Abstract
Recently, we defined a new syndromic form of X-linked mental retardation in a 4-generation family with a unique clinical phenotype characterized by mild mental retardation, choreoathetosis, and abnormal behavior (MRXS10). Linkage analysis in this family revealed a candidate region of 13.4 Mb between markers DXS1201 and DXS991 on Xp11; therefore, mutation analysis was performed by direct sequencing in most of the 135 annotated genes located in the region. The gene (HADH2) encoding l-3-hydroxyacyl-CoA dehydrogenase II displayed a sequence alteration (c.574 C→A; p.R192R) in all patients and carrier females that was absent in unaffected male family members and could not be found in 2,500 control X chromosomes, including in those of 500 healthy males. The silent C→A substitution is located in exon 5 and was shown by western blot to reduce the amount of HADH2 protein by 60%–70% in the patient. Quantitative in vivo and in vitro expression studies revealed a ratio of splicing transcript amounts different from those normally seen in controls. Apparently, the reduced expression of the wild-type fragment, which results in the decreased protein expression, rather than the increased amount of aberrant splicing fragments of the HADH2 gene, is pathogenic. Our data therefore strongly suggest that reduced expression of the HADH2 protein causes MRXS10, a phenotype different from that caused by 2-methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency, which is a neurodegenerative disorder caused by missense mutations in this multifunctional protein.
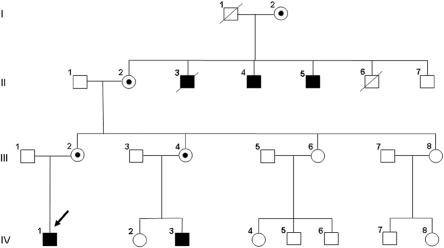
The prevalence of X-linked mental retardation (XLMR) has been estimated to be 1 in 500 males. More than 250 different XLMR entities have been described to date.1–4 Of these entities, ∼165 were classified as syndromic forms characterized by specific biochemical, morphological, or neurological features associated with mental retardation (MR).4 Recently, we described a new XLMR syndrome with a unique clinical phenotype characterized by mild MR, choreoathetosis, and abnormal behavior (MRXS10 [MIM %300220]) in a large Luxembourgian family classified as having MRXS10.5 This 4-generation family included five affected males and four unaffected carrier females (fig. 1). Choreoathetosis, the main distinguishing feature of the disorder, is characterized by involuntary, irregular, purposeless, nonrhythmic, abrupt, and rapid movements, as described in detail by Reyniers and colleagues.5
Figure 1. .
Pedigree of the MRXS10-affected family. Affected males are indicated by black boxes, the obligate carrier females by circles with dots. The index patient (IV-1) is marked with an arrow.
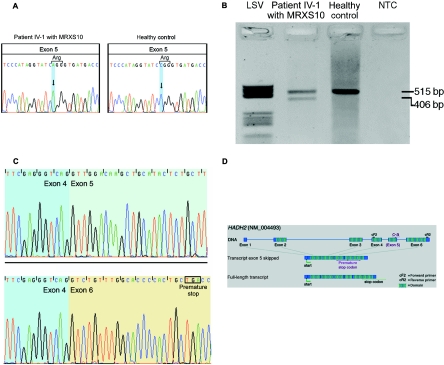
We have refined the linkage interval for the MRXS10-affected family to a region of 13.4 Mb on the short arm of the X chromosome, on Xp11.4-21.5 The establishment of a gene catalogue for the defined interval revealed a total of 135 annotated genes, and mutation screening was performed on genomic DNA and on cDNA. Through use of standard protocols, DNA and RNA were extracted from blood samples and immortalized lymphoblastoid cell lines that were established from peripheral lymphocytes. At the genomic level, the screening was performed by exon sequencing with the adjacent intronic sequences through use of Big Dye kits (Perkin Elmer), with separation of the fragments on an ABI capillary sequencer (ABI 3100). Initially, all 10 known MRX (nonsyndromic forms of MR) and MRXS (syndromic forms of MR) genes within the 13.4-Mb linkage interval between markers DXS1201 and DXS991 were screened for mutations by sequencing genomic DNA and cDNA (appendix A; genes are indicated by a superscript “MRX”). All of them could be excluded as causative of MRXS10. Next, 90 of the 125 remaining annotated genes within the candidate interval were screened for mutations (appendix A; genes are highlighted in bold). These screening efforts revealed a silent C→A substitution in patient IV-1 (fig. 1; indicated with an arrow) in exon 5 (c.574C→A; p.R192R) of the HADH2 gene (MIM *300256; GenBank accession number NM_004493) (fig. 2A). For patient IV-1, in addition to the expected 515-bp fragment, RT-PCR revealed a second visible 406-bp fragment not seen in controls (fig. 2B). Subsequent sequencing of these two fragments demonstrated the loss of exon 5 in the 406-bp fragment, which introduces a premature stop codon in exon 6 (fig. 2C and 2D). A third fragment, obtained by cloning experiments but not visible in fig. 2B, lacks 27 bp of the 3′ end of exon 5. The novel c.574C→A mutation segregates with the disease in the family and was not found in 2,500 control X chromosomes (2,000 X chromosomes from 1,000 healthy white females and 500 X chromosomes from 500 healthy white males, both groups of European origin), as demonstrated by denaturing high-pressure liquid chromatography (WAVE [Transgenomics]). This experiment was done with a 51%–65% gradient of B buffer and a running temperature of 61°C. For PCR amplification, the HADH2-WAVE primer pair 5′-AAG TTC TCC AGG GAT AGT GG-3′ (forward) and 5′-CCC AAT CCC AGG TAT GAT GG-3′ (reverse) was used. Heteroduplex conformation was made possible by adding female wild-type DNA to DNA from hemizygous males.
Figure 2. .
A, Silent c.574C→A substitution in exon 5 of HADH2. Left, Sequence of MRXS10-affected patient IV-1, who harbors the c.574C→A substitution. Right, Wild-type sequence of a healthy control. B, RT-PCR amplification of the HADH2 gene through use of primers HADH2-cF2 and HADH2-cR2. In patient IV-1 (lane 2), the amplification results in a 515-bp wild-type fragment and in a second, shorter fragment of 406 bp. In the healthy control (lane 3), only the wild-type 515-bp fragment is visible. The negative template control is shown in lane 4. Molecular weight marker LSV is shown in lane 1. C, cDNA sequences of the exon boundaries (between exons 4 and 5) of the 515-bp fragment with exon 5 (top) and of the exon boundaries (exons 4–6) of the 416-bp fragment lacking exon 5 (bottom). D, Genomic structure of HADH2. The gene consists of six coding exons, which result in a 261-aa protein with a conserved short-chain dehydrogenase domain at amino acid positions 28–257 (hatched boxes). The c.574C→A substitution is located in exon 5. In the transcript variant lacking exon 5, a premature stop codon is created in exon 6. The position of the primers used for cDNA amplification, as shown in panel B, are indicated by “cF2” and “cR2.'
To quantify the levels of full-length and aberrant transcripts in MRXS10-affected patients IV-1 and IV-3 and in healthy white controls of European origin, real-time experiments were performed using transcript-specific TaqMan primers and probes on RNA extracted from lymphoblastoid cell lines (fig. 3).
Figure 3. .
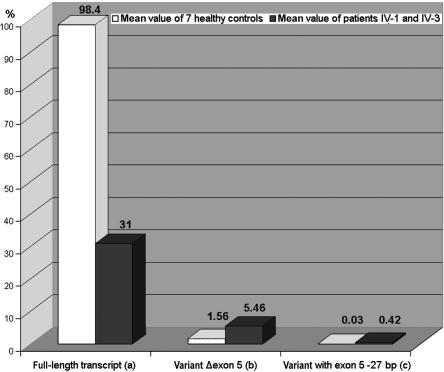
Transcript quantification applying real-time PCR. Whereas the full-length transcript is reduced to one-third in patients IV-1 and IV-3, as compared with the mean value of seven healthy controls (a), the Δexon 5 variant is increased 3.5-fold in the patient (b), and the transcript variant that lacks 27 bp of the 3′ end of exon 5 is increased 14-fold in the patients (c). RNA samples from the two patients and the seven controls were reverse transcribed (cDNA Synthesis Kit [Amersham]).
