Abstract
Cerebral cavernous malformations (CCMs) are vascular abnormalities of the brain that can result in a variety of neurological disabilities, including hemorrhagic stroke and seizures. Mutations in the gene KRIT1 are responsible for CCM1, mutations in the gene MGC4607 are responsible for CCM2, and mutations in the gene PDCD10 are responsible for CCM3. DNA sequence analysis of the known CCM genes in a cohort of 63 CCM-affected families showed that a high proportion (40%) of these lacked any identifiable mutation. We used multiplex ligation-dependent probe analysis to screen 25 CCM1, -2, and -3 mutation–negative probands for potential deletions or duplications within all three CCM genes. We identified a total of 15 deletions: 1 in the CCM1 gene, 0 in the CCM3 gene, and 14 in the CCM2 gene. In our cohort, mutation screening that included sequence and deletion analyses gave disease-gene frequencies of 40% for CCM1, 38% for CCM2, 6% for CCM3, and 16% with no mutation detected. These data indicate that the prevalence of CCM2 is much higher than previously predicted, nearly equal to CCM1, and that large genomic deletions in the CCM2 gene represent a major component of this disease. A common 77.6-kb deletion spanning CCM2 exons 2–10 was identified, which is present in 13% of our entire CCM cohort. Eight probands exhibit an apparently identical recombination event in the CCM2 gene, involving an AluSx in intron 1 and an AluSg distal to exon 10. Haplotype analysis revealed that this CCM2 deletion occurred independently at least twice in our families. We hypothesize that these deletions occur in a hypermutable region because of surrounding repetitive sequence elements that may catalyze the formation of intragenic deletions.
Cerebral cavernous malformations (CCMs) are congenital vascular anomalies of the CNS, with an incidence in the general population of 0.1%–0.5%.1 The lesions are characterized by grossly enlarged blood vessels consisting of a single layer of endothelium and without any intervening neural tissue, ranging in size from a few millimeters to several centimeters.1–3 CCMs usually present clinically during the 3rd to 5th decade of life as hemorrhagic stroke, seizures, recurrent headaches, and/or focal neurologic deficits.4,5 Magnetic resonance imaging (MRI) can detect these lesions and is the only tool available that can detect this clinically silent lesion.
Familial forms of CCM are inherited in an autosomal dominant fashion, with three known loci on chromosomes 7q21.2 (CCM1 [MIM 116860]),6–8 7p13 (CCM2 [MIM 603284]),9 and 3q25.2-q27 (CCM3 [MIM 603285]).9 The disease gene responsible for CCM1 encodes KRIT1 (KREV interaction trapped 1).10,11 The disease gene for CCM2 is MGC4607; it encodes malcavernin.12,13 The disease gene for CCM3 encodes PDCD10 (programmed cell death 10).14
With the recent identification of the CCM3 gene, we noted an apparent discrepancy in the relative frequencies of mutations in the three CCM genes between the values originally predicted by linkage in families and the values subsequently obtained by DNA sequence–analysis screens of probands. On the basis of the linkage results of 20 CCM-affected families, Craig et al. predicted that CCM1 would account for 40% of all familial CCM cases, CCM2 would account for 20%, and CCM3 would account for 40%.9 Although the frequency of identified mutations reported in different patient cohorts for both CCM1 and CCM2 appeared to support the disease frequency based on the initial linkage data,11–13,15–17 the frequency of mutations in CCM3 was found to be lower than expected (<10%). On the basis of the results from separate screens, ∼30% of probands with CCM had no mutation detectable by sequence analysis.18,19 These data suggest either that another CCM gene exists or that a significant fraction of CCM mutations are not found by routine DNA sequence analysis. Potential mutations that would be undetected by sequence analysis include mutations within regulatory regions not included in routine sequencing, as well as larger genomic insertions, deletions, or duplications.
We hypothesized that some or most of the probands with no mutations detected by routine sequencing could be accounted for by large genomic deletions or duplications and that these genomic rearrangements would account for the discrepancy between the predicted and observed genotype frequencies. The multiplex ligation-dependent probe amplification (MLPA) assay is designed to screen for exonic and whole-gene deletion or duplication events. MLPA has been used to identify deletions and duplications in a variety of inherited diseases.20–22 In the present study, we used MLPA to screen a panel of probands for deletions or duplications within the three known CCM genes. The panel consisted of probands who were negative, on DNA sequence analysis, for mutations within the CCM1, CCM2, and CCM3 genes.
