Abstract
Split–hand/foot malformation with long-bone deficiency (SHFLD) is a rare, severe limb deformity characterized by tibia aplasia with or without split-hand/split-foot deformity. Identification of genetic susceptibility loci for SHFLD has been unsuccessful because of its rare incidence, variable phenotypic expression and associated anomalies, and uncertain inheritance pattern. SHFLD is usually inherited as an autosomal dominant trait with reduced penetrance, although recessive inheritance has also been postulated. We conducted a genomewide linkage analysis, using a 10K SNP array in a large consanguineous family (UR078) from the United Arab Emirates (UAE) who had disease transmission consistent with an autosomal dominant inheritance pattern. The study identified two novel SHFLD susceptibility loci at 1q42.2-q43 (nonparametric linkage [NPL] 9.8, P=.000065) and 6q14.1 (NPL 7.12, P=.000897). These results were also supported by multipoint parametric linkage analysis. Maximum multipoint LOD scores of 3.20 and 3.78 were detected for genomic locations 1q42.2-43 and 6q14.1, respectively, with the use of an autosomal dominant mode of inheritance with reduced penetrance. Haplotype analysis with informative crossovers enabled mapping of the SHFLD loci to a region of ∼18.38 cM (8.4 Mb) between single-nucleotide polymorphisms rs1124110 and rs535043 on 1q42.2-q43 and to a region of ∼1.96 cM (4.1 Mb) between rs623155 and rs1547251 on 6q14.1. The study identified two novel loci for the SHFLD phenotype in this UAE family.
Split–hand/foot malformation with long-bone deficiency (SHFLD [MIM %119100]), a rare and severe limb deformity, is also known as “cleft hand and absent tibia,” “aplasia of tibia with ectrodactyly,” “ectrodactyly with aplasia of long bones,” or “tibial aplasia (TA) with split-hand/split-foot deformity.” The clinical manifestations are highly variable and range from virtually no malformation to ectrodactyly and tibial hypoplasia or aplasia with or without associated anomalies.1–3 The incidence of SHFLD has been estimated to be ∼1 per million live births.4 It is characterized by hypoplasia or aplasia of tibia, with relatively intact fibula, associated with split-hand/split-foot deformity that more often affects the upper limb. SHFLD was first described in 1575,5,6 and Otto2 also reported an affected fetus. Families with SHFLD have been reported with autosomal dominant, recessive, and sporadic forms of inheritance.1–3,7–18 A number of malformations have been described in association with SHFLD, including triphalangeal thumbs and polydactyly,8 cleft lip/palate,16 cardiac defects,19 vaginal agenesis,20 cardiovascular defects,21 hypohidrotic ectodermal dysplasia,22 and ectrodactyly.23 We present genomewide linkage analysis of one large multigenerational Arab family with SHFLD, using the GeneChip Mapping EA 10K Array (Affymetrix) containing ∼10,000 SNP markers. The present analysis provided significant evidence for two susceptibility loci, one on a genomic region spanning 8.4 Mb on chromosome 1q42.13-q43 and another on a region of 4.1 Mb on 6q14.1. We hypothesize that SHFLD could fit the model of digenic inheritance.24–26
Material and Methods
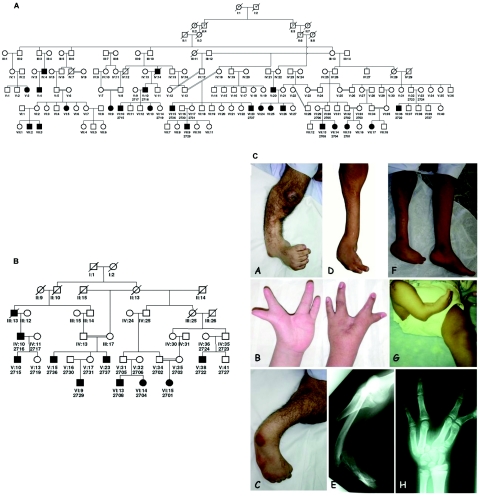
We recently reported a large multigenerational consanguineous family (UR078) from the United Arab Emirates (UAE) with autosomal dominant SHFLD.23 The original eight-generation pedigree (fig. 1A) with 10 consanguineous marriages is much larger than the present partial pedigree used for gene-mapping analysis (fig. 1B). The majority of the family members live in the UAE; however, a few reside in Oman. Of the 145 individuals in this family, 23 (14 males and 9 females) showed an abnormal phenotype, ranging from mild to severe defects involving upper and lower limbs (fig. 1C). The nine affected individuals included in this linkage study all had severe TA, some with additional findings of split hand/foot, syndactyly of fingers/toes, hypoplastic big toes, absence of middle phalanges of some toes, hypoplastic tibiae, and beaked nose. No other abnormalities—in particular, no cleft lip/palate or ectodermal dysplasia—were observed in this family.
Figure 1. .
Complete (A) and partial (B) pedigrees of family UR078 with SHFLD. Affected individuals are shown with blackened symbols, and unaffected individuals are shown with unblackened symbols. Individuals used in the linkage analysis are numbered under their symbols in the pedigree. C, Clinical and x-ray photos of selected individuals (A–H) from family UR078 with SHFLD.