PCR amplification and detection were performed on a Sequence Detection System (ABI PRISM 7000 [Applied Biosystems]), with application of the standard two-step protocol (45 cycles; annealing temperature 61°C) recommended by the company. TaqMan primer transcripts were designed by TibMolbiol for the different HADH2 transcripts. For the full-length transcript, primers 5′-GCC TTC GAG GGT CAG GTT G-3′ (forward) and 5′-GGG AGG CTG GTC AGC AGT-3′ (reverse) and probe 5′-FAM-CCA AAC AGA CCT GGG GCA ATG GTC A-TMR-3′ were used. For the Δexon 5 transcript, primers 5′-CCT TCG AGG GTC AGG TCT GTT-3′ (forward) and 5′-GTG AGC ATA CTC AGC AGG GTC AC-3′ (reverse) and probe 5′-FAM-TGA CCA GCC TCC CAG AGA AAG TGT GC-TMR-3′ were used. For the Δ27-bp transcript, primers 5′-GGT CAT CAT CAA CAC TGC CAG TGT G-3′ (forward) and 5′-CCA AAC AGA CCT ATG GGA GCC AGA T-3′ (reverse) and probe 5′- FAM-CCC CTT GGA AGC AGA GTA TGC AGC TTG TC-TMR-3′ were used.
These experiments revealed that, on one hand, the level of full-length transcript was reduced to one-third in both affected males, whereas, on the other hand, the levels of the two aberrant transcript variants (Δexon 5 and Δ27 bp) were increased 3.5-fold and 14-fold, respectively, as compared with the controls. In relation to the full-length transcript expressed in controls, the Δexon 5 transcript accounts for only ∼5.5%, and the variant lacking 27 bp accounts for only ∼0.4%, which makes a biological relevance for the latter variants unlikely (fig. 3). To demonstrate that the detected C→A mutation is causative of the misregulated splicing process, in vitro splicing and expression experiments were performed. We cloned the genomic HADH2 sequence of exons/introns 4–6 with and without the c.574C→A substitution in a p-Target mammalian expression vector (Promega), followed by an in vitro splicing process by use of the HeLa Scribe nuclear extract in vitro transcription system (Promega) and subsequent mRNA isolation. By sequencing the cloned HADH2 fragments, we made sure that there was no difference between the wild-type and the mutated fragment other than the c.574C→A substitution. Real-time PCR analysis with the mRNA obtained from these in vitro experiments confirmed that the altered amounts of the transcripts in the MRXS10-affected patients were due to the c.574C→A substitution. Here, the full-length transcript was reduced ∼50%, and the amount of the Δexon 5 variant was increased twofold. The Δ27-bp transcript was increased fourfold.
One possible explanation for the observed incomplete and skewed splicing is that the detected C→A substitution is located in a regulatory element, which is important for the correct splicing process. Such additional regulatory elements, apart from the classic GU/AG sites, are, for example, exonic-splice-enhancer (ESE) or exonic-splice-silencer sites or specific stem-loop structures that can impede or promote efficient exon recognition.6 These regulatory elements normally stabilize the correct quantitative ratio of the different transcript variants of a gene. An alteration of such an element leads to a shift of this ratio and therefore to an over- or underexpression of the single variants. For instance, in the tau gene, which is associated with frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17 [MIM #600274]), a destabilized splicing regulatory element is causative of the altered amount of the exon 10–containing variant that is responsible for the aberrant neuronal development.7
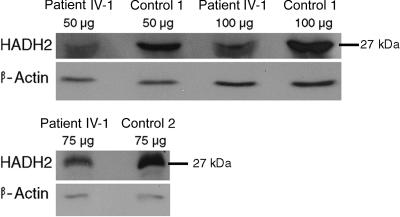
As in the case of a recently described mutation in a putative ESE in the renin receptor gene, which causes MR in association with epilepsy (XMRE [MIM #300423]),8 the mutation in the HADH2 gene results in an altered splicing process. However, in contrast to the XMRE-affected family for which we discussed the generation of aberrant dimeric proteins as causative of the syndrome, we hypothesized that the reduced expression of the HADH2 protein was the cause of MRXS10. To support this hypothesis, we performed western-blot analysis. Proteins were isolated from lymphoblastoid cell lines of an affected male (IV-1) and two control individuals, measured by the method of Bradford,9 were separated on a 12% polyacrylamid gel (SDS-PAGE), and were transferred to a PROTRAN nitrocellulose membrane (Schleicher & Schuell). After blocking the membrane overnight with 5% BSA, the HADH2 protein was detected by a polyclonal antibody (IMG3785 [IMGENEX)]. For normalization, β-actin was used and quantities were calculated by densidometric analysis with the SCION IMAGE system. As shown in figure 4, the amount of the HADH2 protein in the affected male is reduced compared with that of controls. In two independent experiments, densidometric calculation revealed an average decrease of ∼60% in patient IV-1.
