Abstract
Tourette disorder (TD) is a neuropsychiatric disorder with a complex mode of inheritance and is characterized by multiple waxing and waning motor and phonic tics. This article reports the results of the largest genetic linkage study yet undertaken for TD. The sample analyzed includes 238 nuclear families yielding 304 “independent” sibling pairs and 18 separate multigenerational families, for a total of 2,040 individuals. A whole-genome screen with the use of 390 microsatellite markers was completed. Analyses were completed using two diagnostic classifications: (1) only individuals with TD were included as affected and (2) individuals with either TD or chronic-tic (CT) disorder were included as affected. Strong evidence of linkage was observed for a region on chromosome 2p (-logP=4.42, P=3.8×10-5) in the analyses that included individuals with TD or CT disorder as affected. Results in several other regions also provide moderate evidence (−logP >2.0) of additional susceptibility loci for TD.
Tourette disorder (TD, or Gilles de la Tourette syndrome [MIM #137580]) is a childhood-onset chronic neuropsychiatric disorder characterized by multiple motor and phonic tics that wax and wane over time.1 Once thought to be rare, with prevalence estimates ranging from 1:20,000 to 1:2,000,2 it has recently been observed that, in school-age populations, the prevalence may be as high as 1% among boys.3 Family studies4–6 demonstrate that TD is familial, and twin studies provide evidence that genetic factors are important in its transmission.7,8 Initial complex segregation analyses of nuclear family study data were consistent with autosomal dominant inheritance.9,10 However, subsequent studies5,11–13 suggest that the mode of inheritance is more complex. Evidence from several genetic epidemiologic studies has pointed to a common genetic basis for TD, chronic-tic (CT) disorder, and obsessive-compulsive disorder (OCD).14 When these diagnoses are considered together, bilineal transmission is common in families ascertained for probands with TD.5,15–17
The likelihood that some forms of TD, CT disorder, and OCD appear to be variant expressions of the same genetic susceptibility factors complicates linkage studies of TD and may partially explain the equivocal results of such studies, particularly those performed using parametric analyses.14 It is also difficult to compare the results of several independent linkage studies that have been performed in extended pedigrees affected with TD, since these have often employed different parametric models and have used marker sets that were substantially nonoverlapping.
Given the complications in genetic analyses of TD in large pedigrees, the Tourette Syndrome Association International Consortium for Genetics (TSAICG) undertook a complementary approach and started a multisite collection of affected sib pairs (ASPs). An initial linkage analysis of 92 ASPs from 76 nuclear families provided moderate evidence (maximum LOD score [MLS] >2) of linkage in two regions: one on chromosome 4q with a peak MLS ∼3 cM telomeric of D4S1625 and two adjacent segments on chromosome 8 bounded by markers D8S1106, D8S1145, and D8S136.18 The TSAICG has extended this prior work by collecting a larger sample of ASPs for genome screening.
We report here the results of a complete genome screen that used 390 highly polymorphic DNA markers in a sample that includes 304 independent ASPs (from 238 nuclear families) and 18 large multigenerational pedigrees. The major rationale for including both ASPs and large families is that it provides the first uniform analysis of the majority of large pedigrees reported so far for TD. Data from the large pedigrees, collected by members of the TSAICG with the use of similar assessment protocols, have now been evaluated using the same markers and by nonparametric analysis methods that were not available when the initial studies of these pedigrees were conducted. Furthermore, the ASP and large family samples can easily be combined for further exploration of any linkage findings in each of the samples separately.
Material and Methods
Sample
Data from 238 ASP families and 18 large multigenerational families (a total of 2,040 individuals: 1,052 in the ASP sample and 988 in the multigenerational family sample) were analyzed in the genome scan. Of 255 sib-pair families, 17 were excluded from the analyses either because both sibs did not meet the diagnostic criteria for TD after complete assessment and consensus diagnosis (n=12) or because non-Mendelian inheritance was observed in the family (n=5). Two multigenerational families were excluded from the large family sample because they were uninformative for linkage.