Material and Methods
Study Subjects
The population for the present study consists of 63 non-Hispanic probands with CCM, as well as 255 additional affected and unaffected family members. Each of the probands received a positive diagnosis of CCM through MRI and had multiple lesions and/or at least one other family member with a CCM lesion confirmed by MRI. These criteria have been used by us11,12 and others9,13,14,23 as strong indicators of the inherited form of CCM. This particular CCM cohort does not include any probands with the common (founder) Hispanic CCM1 mutation (c.1363C→T; p.Q455X).11
Blood samples were obtained from all individuals in the study after receipt of informed consent, and DNA was prepared from these samples through use of standard methods.
Mutation Analysis
Coding exons of the KRIT1 (CCM1), MGC4607 (CCM2), and PDCD10 (CCM3) genes were amplified using primers described elsewhere.11,12,18 Exons were sequenced using the Big Dye Terminator chemistry version 1.1 and were run on the ABI PRISM 3100 or 3730 (Applied Biosystems). Sequence tracings were analyzed using Sequencher 4.1.4 (Gene Codes).
MLPA Analysis
The MLPA CCM test kit (SALSA P130 and P131) was obtained from MRC-Holland. The P130 probe mix contains MLPA probes for 8 of the 19 exons of CCM1 and for all 10 exons of CCM2, with two probes for exon 2 of CCM2 and one probe located 690 bp proximal to exon 1 of CCM1. The P131 probe mix contains probes for an additional 9 of the 19 exons of CCM1 and for 8 of the 9 exons of CCM3, with two probes for exon 1 of CCM3. There are currently no probes for exons 3 and 7 of CCM1 or exon 8 of CCM3.
MLPA was performed on those probands who were negative, by sequence analysis, for mutations in CCM1, CCM2, and CCM3. Each reaction set included three control samples: two unaffected individuals and one CCM-affected person with a known mutation detectable by sequence analysis. MLPA was performed twice on all deletion-positive probands. For additional confirmation of deletions, MLPA was also performed on a second MRI-positive family member, when available. In the validation screen, only the relevant probe mix was used (P130 for any CCM2 deletions; P130 and P131 for any CCM1 deletions).
MLPA was performed according to the protocol supplied, by use of 100–300 ng DNA sample per reaction. Volumes of all reactions were halved, except in the DNA denaturation step, when samples were diluted to 7 μl with Tris-EDTA. Samples were run on an ABI PRISM 3100 or 3730, and the data were analyzed with GeneMapper version 3.5 (Applied Biosystems). Quantification of copy number was performed as recommended by MRC-Holland, by use of both visual inspection and normalized peak-area calculations. For the visual inspection, peak heights were compared between the sample and controls, to find any alteration in relative peak heights within the test sample (fig. 1). For the normalized peak-area calculations, each peak area was normalized by dividing the individual peak area by the total peak area of all peaks for that sample. Then, for each exon, the normalized peak area of the sample was divided by the averaged normalized peak area of the controls. By definition, ratios with a range of 0.45–0.7 indicate a deletion of that exon, and ratios with a range of 1.3–1.6 indicate a duplication of that exon.
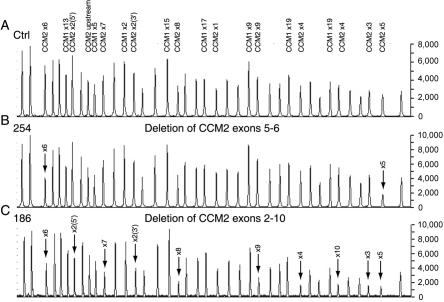
Figure 1. .
Representative chromatograms from MLPA analysis of CCM1 and CCM2. A, Results for control individual with peaks corresponding to CCM1 and CCM2 exonic probes. There are two probes to CCM2 exon 2 (5′ and 3′) and one probe to a region 690 bp upstream of CCM2 exon 1 (“upstream”). All unlabeled peaks represent the control peaks resulting from the amplification of probes located on different chromosomes. B, Chromatogram from CCM-affected proband 254, showing the relative reduction in peak area of probes hybridizing to exons 5 and 6 of CCM2. Arrows mark the deleted CCM2 exons. C, Chromatogram from CCM-affected proband 186, showing the relative reduction in peak area of probes hybridizing to exons 2–10 of CCM2. This represents the common CCM2 deletion spanning CCM2 exons 2–10. Arrows mark the deleted CCM2 exons.
