scholar]
- 8.Johnston JJ, Kelley RI, Crawford TO, Morton DH, Agarwala R, Koch T, Schaffer AA, Francomano CA, Biesecker LG (2000) A novel nemaline myopathy in the Amish caused by a mutation in troponin T1. Am J Hum Genet 67:814–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donner K, Ollikainen M, Ridanpaa M, Christen HJ, Goebel HH, de Visser M, Pelin K, Wallgren-Pettersson C (2002) Mutations in the β-tropomyosin (TPM2) gene—a rare cause of nemaline myopathy. Neuromuscul Disord 12:151–158 10.1016/S0960-8966(01)00252-8 [DOI] [PubMed] [Google Scholar]
- 10.Laing NG, Wilton SD, Akkari PA, Dorosz S, Boundy K, Kneebone C, Blumbergs P, White S, Watkins H, Love DR, et al (1995) A mutation in the α tropomyosin gene TPM3 associated with autosomal dominant nemaline myopathy. Nat Genet 9:75–79 10.1038/ng0195-75 [DOI] [PubMed] [Google Scholar]
- 11.Wattanasirichaigoon D, Swoboda KJ, Takada F, Tong HQ, Lip V, Iannaccone ST, Wallgren-Pettersson C, Laing NG, Beggs AH (2002) Mutations of the slow muscle α-tropomyosin gene, TPM3, are a rare cause of nemaline myopathy. Neurology 59:613–617 [DOI] [PubMed] [Google Scholar]
- 12.Gommans IM, Davis M, Saar K, Lammens M, Mastaglia F, Lamont P, van Duijnhoven G, ter Laak HJ, Reis A, Vogels OJ, et al (2003) A locus on chromosome 15q for a dominantly inherited nemaline myopathy with core-like lesions. Brain 126:1545–1551 10.1093/brain/awg162 [DOI] [PubMed] [Google Scholar]
- 13.Bamburg JR, McGough A, Ono S (1999) Putting a new twist on actin: ADF/cofilins modulate actin dynamics. Trends Cell Biol 9:364–370 10.1016/S0962-8924(99)01619-0 [DOI] [PubMed] [Google Scholar]
- 14.Maciver SK, Hussey PJ (2002) The ADF/cofilin family: actin-remodeling proteins. Genome Biol 3:reviews3007 10.1186/gb-2002-3-5-reviews3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vartiainen MK, Mustonen T, Mattila PK, Ojala PJ, Thesleff I, Partanen J, Lappalainen P (2002) The three mouse actin-depolymerizing factor/cofilins evolved to fulfill cell-type-specific requirements for actin dynamics. Mol Biol Cell 13:183–194 10.1091/mbc.01-07-0331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ono S, Ono K (2002) Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol 156:1065–1076 10.1083/jcb.200110013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thirion C, Stucka R, Mendel B, Gruhler A, Jaksch M, Nowak KJ, Binz N, Laing NG, Lochmuller H (2001) Characterization of human muscle type cofilin (CFL2) in normal and regenerating muscle. Eur J Biochem 268:3473–3482 10.1046/j.1432-1327.2001.02247.x [DOI] [PubMed] [Google Scholar]
- 18.Chan Y, Tong HQ, Beggs AH, Kunkel LM (1998) Human skeletal muscle-specific α-actinin-2 and -3 isoforms form homodimers and heterodimers in vitro and in vivo. Biochem Biophys Res Commun 248:134–139 10.1006/bbrc.1998.8920 [DOI] [PubMed] [Google Scholar]
- 19.Bowman GD, Nodelman IM, Hong Y, Chua NH, Lindberg U, Schutt CE (2000) A comparative structural analysis of the ADF/cofilin family. Proteins 41:374–384 [DOI] [PubMed] [Google Scholar]
- 20.Iida K, Matsumoto S, Yahara I (1992) The KKRKK sequence is involved in heat shock-induced nuclear translocation of the 18-kDa actin-binding protein, cofilin. Cell Struct Funct 17:39–46 [DOI] [PubMed] [Google Scholar]
- 21.Lee YJ, Keng PC (2005) Studying the effects of actin cytoskeletal destabilization on cell cycle by cofilin overexpression. Mol Biotechnol 31:1–10 10.1385/MB:31:1:001 [DOI] [PubMed] [Google Scholar]
- 22.Ono S, Baillie DL, Benian GM (1999) UNC-60B, an ADF/cofilin family protein, is required for proper assembly of actin into myofibrils in Caenorhabditis elegans body wall muscle. J Cell Biol 145:491–502 10.1083/jcb.145.3.491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Payne CM, Curless RG (1976) Concentric laminated bodies: ultrastructural demonstration of muscle fiber type specificity. J Neurol Sci 29:311–322 10.1016/0022-510X(76)90180-5 [DOI] [PubMed] [Google Scholar]
- 24.Bonnemann CG, Thompson TG, van der Ven PF, Goebel HH, Warlo I, Vollmers B, Reimann J, Herms J, Gautel M, Takada F, et al (2003) Filamin C accumulation is a strong but nonspecific immunohistochemical marker of core formation in muscle. J Neurol Sci 206:71–78 10.1016/S0022-510X(02)00341-6 [DOI] [PubMed] [Google Scholar]