Figure 4. .
Detection of the HADH2 protein by western-blot analysis. Amounts between 50 and 100 μg of total protein were loaded. As reference, protein β-actin was detected. The ∼27-kDa bands representing the HADH2 protein of patient IV-1 with MRXS10 show a weaker signal than do the bands of control individuals 1 and 2.
Ofman and colleagues10 reported missense mutations in the HADH2 gene that result in 2-methyl-3-hydroxybutyryl-CoA dehydrogenase (MHBD) deficiency (MIM #300438), an inborn error of isoleucine degradation. It is associated with severe neurological abnormalities, such as the gradual loss of mental and motor skills that progresses to profound developmental regression, choreoathetosis, near blindness, and epilepsy. MHBD catalyzes the transition of 2-methyl-3-hydroxybutyryl-CoA to 2-methyl-acetoacetyl-CoA, which, in turn, is split into acetyl CoA and propionyl CoA. Whereas patients with loss-of-function mutations in the gene encoding MHBD have only very low residual amounts of MHBD activity, our patients show almost normal MHBD activities, amounting to 85% of the level in controls, which indicates no major decrease in MHBD activity (table 1). This result, together with the fact that the phenotype of MHBD deficiency is much more severe than the MRXS10 phenotype, supports our assumption that the clinical phenotype of MRXS10 is distinct from MHBD deficiency and is caused by a different molecular mechanism.
Table 1. .
Determination of MHBD Activity[Note]
| Amount (%) of Activity |
||||
| Protein | Patient IV-1 | Patient IV-3 | Control 1 | Control 2 |
| 2-Methyl-acetoacetyl-CoA | 2.28 (90) | 2.02 (80) | 2.62 (104) | 2.42 (96) |
| Acetoacetyl-CoA | 89 (130) | 76 (111) | 74 (108) | 63 (92) |
| Ratio | .026 (70) | .027 (73) | .035 (95) | .039 (105) |
Note.— The amount is indicated in nmol/min/mg (percentage of control average). The activity in cultured Epstein Barr–virus immortalized lymphoblastoid cell lines from two patients with MRXS10 and from two unaffected control subjects was measured spectrophotometrically in the reverse direction by following the decrease in absorbance at 340 nm at 37°C. Acetoacetyl-CoA was measured as a reference. The standard reaction medium, with a total volume of 250 μl, contained the following components: 50 mM MES/100 mM potassium phosphate buffer (pH 6.16), 0.1% (wt/vol) Triton X-100, 0.2 mM NADH, and 0.32 mg/ml lymphoblastoid cell homogenate protein. Reactions were started by the addition of 2-methyl-acetoacetyl-CoA at a final concentration of .05 mM. Activity is given as a mean of two independent measurements.
To determine whether HADH2 is mutated in other XLMR entities, we screened another 18 syndromic or nonsyndromic entities that are linked to the same genomic interval. However, we failed to detect additional alterations. We also investigated six patients with sporadic MR who present with choreoathetoid movements, and we did not identify additional mutations in HADH2. These results support the expected uniqueness of the phenotype—that is, the MRXS10 syndrome and the genetic aberration.
Here, we report the finding of a novel and unique mutation in the HADH2 gene. We demonstrated that the change is causative of an aberrant transcript-splicing process in the gene, which results in a lower level of wild-type HADH2 and the increase of minor aberrant fragments in the patients as compared with controls. Reduced expression of the HADH2 protein could be demonstrated for the patient with MRXS10 by western-blot analysis. This seems not to impair its role in isoleucine degradation; rather, it impairs one or more other functions attributed to this multifunctional protein. The mutated HADH2 gene is widely expressed and encodes a homotetrameric protein, with a molecular weight of 108 kDa, with multifunctional capacities. Since several researchers have reported different functions of this protein, different acronyms have been used to describe it, such as ERAB (endoplasmic reticulum–associated amyloid-β–binding protein), ABAD (amyloid β peptide–binding alcohol dehydrogenase type II), HSD10 (17 β-hydroxysteroid dehydrogenase type 10), MHBD, and SCHAD (3-α hydroxyacyl-CoA dehydrogenase, short chain).