PCR Analysis of CCM2 Deletions
For deletions spanning regions of the CCM2 gene, long-range PCR was performed using the Expand 20 kb PLUS PCR System (Roche Applied Science) following the protocol supplied. A series of forward primers mapping within intron 1 and a series of reverse primers in the intergenic region distal to exon 10 were used to map the precise breakpoints for the deletions spanning exons 2–10.
The primers CCM2i1 J F (5′-GGGACCTCTGTTTTACAGGATATAGAAT-3′) and CCM2outside R4 (5′-AGATACCAGACTATATCATGCTGCTACAAC-3′) generated an 839-bp product that spanned the deletion of exons 2–10 of the CCM2 gene (figs. 2 and 3). For this specific amplification, a control set of primers (CCM1 exon 2) was included in the reaction, to ensure appropriate DNA quality and quantity and to confirm that any negative result from the J F and R4 primers was not due to failure of the PCR itself. The PCR was performed using standard conditions (200 mM dNTPs, 10 mM tris-HCl [pH 9.0], 50 mM KCl, 0.1% Triton X-100, 0.01% [w/v] gelatin, 1.5 mM MgCl2, 0.4 mM each primer, 0.1 U Taq [Invitrogen]), with 35 cycles of 10 s at 94°C, 30 s at 60°C, and 3 min at 72°C.
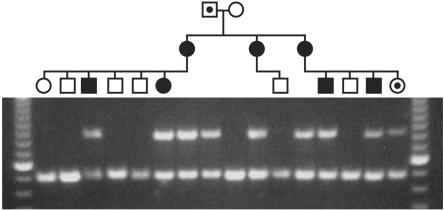
Figure 2. .
Segregation of deletion with disease in family 186. Blackened symbols show affected individuals; blackened dots indicate carrier status (by microsatellite marker disease haplotype); unblackened symbols depict healthy individuals with normal MRI results. The top band is a PCR product through use of primers spanning the CCM2 exons 2–10 deletion, and only those individuals who harbor the deletion will show this band. The bottom band is a control PCR product (CCM1 exon 2), to ensure appropriate DNA quality and quantity in the multiplex PCR. A 100-bp ladder was used for sizing, with the brighter band indicating 600 bp.
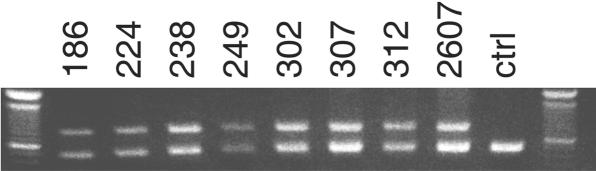
Figure 3. .
PCR across the deletion of CCM2 exons 2–10. PCR by use of primers in intron 1 and distal to exon 10 resulted in the same size band in all eight probands who had a deletion of CCM2 exons 2–10 as detected by MLPA. The top band (839 bp) is a PCR product through use of primers spanning the deletion, and only those individuals who harbor the deletion will show this band. The bottom band is a control PCR product (CCM1 exon 2), to ensure appropriate DNA quality and quantity in the multiplex PCR. A 100-bp ladder was used for sizing, with the brighter band indicating 600 bp. ctrl = Control.
The primers CCM2i4 F (5′-CTAAGGAGTACATGTGTGAATTTTATGAAG-3′) and CCM2i6 R (5′-AAAACTAAGATGACTTTCAGGGACATAGT-3′) generated an ∼4-kb product that spanned the deletion of exons 5–6 of the CCM2 gene. The PCR and cycling conditions were performed, as recommended by Roche, by use of the Expand 20 kb PLUS PCR System.
Microsatellite Analysis of CCM2 Haplotypes
Haplotypes were generated for all available members of the eight families with the CCM2 exons 2–10 deletion. Genotypes for each marker were performed using standard methods. Marker CL7N4 is a (GT)n dinucleotide repeat and was amplified using the forward primer 5′-ATCACCTTGGGGGTTAGGAT-3′ and the reverse primer 5′-GTGATGCAATGGGAAGGAGT-3′. Marker CL7N5 is a (GA)n dinucleotide repeat and was amplified using the forward primer 5′-CCACCTTCTTGAGGGAACAG-3′ and the reverse primer 5′-ACTCATGTGGGCCAATCTCT-3′.