Blood samples were collected from all cooperative and informative family members after informed consent for the genetic studies. DNA was not collected from individuals in the pedigree who could not be reliably defined as “affected,” because they had minimal symptoms, such as short big toe or syndactyly without TA. DNA from peripheral blood samples was isolated using standard procedures (Gentra Kit).
A genomewide search was undertaken using a GeneChip Mapping 10K XbaI Array containing 10,555 SNPs. These SNP markers are equally distributed in the genome, with a mean intermarker distance of 210 kb and an average heterozygosity of 0.38 (Affymetrix). The assay was done using 250 ng of genomic DNA, and >99% of the SNPs were determined unequivocally for each sample. Scanned images were processed with Affymetrix Micro Array Suite software. Data were analyzed with GDAS v2 software. PedCheck was used for detection of Mendelian errors.27
SNP genotype data were imported into the linkage analysis programs GENEHUNTER28 and MERLIN.29 Since the parameters of the disease model were uncertain, in the initial genome scan we assessed the evidence of linkage with nonparametric, penetrance-independent, affected-only, and allele-sharing models. Owing to the size of the family being studied, the SNP data were initially analyzed by splitting the entire family into two separate families (UR078A and UR078B), with minimal overlap between them. On finding significant evidence of linkage by exceeding the predetermined threshold (P<.01), we performed two-point, as well as multipoint (four-point), LOD scores maximized over various plausible genetic model parameters (MOD-score analysis) on the entire pedigree, using the LINKAGE analysis package. For each marker, we assessed our linkages with the white and Asian allele frequencies provided by Affymetrix. Linkage analysis using Asian or white allele frequencies for the Arab population may not be appropriate, and it may impact the parametric linkage results. However, since we have only a few founders available from this family, we could not estimate the marker-allele frequencies from the family data. The map order and intermarker distances between SNPs were based on the National Center for Biotechnology Information (NCBI) build 35.1.
To assess the false-positive evidence of linkage, we performed a simulation experiment to evaluate our results, using empirical P values. The simulations were designed to match our observed data in marker density, marker informativeness, pedigree structure, and individual phenotypes. We generated 10,000 replicate data sets, under the null hypothesis of no linkage, to estimate the empirical P value. Putative haplotypes containing the disease-causing loci were determined by using the critical recombinants across the family members.
It has been demonstrated that applying linkage analyses that assume linkage equilibrium to dense markers may lead to bias,30,31 especially when analyzing SNP linkage maps in data sets in which some parental genotypes are missing. Therefore, we assessed the impact of linkage disequilibrium (LD) on linkage at the linked regions. We used MERLIN to accommodate marker-marker LD in both parametric and nonparametric analyses, by organizing closely located adjacent markers into clusters. Although many empirical studies have shown that the extent and distribution of LD is extremely variable throughout the genome, in most cases significant LD does not influence markers separated by >0.1 cM in outbred populations.32–34 However, to be conservative, we used markers within 0.3 cM of one other in a cluster.
Results
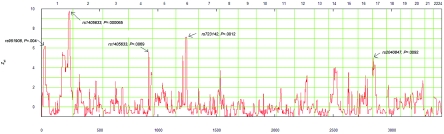
Before this genomewide genotyping, we had excluded the published candidate genomic regions on chromosomes 7p13, 7q36, 8q24.1, and 10q24 by linkage and haplotype analysis.23 Initial analysis with GENEHUNTER revealed six genomic regions (P<.01) on chromosomes 1p36, 1q31, 1q42, 4q34, 6q14, and 17p13 that may harbor the putative SHFLD susceptibility loci (table 1) (fig. 2). Subsequent analyses with MERLIN indicated similar results in these regions. Among these six linked regions, two linkages were found to be the most interesting—one on chromosome 1q42 and the other on 6q41. The maximum multipoint NPL yielded significant evidence (NPL 9.8, P=.000065) for SNP marker rs966302 (physical map position 232,585,612 bp) on chromosome 1q42.13-q43 (table 2). The locus, which was identified on chromosome 6q14.1, yielded the second-highest NPL results (NPL 7.12, P=.00089) at marker rs688867 (80,995,947 bp) (table 2). With the use of 10,000 simulations, the empirical P values for 1q42-43 and 6q41 were .0004 and .006, respectively. These genomic regions were also supported by parametric linkage analysis that used the entire family. The multipoint (four-point) LOD scores for 1q42.13-43 and 6q41.1 loci were 3.20 and 3.78, respectively (table 3). The best-fitted model for the 1q42-43 linkage was incomplete dominance with 70% penetrance and disease-allele frequency 0.01, and for 6q41 it was incomplete dominance with 50% penetrance and disease-allele frequency 0.0001. Evidence of linkage for the remaining candidate regions was not supported by the parametric linkage analyses.
Table 1. .