Tieu et al.11 showed, in a mouse-model system, that increased expression of HADH2 protects against specific hallmarks of Parkinson disease (PD [MIM #168600]); in accordance with this, the HADH2 expression was shown to be reduced in midbrains of patients with PD. However, reexamination of the clinical phenotype of the patients with MRXS10, especially the two who had reached ages 66 and 74 years, did not reveal any clinical features corresponding to those of PD.
Another described function of HADH2 is its interaction with beta-amyloid (Abeta), a protein that builds up in the brain of patients with Alzheimer disease (AD [MIM #104300]) and collects in clumps or plaques. Lustbader et al.12 demonstrated that HADH2 and the Abeta protein interact directly in the mitochondria of patients with AD and increases the mitochondrial toxicity of Abeta. Furthermore, He and coworkers13 described HADH2 as a mitochondrial protein that is able to oxidize and therefore to inactivate the sex steroid hormones 17β-estradiol and dihydroandrosterone. He et al.13 proposed that it could weaken the protective effects of estrogen against AD and generate aldehyds in neurons in the brain, which would mean that high concentrations of HADH2 could be a risk factor for AD. So far, such conclusions are in agreement with our observations, since patients with MRXS10 have not yet developed symptoms of AD. However, additional functional studies are required to clear up our understanding of the disturbed neuronal regulations caused by the reduced protein expression of HADH2 in MRXS10.
Acknowledgments
We thank the members of the MRXS10-affected family. This work was supported by German Ministry for Research and Education grant BMBF 01KW9974 (to A.M.), European Community grant EC QLG2-CT-1999-00791 (to A.M.), Belgian National Fund for Scientific Research–Flanders and an Interuniversity Attraction Poles Program grant (to B.W. and R.F.K.), and National Institute of Child Health & Human Development grant HD26202 (to C.E.S.). C.L. was supported by the FAZIT-Stiftung, Frankfurt/Main, Germany. We are indebted to Dr. Ute Reuning for her advice in protein analysis and critical reading of the manuscript.
Appendix A: Annotated Genes Located in the Candidate Interval
Annotated genes in the candidate interval are PPP1R2P9, MAOAMRX, MAOB, NDPMRX, EFHC2, FUNDC1, DUSP21, UTX, Cxorf36, ZNF673, ZNF674MRX, CHST7, SLC9A7, RP2, PHF16, RGN, NDUFB11, RBM10, UBE1, INE1, PP3895, PCTK1, USP11, ZNF157, ZNF41MRX, ARAF, SYN1, TIMP1, CFP, ELK1, UXT, ZNF81MRX, ZNF182, ZNF630, SSX-Cluster (7 genes: SSX1, -3, -4, -4B, -5, -6, and -9), PNPK6288*, SLC38A5, FTSJ1MRX, PORCN, EBP, OATL1, RBM3, WDR13, WAS, SUV39H1, GATA1, HDAC6, ERAS, PCSK1N, TIMM17B, PQBP1MRX, SLC35A2, PIM2, OTUD5, KCND1, GRIPAP1, TFE3, CCDC120, PRAF2, WDR45, GPKOW, FLJ21687*, PLP2, LMO6, SYP, CACNA1F, CCDC22, FOXP3, PPP1R3F, GAGE-Cluster (8 genes: GAGE2, -8, -4, -6, -5, -7B, -7, and -1), PAGE1, PAGE4, LOC158572*, CLCN5, AKAP4, CCNB3, DGKK, SHROOM4MRX, BMP15, NUDT10, LOC340602*, NUDT11, GSPT2, MAGED1, MAGED4, LOC401589*, XAGE2, XAGE1, SSX8, SSX7, SSX2, SPANXN5, XAGE5, XAGE3, TMEM29, GPR173, TSPYL2, JARID1CMRX, IQSEC2, SMC1A, RIBC1, HADH2, HUWE1, PHF8MRX, FAM120C, WNK3, TSR2, FGD1, GNL3L, ITIH5L, MAGED2, TRO, PFKFB1, APEX2, ALAS2, PAGE2B, PAGE2, FAM104B, PAGE5, PAGE3, MAGEH1, and USP51. Screened genes are indicated in bold. Genes excluded from screening comprise SSX-, MAGE-, XAGE-, and PAGE- genes, as well as five genes associated with other diseases. Genes associated with other diseases are indicated in italics. Genes already associated with MR are indicated by a superscript “MRX.” Genes annotated by National Center for Biotechnology Information but still awaiting an approved symbol given by the HUGO Nomenclature Committee are marked with an asterisk (*).