Results
Sequence Analysis of Probands with CCM
We initially performed DNA sequence analysis of the three known CCM genes—CCM1 (KRIT1), CCM2 (MGC4607), and CCM3 (PDCD10)—on a cohort of 63 probands with CCM. We identified 24 mutations in the CCM1 gene, 10 mutations in the CCM2 gene, and 4 mutations in the CCM3 gene. Most of these results have been reported elsewhere.11,12,15,18 The results of this mutation survey showed that the relative mutation frequencies of the three CCM genes were 38% for CCM1, 16% for CCM2, and 6% for CCM3. The remaining 25 probands (40%) lacked any mutation identifiable through use of DNA sequence analysis. In light of this observation, we wanted to determine whether any of these 25 “mutation-negative” probands had other types of mutations not detectable by sequence analysis, such as large deletions or duplications, which might reconcile the discrepancy between predicted and observed genotype frequencies.
Detection of CCM Deletions by MLPA
Using MLPA, we screened our panel of 25 mutation-negative probands for deletions and duplications within the three known CCM genes (table 1). No duplications and 15 deletions were identified (fig. 1 and table 1). One deletion was in the CCM1 gene, and 14 deletions were in the CCM2 gene. When appropriate samples were available, each of the deletions was confirmed by MLPA in an additional affected family member. We used long-range PCR for confirmation of the deletions spanning CCM2 exons 2–10 and spanning CCM2 exons 5–6. The PCR also demonstrated that the deletion cosegregated with the disease in families 186, 224, 238, 249, 302, and 312 (fig. 2 and data not shown). Combining the deletion screen results with our previous DNA sequencing results, in our panel of 63 probands, we found a mutation frequency of 40% (25 of 63) for CCM1, 38% (24 of 63) for CCM2, 6% (4 of 63) for CCM3, and 16% (10 of 63) who lacked any identifiable mutation (table 2).
Table 1. .
Results of MLPA Analysis
| Family | Family History | Multiple Lesionsa | Gene with Deletion | Exon(s) Deleted |
| 177 | Yes | Yes | None | |
| 201 | Yes | Yes | None | |
| 219b | Yes | Yes | CCM2 | 1 |
| 238b,c | Yes | Yes | CCM2 | 2–10 |
| 249b,c | Yes | Yes | CCM2 | 2–10 |
| 254b,c | Yes | Yes | CCM2 | 5–6 |
| 258 | Yes | Yes | None | |
| 290 | Yes | Yes | None | |
| 302c | Yes | Yes | CCM2 | 2–10 |
| 307c | Yes | Yes | CCM2 | 2–10 |
| 312b,c | Yes | Yes | CCM2 | 2–10 |
| 326b | Yes | Yes | CCM2 | 2 |
| 2567 | Yes | Yes | None | |
| 2696 | Yes | Yes | None | |
| 32 | Yes | NA | CCM2 | 1–2 |
| 207 | Yes | NA | None | |
| 1000 | Yes | NA | None | |
| 5030b | Yes | NA | CCM1 | 5–19 |
| 186b,c | Yes | No | CCM2 | 2–10 |
| 224c | Yes | No | CCM2 | 2–10 |
| 202 | No | Yes | CCM2 | 2 |
| 239 | No | Yes | None | |
| 245 | No | Yes | CCM2 | 1–2 |
| 261 | No | Yes | None | |
| 2607c | No | NA | CCM2 | 2–10 |
NA=not available.
Results confirmed by MLPA of a second affected family member.
Results confirmed by PCR on proband.
Table 2. .
Frequency of CCM Mutations[Note]
| Frequency of Mutation |
||||||
|
CCM1 |
CCM2 |
CCM3 |
||||
| Analysis | % | No. | % | No. | % | No. |
| Sequence | 38.1 | 24 | 15.9 | 10 | 6.3 | 4 |
| Deletion | 1.6 |
1 |
22.2 |
14 |
0 |
0 |
| Total | 39.7 | 25 | 38.1 | 24 | 6.3 | 4 |
Note.— Results from a panel of 63 subjects.
A Common Deletion in CCM2
Interestingly, 12 of the 14 identified CCM2 deletions included exon 2, with 8 of these spanning exons 2–10 (fig. 4A). To determine whether the deletions spanning exons 2–10 represent a common deletion in the CCM2 gene, we attempted to map the precise breakpoints for each deletion. We employed long-range PCR, using a series of forward primers mapping within intron 1 and a series of reverse primers in the intergenic region distal to exon 10. One pair of primers amplified a 6.5-kb band that provided the approximate location of the deletion breakpoints. Further PCR experiments through use of additional primers that more closely flanked the predicted breakpoints resulted in an amplification product of the same size for each of the eight probands with the exons 2–10 deletion (fig. 3). The deletion in all eight families spans 77.6 kb. Sequence analysis of the resulting PCR products identified the proximal breakpoint within an AluSx repeat mapping in intron 1 (chr7:45024885–45025183) and the distal breakpoint within an AluSg repeat in the intragenic region (chr7:45101654–45101952) (fig. 4A and 4B). DNA sequence analysis confirmed that each of the eight probands harbored an apparently identical deletion, since the sequence within the “hybrid” Alu repeat at the deletion junction was identical in each (fig. 4B).