Initial Genomewide Linkage Analysis Results Using NPL (P<.01), Revealing Six Genomic Regions that May Harbor the Putative SHFLD Susceptibility Loci
| Position |
NonparametricAnalysis |
|||
| SNP Marker | Cytogenetic | Build 35.1 | NPL | P |
| rs951908 | 1p36.13 | 19372422 | 6.20 | .004000 |
| rs530157 | 1q31.1 | 183800525 | 5.58 | .008400 |
| rs1405633 | 1q42.3 | 234486121 | 9.79 | .000065 |
| rs720096 | 4q34.3 | 179788065 | 5.63 | .006900 |
| rs723142 | 6q14.1 | 83094274 | 7.09 | .001200 |
| rs2040847 | 17p13.1 | 6498735 | 5.01 | .009200 |
Figure 2. .
Multipoint linkage analysis using NPL in the genomewide scan for the Arab family with SHFLD. The X-axis represents the chromosome locations for all the autosomes, and the Y-axis represents the Zall. P values are derived from the NPLall statistics. The highest peak is on chromosome 1 with P=.000065 (NPL Z=9.8), and the second highest is on chromosome 6 (NPL 7.12, P=.000897). Arrows indicate the SNP markers with the highest NPL peaks.
Table 2. .
SHFLD Loci in Family UR078 Mapped by Linkage Analysis to Chromosome 1q42.13-1q43 and 6q14.1 Regions[Note]
| Position |
NPL Multipoint LinkageGenome Scan |
||||
| SNP Marker |
Genetic (cM) |
Cytogenetic | Physical (bp) |
NPL | P |
| rs1124110 | 233.32 | 1q42.13 | 226,617,707 | 4.61 | .009373 |
| rs1933633 | 234.32 | 1q42.2 | 227,231,111 | 6.25 | .003083 |
| rs953244 | 235.28 | 1q42.2 | 228,130,318 | 7.76 | .000215 |
| rs4333837 | 235.37 | 1q42.2 | 228,331,909 | 7.84 | .000201 |
| rs967433 | 235.38 | 1q42.2 | 228,338,550 | 7.85 | .000201 |
| rs720806 | 237.54 | 1q42.2 | 228,945,997 | 8.94 | .000097 |
| rs965917 | 238.86 | 1q42.2 | 229,690,846 | 9.38 | .000086 |
| rs1294330 | 238.97 | 1q42.2 | 229,755,343 | 9.45 | .000084 |
| rs780245 | 239.42 | 1q42.2 | 230,004,265 | 9.48 | .000084 |
| rs923975 | 239.95 | 1q42.2 | 230,304,605 | 9.58 | .000084 |
| rs923976 | 239.95 | 1q42.2 | 230,304,802 | 9.58 | .000084 |
| rs955612 | 241.23 | 1q42.3 | 231,023,328 | 9.58 | .000084 |
| rs1416473 | 244.23 | 1q42.3 | 231,889,300 | 9.74 | .000084 |
| rs966302 | 245.54 | 1q42.3 | 232,585,612 | 9.79 | .000065 |
| rs1405633 | 246.04 | 1q42.3 | 232,745,539 | 9.79 | .000065 |
| rs1749569 | 246.04 | 1q42.3 | 232,745,622 | 9.79 | .000065 |
| rs966364 | 246.48 | 1q42.3 | 232,883,969 | 9.75 | .000084 |
| rs959175 | 247.01 | 1q43 | 233,078,923 | 9.71 | .000084 |
| rs1337797 | 248.33 | 1q43 | 233,625,390 | 9.44 | .000084 |
| rs950964 | 248.62 | 1q43 | 233,746,722 | 9.33 | .000086 |
| rs1074189 | 249.44 | 1q43 | 234,087,534 | 8.74 | .000109 |
| rs1361358 | 250.12 | 1q43 | 234,370,458 | 7.80 | .000215 |
| rs535043 | 251.70 | 1q43 | 235,022,077 | 5.23 | .009182 |
| rs623155 | 90.01 | 6q14.1 | 79,324,200 | 7.10 | .000897 |
| rs1415863 | 90.24 | 6q14.1 | 79,756,878 | 7.10 | .000897 |
| rs1414280 | 90.36 | 6q14.1 | 79,985,449 | 7.11 | .000897 |
| rs721265 | 90.63 | 6q14.1 | 80,549,653 | 7.10 | .001173 |
| rs719172 | 90.65 | 6q14.1 | 80,611,300 | 7.11 | .000897 |
| rs688867 | 90.78 | 6q14.1 | 80,995,947 | 7.12 | .000897 |
| rs723587 | 90.82 | 6q14.1 | 81,113,079 | 7.12 | .000897 |
| rs1902066 | 90.93 | 6q14.1 | 81,402,752 | 7.10 | .000897 |
| rs1377986 | 90.96 | 6q14.1 | 81,463,288 | 7.10 | .001173 |
| rs1584896 | 90.96 | 6q14.1 | 81,463,350 | 7.10 | .001173 |
| rs1584897 | 90.96 | 6q14.1 | 81,463,381 | 6.99 | .001266 |
| rs724993 | 91.08 | 6q14.1 | 81,799,004 | 7.02 | .001217 |
| rs962984 | 91.19 | 6q14.1 | 82,120,869 | 7.04 | .001217 |
| rs962983 | 91.19 | 6q14.1 | 82,121,184 | 7.04 | .001217 |
| rs1343232 | 91.21 | 6q14.1 | 82,187,051 | 7.04 | .001207 |
| rs719144 | 91.4 | 6q14.1 | 82,388,507 | 7.07 | .001186 |
| rs733413 | 91.6 | 6q14.1 | 82,587,633 | 7.09 | .001173 |
| rs72968 | 91.6 | 6q14.1 | 82,587,967 | 7.09 | .001173 |
| rs1931621 | 91.61 | 6q14.1 | 82,596,847 | 7.09 | .001173 |
| rs1342196 | 91.75 | 6q14.1 | 82,862,011 | 7.10 | .001173 |
| rs2226121 | 91.75 | 6q14.1 | 82,862,272 | 7.10 | .001173 |
| rs1556778 | 91.75 | 6q14.1 | 82,862,533 | 7.10 | .001173 |
| rs950611 | 91.83 | 6q14.1 | 83,023,894 | 7.10 | .001173 |
Note.— Recombination narrowed the candidate region to an interval flanked by rs1294330 and rs1361358 for chromosome 1q and to an interval flanked by rs623155 and rs1547251 for chromosome 6q.