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for HADH2 [accession number NM_004493])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for MRXS10, HADH2, FTDP-17, XMRE, MHBD deficiency, PD, and AD)
References
- 1.Chelly J, Mandel JL (2001) Monogenic causes of X-linked mental retardation. Nat Rev Genet 2:669–680 10.1038/35088558 [DOI] [PubMed] [Google Scholar]
- 2.Stevenson RE, Schwartz, CE (2002) Clinical and molecular contributions to the understanding of X-linked mental retardation. Cytogenet Genome Res 99:265–275 10.1159/000071603 [DOI] [PubMed] [Google Scholar]
- 3.Ropers HH, Hamel BC (2005) X-linked mental retardation. Nat Rev Genet 6:46–57 10.1038/nrg1501 [DOI] [PubMed] [Google Scholar]
- 4.Stevenson RE (2005) Advances in X-linked mental retardation. Curr Opin Pediatr 17:720–724 10.1097/01.mop.0000184290.57525.fb [DOI] [PubMed] [Google Scholar]
- 5.Reyniers E, Van Bogaert NP, Vits L, Pauly F, Fransen E, Van Regemorter V, Kooy FR (1999) A new neurological syndrome with mental retardation, choreoathetosis, and abnormal behavior maps to chromosome Xp11. Am J Hum Genet 65:1406–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cartegni L, Chew SL, Krainer AR (2002) Listening to silence and understanding nonsense: exonic mutations that effect splicing. Nat Rev Genet 3:285–298 10.1038/nrg775 [DOI] [PubMed] [Google Scholar]
- 7.Varani L, Hasegawa M, Spillantini MG, Smith MJ, Murrell JR, Ghetti B, Klug A, Goedert M, Varani G (1999) Structure of tau exon 10 splicing regulatory element RNA and destabilization by mutations of frontotemporal dementia and parkinsonism linked to chromosome 17. Proc Natl Acad Sci USA 96:8229–8234 10.1073/pnas.96.14.8229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramser J, Abidi FE, Burckle CA, Lenski C, Toriello H, Wen G, Lubs HA, Engert S, Stevenson RE, Meindl A, et al (2005) A unique exonic splice enhancer mutation in a family with X-linked mental retardation and epilepsy points to a novel role of the renin receptor. Hum Mol Genet 14:1019–1027 10.1093/hmg/ddi094 [DOI] [PubMed] [Google Scholar]
- 9.Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 10.Ofman R, Ruiter JPN, Feenstra M, Duran M, Poll-The BT, Zschocke J, Ensenauer R, Lehnert W, Sass JO, Sperl W, et al (2003) 2-Methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency is caused by mutations in the HADH2 gene. Am J Hum Genet 72:1300–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tieu K, Perier C, Vila M, Caspersen C, Zhang HP, Teismann P, Jackson-Lewis V, Stern DM, Yan SD, Przedborski S (2004) l-3-Hydroxyacyl-CoA dehydrogenase II protects in a model of Parkinson’s disease. Ann Neurol 56:51–60 10.1002/ana.20133 [DOI] [PubMed] [Google Scholar]
- 12.Lustbader JW, Cirilli M, Lin C, Xu HW, Takuma K, Wang N, Caspersen C, Chen X, Pollak S, Chaney M, et al (2004) ABAD directly links Aβ to mitochondrial toxicity in Alzheimer’s disease. Science 304:448–452 10.1126/science.1091230 [DOI] [PubMed] [Google Scholar]
- 13.He XY, Merz G, Mehta P, Schulz H, Yang SY (1999) Human brain short chain l-3-hydroxyacyl coenzyme A dehydrogenase is a single-domain multifunctional enzyme: characterization of a novel 17β-hydroxysteroid dehydrogenase. J Biol Chem 274:15014–15019 10.1074/jbc.274.21.15014 [DOI] [PubMed] [Google Scholar]