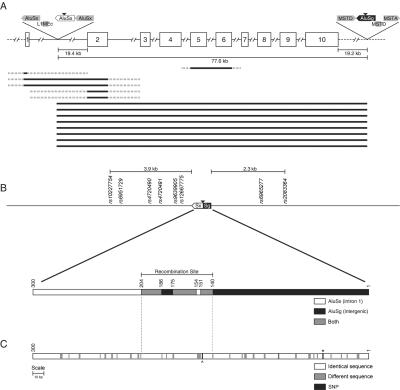
Figure 4. .
Large deletions within CCM2. A, Schematic diagram of the CCM2 gene and the 14 CCM2 deletions that were identified. The CCM2 gene is depicted with each exon shown as a white box with the corresponding exon number and each intron shown as a line. Thick black lines below the gene depict each of the CCM2 deletions, with the solid portion showing the minimal extent of the deletion and the broken portion showing the maximal extent of the deletion. SINE and LINE elements located near the breakpoints of the common exons 2–10 deletion are depicted as gray boxes above the CCM2 gene, with orientation of each element indicated by the arrowhead. The black triangles indicate the location of the breakpoints in the common deletion spanning CCM2 exons 2–10. Not drawn to scale. B, Schematic diagram of the recombinant Alu element in the common CCM2 deletion and surrounding region. The recombinant Alu element is shown as a box, with the AluSx and AluSg halves indicated. Nearby SNPs are also shown. A more detailed version of the recombinant Alu element is shown below. White indicates regions where the sequence is specific to the AluSx, black indicates regions where the sequence is specific to the AluSg, and gray indicates regions where the sequence is identical between the AluSx and the AluSg. Base-pair positions are shown above the box. C, Sequence homology between the parental AluSx (chr7:45024885–45025183) and AluSg (chr7:45101654–45101952). White boxes indicate identical base pairs, gray boxes indicate different base pairs, and black boxes indicate the location of two SNPs. The carat represents a SNP in AluSx (chr7:45025035G→A), and the asterisk (*) represents SNP rs6955183 in AluSg.
All eight of these probands/families are reported to be non-Hispanic whites, without any obvious shared ethnicity. We performed SNP haplotype analysis to determine whether this identical deletion was due to a founder effect. We used long-range PCR to amplify eight known SNPs in cis with the deletion: six mapping within a 4-kb region proximal to the deletion and two mapping within a 2.3-kb region distal to the deletion (fig. 4B). All of the probands shared the same haplotype immediately flanking the deletion, but this haplotype (GGAAGCGC) included the most common allele for each SNP. Furthermore, these SNPs are in nearly complete linkage disequilibrium with each other, with D′ values ranging from 0.86 to 1 (International HapMap Project). Thus, these data could not formally distinguish between a common founder for all eight families and the possibility that the deletion had occurred more than once on a relatively common SNP haplotype. To further investigate the possibility of a common founder, we examined the eight families for the more informative class of microsatellite repeats, choosing simple sequence repeats that mapped very near the deletion breakpoints. These data were consistent with at least two separate microsatellite haplotypes flanking the deletion (fig. 5).
Figure 5. .
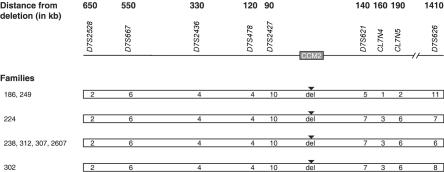
Schematic representation of affected haplotypes for families with the CCM2 exons 2–10 deletion. Each haplotype is represented by an unblackened bar, with the affected alleles for each marker shown within the bar. The location of the deletion is shown as “del” with a blackened triangle above the bar. Marker names are listed above their associated alleles, and family numbers are listed to the left of their associated haplotypes. For marker D7S2528, allele 2 is 306 bp. For marker D7S2427, allele 10 is 222 bp. For marker D7S2436, allele 4 is 262 bp. For marker D7S621, allele 5 is 274 bp and allele 7 is 266 bp. For marker CL7N4, allele 1 is 240 bp and allele 3 is 236 bp. For marker CL7N5, allele 2 is 191 bp and allele 6 is 183 bp. For marker D7S626, allele 6 is 371 bp, allele 7 is 367 bp, allele 8 is 363 bp, and allele 11 is 351 bp. The distance (in kb) of each marker from the deletion is shown at the top. Not drawn to scale.