Table 3. .
Multipoint Linkage Data for Markers on Chromosomes 1q42.13-1q43 and 6q14.1[Note]
| Four-Point Analysis(Recombination Fraction) |
|||||||
| SNP Marker Order | 0 | .01 | .05 | .1 | .2 | .3 | .4 |
| 1q42.2-q43: | |||||||
| rs1749569-rs966364-rs959175 | −1.40 | −.57 | .03 | .23 | .26 | .17 | .07 |
| rs966364-rs959175-rs1337797 | 1.70 | 1.66 | 1.50 | 1.30 | .91 | .54 | .24 |
| rs959175-rs1337797-rs950964 | 2.45 | 2.39 | 2.14 | 1.84 | 1.23 | .68 | .26 |
| rs1337797-rs950964-rs1074189 | 3.20 | 3.10 | 2.80 | 2.40 | 1.60 | .90 | .40 |
| rs950964-rs1074189-rs1361358 | 2.70 | 2.70 | 2.40 | 2.00 | 1.30 | .70 | .30 |
| rs1074189-rs1361358-rs535043 | .40 | .50 | .60 | .60 | .40 | .20 | .00 |
| 6q14.1: | |||||||
| rs1414280-rs721265-rs719172 | 1.26 | 1.72 | 2.02 | 1.95 | 1.52 | .96 | .41 |
| rs721265-rs719172-rs688867 | .10 | 1.80 | 2.20 | 2.00 | 1.50 | .90 | .40 |
| rs719172-rs688867-rs723587 | 3.78 | 3.70 | 3.38 | 2.96 | 2.10 | 1.25 | .51 |
| rs688867-rs723587-rs1902066 | 3.38 | 3.31 | 3.00 | 2.61 | 1.83 | 1.08 | .45 |
| rs723587-rs1902066-rs1377986 | 1.54 | 1.50 | 1.35 | 1.15 | .78 | .46 | .19 |
| rs1902066-rs1377986-rs1584896 | 1.93 | 1.89 | 1.69 | 1.46 | .99 | .56 | .22 |
| rs1342196-rs2226121-rs1556778 | 3.17 | 3.10 | 2.83 | 2.48 | 1.77 | 1.08 | .47 |
Note.— Parametric linkage analysis using whole-family data.
In our initial linkage analysis, we used the Asian allele frequencies provided by Affymetrix, which may not truly represent the genetic makeup of the Arab population. To assess the impact of allele frequencies on our linkage findings, we used the white allele frequencies and recomputed the parametric linkage. Interestingly, we reproduced our initial linkage findings. The LOD score at 1q42.13-43 increased from 3.2 to 4.5, and at 6q41.1 the LOD score was reduced slightly, from 3.78 to 3.2. The multipoint linkage was reanalyzed to accommodate marker-marker LD in both nonparametric and parametric analyses, by the organization of closely located adjacent markers into clusters. Several clusters of two to six SNPs demonstrated LD. With the assumption of no LD within the cluster, MERLIN uses population haplotype frequencies while calculating linkage. The NPL scores at 1q42.13-43 and 6q41 were reduced from 9.8 to 8.5 and from 7.12 to 5.15, respectively. However, this reduction of linkage scores might be due to both the effect of LD and the reduction of information content (IC). Because of the clustering (hence, the reduction of the number of markers), the IC was reduced from 89% to 74% at the peak region at 1q42 and from 80% to 69% at 6q14.1. Nonetheless, evidence of linkage at both peaks is consistent.