The microsatellite haplotype data demonstrated that this common deletion spanning CCM2 exons 2–10 had occurred independently at least twice in this limited cohort of CCM-affected families, suggesting that, in the history of CCM across the globe, this deletion may have occurred multiple times. Since the haplotypes differ only on the distal side of the deletion, an alternate explanation would be that there is an ancient founder and that these differences are due to ancestral recombination events distal to the deletion. According to the sequence deposited in the University of California–Santa Cruz Human Genome Bioinformatics database, these particular AluSx (chr7:45024885–45025183) and AluSg (chr7:45101654–45101952) repeats share 83% homology. Alignment of these particular repeat elements shows stretches of identity that might catalyze the recombination event (fig. 4C), with the central region of predicted recombination containing two of the longest stretches of homology.
We further hypothesized that sequence variation might exist within these AluSx and AluSg repeats that would increase their sequence homology and thus make them even more prone to the deletion-causing recombination event. Sequencing of 58 control alleles resulted in the identification of a novel SNP within the AluSx element and confirmation of a SNP (rs6955183), noted elsewhere (dbSNP), within the AluSg element. The alleles at these SNPs are in nearly complete linkage disequilibrium with each other, and the haplotype containing the minor alleles decreases the homology between the two Alu elements. The CCM2 deletion occurs on the more common haplotype, which increases the overall homology between the two elements, albeit only slightly.
Discussion
Using MLPA, we determined that many of the CCM mutations undetected by routine DNA-based sequence analysis could be accounted for by large genomic CCM deletions. We identified deletions in 60% (15 of 25) of our probands in whom mutations had not been previously identified. Deletions, especially in the CCM2 gene, account for a significant portion of the undetected mutations in our panel. It is possible that a few of these unidentified mutations are single-exon deletions of either CCM1 exon 3, CCM1 exon 7, or CCM3 exon 8, since these exons are not included in the MLPA assay. The fact that 16% (10 of 63) of CCM mutations still remain unidentified suggests either that the remaining mutations are in regions not examined in routine DNA sequencing or that there may be an additional CCM gene(s).
The inclusion of the deletion-screen results with our previous DNA sequencing results altered the observed frequencies of CCM1, -2, and -3 mutations. The discrepancy between predicted and observed mutation frequencies is now even more pronounced. With the identification of only one deletion in the CCM1 gene, the frequency (40%) of CCM1 mutations in our panel matches the frequency predicted by linkage (40%).9 Although deletions had been identified in the CCM3 gene,14 we did not find any CCM3 deletions or duplications within our panel. Thus, the frequency of CCM3 mutations (6%) in our cohort is much lower than the 40% predicted in the initial linkage screen.9 The overwhelming majority (93%) of the deletions in our panel were found in the CCM2 gene, indicating that the prevalence of CCM2 may be much higher than previously suspected. Deletions had also been identified in the CCM2 gene.13 The inclusion of the deletion data increases the frequency of CCM2 mutations in our panel to 38%, nearly double that of the original prediction (20%)9 and approximately equal to the frequency of CCM1 mutations. Of the patients in our panel, ∼78% have mutations in either the CCM1 or CCM2 genes, whereas mutations in the CCM3 gene appear to be relatively rare.
We identified 8 probands with a deletion spanning CCM2 exons 2–10, and this specific mutation is found in 13% (8 of 63) of our CCM cohort. All eight probands with this deletion have the same breakpoints within an AluSx in CCM2 intron 1 and an AluSg distal to CCM2 exon 10. This common CCM2 deletion can be identified by a simple PCR assay through use of primers that flank the deletion breakpoints. The deletion occurred independently at least twice in our limited cohort, suggesting that it may have occurred many more times worldwide. Nonetheless, we cannot rule out the existence of a founder effect in specific populations. The deletion is likely catalyzed by a hypermutable region because of surrounding repetitive sequence elements.
Two SNPs within these Alu elements decrease the homology of the Alu sequences to each other and may reduce the chance of recombination between these two elements. The large CCM2 deletion in each of the eight families in this panel occurred on the more common haplotype, which contained the alleles with increased homology between the two Alu elements. It is interesting to note that, whereas there are three AluSx elements in the immediate vicinity of the intron 1 breakpoint, the AluSx (chr7:45024885–45025183) is more homologous to the AluSg (chr7:45101654–45101952) distal to exon 10 than to any of its neighboring AluSx elements.