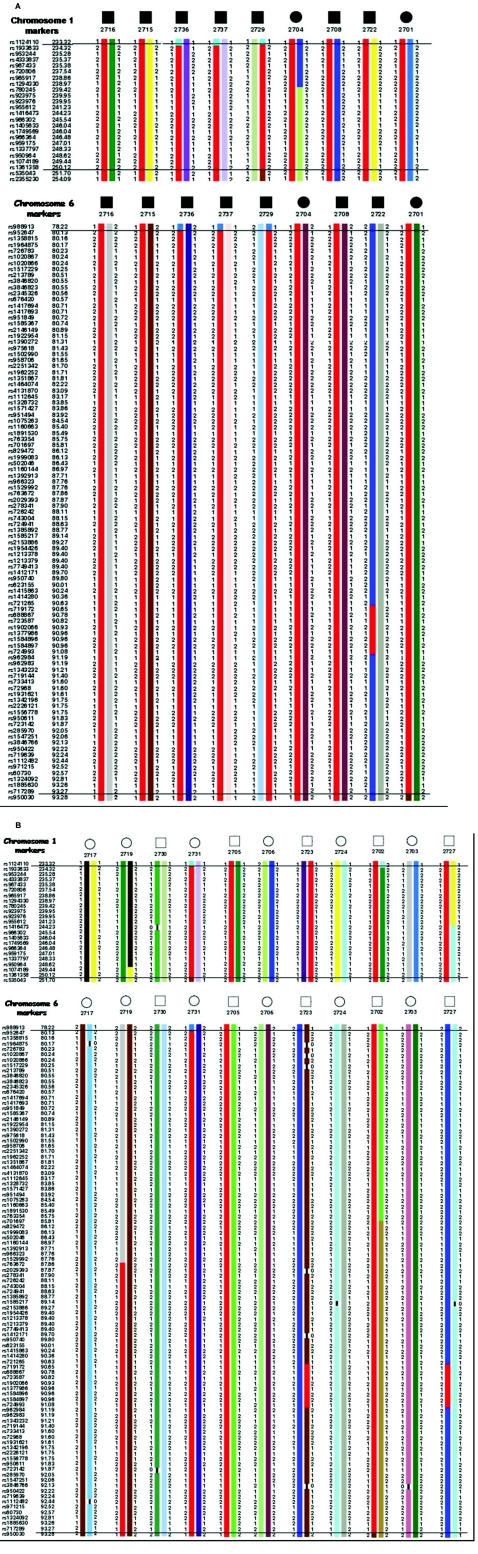
Haplotype analysis was constructed using 28 informative SNP markers on 1q42.13-q43 and revealed informative recombination events in affected individual 2729, which confined the SHFLD candidate region to ∼18.38 cM (8.4 Mb) between SNPs rs1124110 (226,617,707 bp) and rs535043 (235,022,077 bp). Similar haplotype analysis and critical recombination events across the affected family members (except 2722) on chromosome 6q14.1 narrowed the genomic region to ∼1.96 cM (4.1 Mb). The area is bordered by proximal marker rs623155 (79,324,200 bp) and distal marker rs1547251 (83,462,826 bp) (fig. 3).
Figure 3. .
Genotypes and haplotypes of chromosomes 1q and 6q. SNP markers are shown below individuals used in the linkage analysis. Haplotypes associated with affected status are shown in red. Haplotype analysis indicated that the cosegregating segment of the SHFLD locus is flanked proximally by marker rs1294330 and distally by marker rs1361358 on chromosome 1q42.13-1q43, and on chromosome 6q14.1 it was between markers rs623155 and rs1547251.
Discussion
We identified novel genomic regions on 1q42.13-q43 and 6q14.1 that harbor high-risk variants for SHFLD in this UAE family. The 8.4-Mb genomic interval on 1q42.13-q43 contains 17 known putative transcripts, whereas the 4.1-Mb genomic region on chromosome 6q14.1 contains six annotated transcripts (Ensembl). Logical candidate genes include homeobox-like protein 1 (MIXL1 [MIM 609852]), ectodysplasin A receptor–associated death domain (EDARADD [MIM 606603]), a human galectin-8–related gene (LGALS8 [MIM 606099]), alpha-actinin gene (ACTN2 [MIM 102573]), protein related to differential screening-selected gene abberative in neuroblastoma and cerberus (GREM2 [MIM 608832]), and choroideremia-like (CHML [MIM 118825]). There is no previous evidence of linkage that would indicate that 1q or 6q regions are involved in either TA or split–hand/foot malformation; on the other hand, 3q27, 7q21, 10q24, and Xq2635–38 have been implicated in familial split–hand/foot malformation. Interestingly, the 1q42-qter region harbors genetic variation related to several developmental phenotypes. Distal trisomy 1q39–41 or subtelomeric 1qter deletion42 are associated with multiple developmental anomalies, including malformed fingers and toes.