This study documents that large deletions in the CCM2 gene represent a major cause of CCM. These deletions have heretofore been missed by routine DNA sequence-based mutation screens. In our panel, deletions within the CCM2 gene account for 22% of all identified CCM mutations (table 2). In addition, the common exons 2–10 deletion itself accounts for 13% of all identified CCM mutations in our panel. Thus, deletion screening should be performed on all probands with CCM, to increase the detection rate of CCM mutations. The common CCM2 exons 2–10 deletion can be identified using the simple PCR-based amplification assay.
Acknowledgments
We are grateful to the patients with CCM and their families, for participation in this study. This work was supported by National Institutes of Health grant NS43543 (to D.A.M.) and National Research Service Award fellowship 1 F32 NS51082 (to C.L.L.).
Web Resources
The URLs for data presented herein are as follows:
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
- International HapMap Project, http://www.hapmap.org/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CCM1, CCM2, and CCM3)
- University of California–Santa Cruz Human Genome Bioinformatics, http://genome.ucsc.edu/ (for the March 2006 assembly)
References
- 1.Rigamonti D, Hadley MN, Drayer BP, Johnson PC, Hoenig-Rigamonti K, Knight JT, Spetzler RF (1988) Cerebral cavernous malformations: incidence and familial occurrence. N Engl J Med 319:343–347 [DOI] [PubMed] [Google Scholar]
- 2.Tomlinson FH, Houser OW, Scheithauer BW, Sundt TM Jr, Okazaki H, Parisi JE (1994) Angiographically occult vascular malformations: a correlative study of features on magnetic resonance imaging and histological examination. Neurosurgery 34:792–799 [DOI] [PubMed] [Google Scholar]
- 3.Gil-Nagel A, Wilcox KJ, Stewart JM, Anderson VE, Leppik IE, Rich SS (1995) Familial cerebral cavernous angioma: clinical analysis of a family and phenotypic classification. Epilepsy Res 21:27–36 10.1016/0920-1211(95)00005-U [DOI] [PubMed] [Google Scholar]
- 4.Zabramski JM, Wascher TM, Spetzler RF, Johnson B, Golfinos J, Drayer BP, Brown B, Rigamonti D, Brown G (1994) The natural history of familial cavernous malformations: results of an ongoing study. J Neurosurg 80:422–432 [DOI] [PubMed] [Google Scholar]
- 5.Zabramski JM, Henn JS, Coons S (1999) Pathology of cerebral vascular malformations. Neurosurg Clin N Am 10:395–410 [PubMed] [Google Scholar]
- 6.Dubovsky J, Zabramski JM, Kurth J, Spetzler RF, Rich SS, Orr HT, Weber JL (1995) A gene responsible for cavernous malformations of the brain maps to chromosome 7q. Hum Mol Genet 4:453–458 [DOI] [PubMed] [Google Scholar]
- 7.Günel M, Awad IA, Anson J, Lifton RP (1995) Mapping of a gene causing cerebral cavernous malformation to 7q11.2-q21. Proc Nat Acad Sci USA 92:6620–6624 10.1073/pnas.92.14.6620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchuk DA, Gallione CJ, Morrison LA, Clericuzio CL, Hart BL, Kosofsky BE, Louis DN, Gusella JF, Davis LE, Prenger VL (1995) A locus for cerebral cavernous malformations maps to chromosome 7q in two families. Genomics 28:311–314 10.1006/geno.1995.1147 [DOI] [PubMed] [Google Scholar]
- 9.Craig HD, Gunel M, Cepeda O, Johnson EW, Ptacek L, Steinberg GK, Ogilvy CS, Berg MJ, Crawford SC, Scott RM, et al (1998) Multilocus linkage identifies two new loci for a Mendelian form of stroke, cerebral cavernous malformation, at 7p15-13 and 3q25.2-27. Hum Mol Genet 7:1851–1858 10.1093/hmg/7.12.1851 [DOI] [PubMed] [Google Scholar]
- 10.Laberge S, Labauge P, Marechal E, Maciazek J, Tournier-Lasserve E (1999) Genetic heterogeneity and absence of founder effect in a series of 36 French cerebral cavernous angiomas families. Eur J Hum Genet 7:499–504 10.1038/sj.ejhg.5200324 [DOI] [PubMed] [Google Scholar]