Inter- and intrafamilial variability is common in families with SHFLD anomalies.2,3,12,23,43,44 Autosomal dominant inheritance with reduced penetrance was reported by Marioni et al.10 In the present family, individuals 2702, 2705, 2723, and 2731, who had apparently normal phenotypes, were parents of affected children who carried only affected haplotypes for both linked regions. All parents who had affected children shared affected haplotypes from both chromosomes 1 and 6. Therefore, we speculate that individual 2727, with normal phenotype and affected genotypes from both the chromosomes, may produce children with affected phenotypes. The haplotype data also supported the autosomal dominant mode of inheritance in this family. None of the unaffected spouses analyzed (i.e., 2717, 2706, 2703, and 2724) showed affected genotypes for 1q42-43 and 6q14.1 markers. An affected individual (2722), his unaffected brother (2727), and his phenotypically unaffected parent (2723), all of whom carried affected haplotypes for chromosome 1q, also shared a small portion of affected haplotypes for chromosome 6q (i.e., 0.56 cM), which was bordered by SNP markers rs721265 and rs962984. We reconstructed haplotypes for chromosome 6q markers of individuals whose DNA was unavailable and found that the small portion of affected haplotypes in these individuals was transmitted from an apparently unrelated grandparent. It is possible that this grandparent may be indeed related to this family, since consanguinity is common in Arab communities. Moreover, the observed 10 consanguineous marriages suggest the possibility of pseudodominance in this family due to the high frequency of mutant alleles. The reported rate of consanguinity in the UAE population exceeds 50%.45
The data from the linkage analysis indicate that more than one locus contributes to SHFLD. The present family (UR078) provided significant linkage at chromosome 1q42.13-q43 and strong evidence of linkage at 6q14.1. We thus hypothesize that the phenotype in this family could be due to digenic inheritance24; however, it is difficult to prove this hypothesis until we identify the pathologic mutations. The hypothesis of digenic inheritance is supported by a detailed analysis of the haplotypes and the segregating phenotype of all the family members. For example, the phenotypically unaffected 10-year-old individual (2719) inherited the disease-linked haplotype from her affected father only on chromosome 6, whereas, for chromosome 1, the normal haplotype was inherited. A crossover event in this individual (V-13) on chromosome 6 (rs1529992/rs763672) further reduced the proximal risk haplotype. However, phenotypically unaffected individuals should not be used for defining the susceptibility region, since SHFLD has demonstrated incomplete penetrance.
Phenotypically unaffected or affected parents who have affected haplotypes produced affected children who have affected haplotypes from chromosomes 1 and 6. This indicates that the loci on both chromosomes are essential for the phenotypic expression. We also, however, observed individuals with no manifestation of the phenotype (i.e., 2702, 2705, 2723, 2727, and 2731) who carry the two disease-related haplotypes. Reduced penetrance at each locus is a possible explanation for this normal phenotype; alternatively, additional modifier loci may be required for full disease expression. It is also possible that one of these linked loci is a major dominant determinant and that the other is a modifier genomic variant. Similar suggestions were made by Zlotogora24 with reference to the split-hand/split-foot–malformation phenotype. The present study provides evidence of SHFLD susceptibility loci on 1q42.13-q43 and 6q14.1. Further studies are needed to delineate the role of other potential loci involved in SHFLD in the families of different geographic origins. However, the statistically significant evidence of the linkage to 1q and 6q is of interest and should facilitate efforts to identify the underlying susceptibility genes.
Acknowledgments
We thank the patients for their cooperation in the study. The study was supported by the Sheikh Hamdan Awards for Medical Sciences, UAE. The authors from the UAE are thankful to the project advisory board of CAGS, for its constant support and encouragement. We also thank D. S. Krishnamurthy for his assistance with data checking and pedigree analysis. The laboratory of S.E.A. is supported by the Swiss National Science Foundation. S.K.N. was supported by Oklahoma Medical Research Foundation institutional grant 9124, for linkage analysis. U.R. was supported by Green Cross Blood Bank, Ahmedabad, India.
Web Resources
The URLs for data presented herein are as follows:
- Affymetrix, http://www.affymetrix.com/products/arrays/specific/10k.affx
- Ensembl, http://www.ensembl.org/
- MERLIN, http://www.sph.umich.edu/csg/abecasis/Merlin/
- NCBI, http://www.ncbi.nlm.nih.gov/ (for build 35.1)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SHFLD, MIXL1, EDARADD, LGALS8, ACTN2, GREM2, and CHML)
References
- 1.Witters I, Devriendt K, Moerman P, Caudron J, Van Hole C, Fryns JP (2001) Bilateral tibial agenesis with ectrodactyly (OMIM 119100): further evidence for autosomal recessive inheritance. Am J Med Genet 104:209–213 10.1002/ajmg.10056 [DOI] [PubMed] [Google Scholar]
- 2.Majewski F, Kuster W, ter Haar B, Goecke T (1985) Aplasia of tibia with split-hand/split-foot deformity: report of six families with 35 cases and considerations about variability and penetrance. Hum Genet 70:136–147 10.1007/BF00273072 [DOI] [PubMed] [Google Scholar]
- 3.Majewski E, Goecke T, Meinecke P (1996) Ectrodactyly and absence (hypoplasia) of the tibia: are there dominant and recessive types? Am J Med Genet 63:185–189 [DOI] [PubMed] [Google Scholar]
- 4.Fernandez-Palazzi F, Bendahan J, Rivas S (1998) Congenital deficiency of the tibia: a report on 22 cases. J Pediatr Orthop B 7:298–302 [DOI] [PubMed] [Google Scholar]
- 5.Pare A (1575) Chap 7 in: Les oeuvres de M Ambroise Pare, livre 14. Maisson et Cie, Paris [Google Scholar]
- 6.Shenoy R, Kamath N (2004) Bilateral congenital split hand with tibial aplasia. Indian J Pediatr 71:948 [PubMed] [Google Scholar]
- 7.Der Kaloustian VM, Mnaymneh WA (1973) Bilateral tibial aplasia with lobster-claw hands: a rare genetic entity. Acta Paediatr Scand 62:77–78 [DOI] [PubMed] [Google Scholar]
- 8.Canun S, Lomeli RM, Martinez R, Carnevale A (1984) Absent tibiae, triphalangeal thumbs and polydactyly: description of a family and prenatal diagnosis. Clin Genet 25:182–186 [DOI] [PubMed] [Google Scholar]
- 9.Hoyme HE, Jones KL, Nyhan WL, Pauli RM, Robinow M (1987) Autosomal dominant ectrodactyly and absence of long bones of upper or lower limbs: further clinical delineation. J Pediatr 111:538–543 10.1016/S0022-3476(87)80114-2 [DOI] [PubMed] [Google Scholar]
- 10.Marinoni JC, Boyd E, Sherman S, Schwartz C (1994) Familial split hand/split foot long bone deficiency does not segregate with markers linked to the SHFD1 locus in 7q21.3-q22.1. Hum Mol Genet 3:1355–1357 [DOI] [PubMed] [Google Scholar]
- 11.Sener RN, Isikan E, Diren HB, Sayli BS, Sener F (1989) Bilateral split-hand with bilateral tibial aplasia. Pediatr Radiol 19:258–260 10.1007/BF02386848 [DOI] [PubMed] [Google Scholar]
- 12.Bohring A, Wetz HH, Horst J (2005) Intrafamilial variability in autosomal dominant tibial aplasia with ectrodactyly. Eur J Hum Genet 13:P1369 [Google Scholar]
- 13.Kohn G, el Shawwa R, Grunebaum M (1989) Aplasia of the tibia with bifurcation of the femur and ectrodactyly: evidence for an autosomal recessive type. Am J Med Genet 33:172–175 10.1002/ajmg.1320330206 [DOI] [PubMed] [Google Scholar]
- 14.Jones D, Barnes J, Lloyd-Roberts GC (1978) Congenital aplasia and dysplasia of the tibia with intact fibula: classification and management. J Bone Joint Surg Br 60:31–39 [DOI] [PubMed] [Google Scholar]
- 15.McKay M, Clarren SK, Zorn R (1984) Isolated tibial hemimelia in sibs: an autosomal-recessive disorder? Am J Med Genet 17:603–607 10.1002/ajmg.1320170308 [DOI] [PubMed] [Google Scholar]
- 16.Richieri-Costa A (1987) Tibial hemimelia-cleft lip/palate in a Brazilian child born to consanguineous parents. Am J Med Genet 28:325–329 10.1002/ajmg.1320280209 [DOI] [PubMed] [Google Scholar]
- 17.Sener RN, Sayli BS, Isikan UE, Ormeci AR, Unsal M, Tigdemir M (1990) Tetra-oligodactyly with bilateral aplasia and hypoplasia of long bones of upper and lower limbs: a variable manifestation of the syndrome of ectrodactyly with tibial aplasia. Pediatr Radiol 21:57–61 10.1007/BF02010817 [DOI] [PubMed] [Google Scholar]
- 18.Managoli SS, Chaturvedi P (2005) Tibial hemimelia-split hand/foot syndrome with rare anomalies. Indian Pediatr 42:190–191 [PubMed] [Google Scholar]
- 19.Pratt AD, Jr (1971) Apparent congenital absence of the tibia with lethal congenital cardiac disease. Am J Dis Child 122:452–454 [DOI] [PubMed] [Google Scholar]
- 20.Steinkampf MP, Dharia SP, Dickerson RD (2003) Monozygotic twins discordant for vaginal agenesis and bilateral tibial longitudinal deficiency. Fertil Steril 80:643–645 10.1016/S0015-0282(03)00758-1 [DOI] [PubMed] [Google Scholar]
- 21.Evans JA, Greenberg CR (2002) Tibial agenesis with radial ray and cardiovascular defects. Clin Dysmorphol 11:163–169 10.1097/00019605-200207000-00002 [DOI] [PubMed] [Google Scholar]
- 22.Kaissi AA, Ghachem MB, Necib MN, Chehida FB, Karoui H, Baraitser M (2002) Hypohidrotic ectodermal dysplasia with tibial aplasia. Clin Dysmorphol 11:175–178 10.1097/00019605-200207000-00004 [DOI] [PubMed] [Google Scholar]