- 11.Sahoo T, Johnson EW, Thomas JW, Kuehl PM, Jones TL, Dokken CG, Touchman JW, Gallione CJ, Lee-Lin SQ, Kosofsky B, et al (1999) Mutations in the gene encoding KRIT1, a Krev-1/rap1a binding protein, cause cerebral cavernous malformations (CCM1). Hum Mol Genet 8:2325–2333 10.1093/hmg/8.12.2325 [DOI] [PubMed] [Google Scholar]
- 12.Liquori CL, Berg MJ, Siegel AM, Huang E, Zawistowski JS, Stoffer T, Verlaan D, Balogun F, Hughes L, Leedom TP, et al (2003) Mutations in a gene encoding a novel protein containing a phosphotyrosine-binding domain cause type 2 cerebral cavernous malformations. Am J Hum Genet 73:1459–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denier C, Goutagny S, Labauge P, Krivosic V, Arnoult M, Cousin A, Benabid AL, Comoy J, Frerebeau P, Gilbert B, et al (2004) Mutations within the MGC4607 gene cause cerebral cavernous malformations. Am J Hum Genet 74:326–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergametti F, Denier C, Labauge P, Arnoult M, Boetto S, Clanet M, Coubes P, Echenne B, Ibrahim R, Irthum B et al (2005) Mutations within the programmed cell death 10 gene cause cerebral cavernous malformations. Am J Hum Genet 76:42–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahoo T, Goenaga-Diaz E, Serebriiskii IG, Thomas JW, Kotova E, Cuellar JG, Peloquin JM, Golemis E, Beitinjaneh F, Green ED, et al (2001) Computational and experimental analyses reveal previously undetected coding exons of the KRIT1 (CCM1) gene. Genomics 71:123–126 10.1006/geno.2000.6426 [DOI] [PubMed] [Google Scholar]
- 16.Verlaan DJ, Davenport WJ, Stefan H, Sure U, Siegel AM, Rouleau GA (2002) Cerebral cavernous malformations: mutations in Krit1. Neurology 58:853–857 [DOI] [PubMed] [Google Scholar]
- 17.Verlaan DJ, Laurent SB, Rochefort DL, Liquori CL, Marchuk DA, Siegel AM, Rouleau GA (2004) CCM2 mutations account for 13% of cases in a large collection of kindreds with hereditary cavernous malformations. Ann Neurol 55:757–758 10.1002/ana.20112 [DOI] [PubMed] [Google Scholar]
- 18.Liquori CL, Berg MJ, Squitieri F, Ottenbacher M, Sorlie M, Leedom TP, Cannella M, Maglione V, Ptacek L, Johnson EW, et al (2005) Low frequency of PDCD10 mutations in a panel of CCM3 probands: potential for a fourth CCM locus. Hum Mutat 27:118 10.1002/humu.9389 [DOI] [PubMed] [Google Scholar]
- 19.Verlaan DJ, Roussel J, Laurent SB, Elger CE, Siegel AM, Rouleau GA (2005) CCM3 mutations are uncommon in cerebral cavernous malformations. Neurology 65:1982–1983 10.1212/01.wnl.0000188903.75144.49 [DOI] [PubMed] [Google Scholar]
- 20.Hearle NC, Rudd MF, Lim W, Murday V, Lim AG, Phillips RK, Lee PW, O’Donohue J, Morrison PJ, Norman A, et al (2006) Exonic STK11 deletions are not a rare cause of Peutz-Jeghers syndrome. J Med Genet 43:e15 10.1136/jmg.2005.036830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai KK, Lo IF, Tong TM, Cheng LY, Lam ST (2006) Detecting exon deletions and duplications of the DMD gene using multiplex ligation-dependent probe amplification (MLPA). Clin Biochem 39:367–372 10.1016/j.clinbiochem.2005.11.019 [DOI] [PubMed] [Google Scholar]
- 22.Wimmer K, Yao S, Claes K, Kehrer-Sawatzki H, Tinschert S, De Raedt T, Legius E, Callens T, Beiglbock H, Maertens O, et al (2006) Spectrum of single- and multiexon NF1 copy number changes in a cohort of 1,100 unselected NF1 patients. Genes Chromosomes Cancer 45:265–276 10.1002/gcc.20289 [DOI] [PubMed] [Google Scholar]
- 23.Squitieri F, Maglione V, Buzzi MG, Nargi E, Novelletto A, Cannella M, Simonelli M, Colonnese C, Simonelli P, Innocenzi G, et al (2000) Cavernous angiomas of the nervous system in Italy: clinical and genetic study. Neurol Sci 21:129–134 10.1007/s100720070087 [DOI] [PubMed] [Google Scholar]