- 23.Naveed M, Al-Ali MT, Murthy SK, Al-Hajali S, Al-Khaja N, Deutsch S, Bottani A, Antonarakis SE, Nath SK, Radhakrishna U (2006) Ectrodactyly with aplasia of long bones (OMIM; 119100) in a large inbred Arab family with an apparent autosomal dominant inheritance and reduced penetrance: clinical and genetic analysis. Am J Med Genet A 140:1440–1446 [DOI] [PubMed] [Google Scholar]
- 24.Zlotogora J (1994) On the inheritance of the split hand/split foot malformation. Am J Med Genet 53:29–32 10.1002/ajmg.1320530107 [DOI] [PubMed] [Google Scholar]
- 25.Zlotogora J (1995) Heterogeneity of the autosomal dominant split hand/split foot malformation. Am J Hum Genet 56:341–343 [PMC free article] [PubMed] [Google Scholar]
- 26.Zlotogora J, Nubani N (1989) Is there an autosomal recessive form of the split hand and split foot malformation? J Med Genet 26:138–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- 29.Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- 30.Schaid DJ, McDonnell SK, Wang L, Cunningham JM, Thibodeau SN (2002) Caution on pedigree haplotype inference with software that assumes linkage equilibrium. Am J Hum Genet 71:992–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Q, Shete S, Amos CI (2004) Ignoring linkage disequilibrium among tightly linked markers induces false-positive evidence of linkage for affected sib pair analysis. Am J Hum Genet 75:1106–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dawson E, Abecasis GR, Bumpstead S, Chen Y, Hunt S, Beare DM, Pabial J, Dibling T, Tinsley E, Kirby S, et al (2002) A first-generation linkage disequilibrium map of human chromosome 22. Nature 418:544–548 10.1038/nature00864 [DOI] [PubMed] [Google Scholar]
- 33.Phillips MS, Lawrence R, Sachidanandam R, Morris AP, Balding DJ, Donaldson MA, Studebaker JF (2003) Chromosome-wide distribution of haplotype blocks and the role of recombination hot spots. Nat Genet 33:382–387 10.1038/ng1100 [DOI] [PubMed] [Google Scholar]
- 34.Ke X, Hunt S, Tapper W, Lawrence R, Stavrides G, Ghori J, Whittaker P, Collins A, Morris AP, Bentley D, et al (2004) The impact of SNP density on fine-scale patterns of linkage disequilibrium. Hum Mol Genet 13:577–588 10.1093/hmg/ddh060 [DOI] [PubMed] [Google Scholar]
- 35.Ahmad M, Abbas H, Haque S, Flatz G (1987) X-chromosomally inherited split-hand/split-foot anomaly in a Pakistani kindred. Hum Genet 75:169–173 10.1007/BF00591081 [DOI] [PubMed] [Google Scholar]
- 36.Ianakiev P, Kilpatrick MW, Toudjarska I, Basel D, Beighton P, Tsipouras P (2000) Split-hand/split-foot malformation is caused by mutations in the p63 gene on 3q27. Am J Hum Genet 67:59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scherer SW, Poorkaj P, Massa H, Soder S, Allen T, Nunes M, Geshuri D, Wong E, Belloni E, Little S, et al (1994) Physical mapping of the split hand/split foot locus on chromosome 7 and implication in syndromic ectrodactyly. Hum Mol Genet 3:1345–1354 [DOI] [PubMed] [Google Scholar]
- 38.de Mollerat XJ, Gurrieri F, Morgan CT, Sangiorgi E, Everman DB, Gaspari P, Amiel J, Bamshad MJ, Lyle R, Blouin JL, et al (2003) A genomic rearrangement resulting in a tandem duplication is associated with split hand-split foot malformation 3 (SHFM3) at 10q24. Hum Mol Genet 12:1959–1971 10.1093/hmg/ddg212 [DOI] [PubMed] [Google Scholar]
- 39.Flatz S, Fonatsch C (1979) Partial trisomy 1q due to tandem duplication. Clin Genet 15:541–542 [DOI] [PubMed] [Google Scholar]
- 40.Steffensen DM, Chu EH, Speert DP, Wall PM, Meilinger K, Kelch RP (1977) Partial trisomy of the long arm of human chromosome 1 as demonstrated by in situ hybridization with 5S ribosomal RNA. Hum Genet 36:25–33 10.1007/BF00390432 [DOI] [PubMed] [Google Scholar]
- 41.Clark BJ, Lowther GW, Lee WR (1994) Congenital ocular defects associated with an abnormality of the human chromosome 1: trisomy 1q32-qter. J Pediatr Ophthalmol Strabismus 31:41–45 [DOI] [PubMed] [Google Scholar]
- 42.van Bever Y, Rooms L, Laridon A, Reyniers E, van Luijk R, Scheers S, Wauters J, Kooy RF (2005) Clinical report of a pure subtelomeric 1qter deletion in a boy with mental retardation and multiple anomalies adds further evidence for a specific phenotype. Am J Med Genet A 135:91–95 [DOI] [PubMed] [Google Scholar]
- 43.Richieri-Costa A, Ferrareto I, Masiero D, da Silva CR (1987) Tibial hemimelia: report on 37 new cases, clinical and genetic considerations. Am J Med Genet 27:867–884 10.1002/ajmg.1320270414 [DOI] [PubMed] [Google Scholar]
- 44.Matsuyama J, Mabuchi A, Zhang J, Iida A, Ikeda T, Kimizuka M, Ikegawa S (2003) A pair of sibs with tibial hemimelia born to phenotypically normal parents. J Hum Genet 48:173–176 10.1007/s10038-003-0003-9 [DOI] [PubMed] [Google Scholar]
- 45.Abdulrazzaq YM, Bener A, al-Gazali LI, al-Khayat AI, Micallef R, Gaber T (1997) A study of possible deleterious effects of consanguinity. Clin Genet 51:167–173 [DOI] [PubMed] [Google Scholar]