All 238 ASP families included at least two offspring affected with TD and/or CT disorder. All diagnoses were made using DSM-IV-TR criteria.1 The ASP families consisted of 182 with two affected offspring, 47 with three affected offspring, 8 with four affected offspring, and 1 with five affected offspring, yielding a sample equivalent to 304 pairwise “independent” sib pairs. An additional 34 unaffected siblings were included in the sample. These individuals were used to obtain allele-frequency estimates but were not included in the linkage analyses. As noted above, there were 1,052 individuals in the ASP sample. DNA samples were not obtained from 6 mothers and 27 fathers, which yielded a total of 1,019 individuals for whom genotypes were available for most loci. There were no families in which DNA was unavailable for both parents. Of the 33 parents with unavailable DNA, 20 were also missing phenotypic data. There were 647 individuals (including offspring and parents) with TD and 69 with CT disorder in these families. Of the 716 with a diagnosis of either TD or CT disorder, 311 also had a diagnosis of OCD. Only 31 individuals (all parents) received a diagnosis of OCD without TD or CT disorder. The 18 multigenerational families range in size from 14 individuals with 3 members affected with TD or CT disorder to 272 individuals with 91 members affected with TD or CT disorder. Altogether, there were 214 individuals with TD or CT disorder in these 18 multigenerational families.
This research was approved by the ethics committees of each participating site. All individuals 18 years and older signed informed consent forms. For those individuals younger than 18, parents signed consent forms, and the children signed assent forms before participating.
Phenotypic evaluation
When a family entered the study, information concerning diagnosis was collected as follows. Initially, information regarding symptoms associated with TD, OCD, and attention-deficit hyperactivity disorder (ADHD) was obtained with an interview that included a tic inventory and ordinal severity scales modified from the Yale Global Tic Severity Scale,19 the Diagnostic Confidence Index,20 a Modified Yale-Brown Obsessive-Compulsive Scale,21,22 and the Conners Rating Scales (parent and adult versions).23,24 This interview was then followed by a comprehensive assessment of other psychopathology, with use of the Kiddie-SADS–Lifetime Version (K-SADS-PL)25 and the Structured Clinical Interview for DSM (SCID).26
Earlier versions of the TD and OCD instruments have been shown to have a high level of agreement with expert clinician ratings of tic and obsessive-compulsive symptom severity.27–30 These instruments, when compared with clinician diagnoses, have been shown to be both valid (the rate of agreement between interview-derived diagnoses and clinical diagnoses was 0.98 for TD and 0.97 for OCD) and reliable for the diagnoses of TD (κ=1.00) and OCD (κ=0.97).31 As noted above, for the assessment of other psychopathology, the K-SADS-PL25 was used for children younger than 18, and the SCID26 was used for adults. Both interviews have established reliability. For this report, only individuals with a diagnosis of TD or CT disorder were considered to be affected. Future analyses will include information about OCD, ADHD, and other comorbid conditions.
Best-estimate diagnoses
All diagnoses were made using the best-estimate approach.32 Before the initial diagnostic estimate was made, separate files for each individual were prepared. These files contain all available information about the individual, including the completed interview packet, medical records (if available), and all reports from relatives. Web-based document access and data input facilitated the best-estimate consensus diagnostic process. De-identified records for all families were scanned into PDF files and made available via password-protected login to clinicians at each of the individual clinical sites on three continents. Assessment instruments include all materials in the assessment battery.
Diagnosticians used DSM-IV-TR1 criteria for all disorders. For each subject in the sample, two diagnosticians—neither from the originating site of the subject—independently reviewed the assessment materials and recorded diagnoses. A software algorithm compared diagnoses and notified the clinicians if discrepancies were found. Clinicians then used e-mail to resolve differences or to schedule telephone calls for discussion of the differences. In the rare event of an unresolved disagreement, a third diagnostician was asked to review the case and bring resolution. For cases in which consensus was not possible because of missing or ambiguous data, further clinical data were requested from the site of origin. For the diagnosis of TD and CT disorder, the initial independent diagnoses of the two consensus diagnosticians were in agreement 94% of the time (i.e., only 6% of the time was there a disagreement between the two independent diagnosticians that required discussion). Failure to reach consensus occurred in only 21 cases, primarily because of the lack of adequate case data. No individual was included for the linkage study unless consensus diagnosis was achieved.
DNA Markers
Genotyping was completed at the Centre National de Génotypage in Evry, France. A total of 390 STR markers were genotyped (LMS2 Applied Biosystems). These markers had an average heterozygosity of 0.78. Standard DNA amplification protocols were used and were optimized to amplify an average of four markers in the same well.
Fine-mapping genotyping also used standard protocols. Amplified products were pooled, depending on the fluorescent dye label and the size of the products, and were combined with a size standard before being analyzed on an ABI3730. GeneMapper v3.5 was used to analyze the raw results from the ABI3730. However, a genotype was not considered final until two laboratory personnel had independently checked (and verified or corrected) the GeneMapper results and had agreed. The rate of missing genotypes was <4% in both the initial genome scan and the fine-mapping experiments.
Data Analysis
Automatic genotyping was performed on the basis of a series of software processes, trace processing, fragment sizing, allele calling, and the assignment of genotype quality scores (QS) that are implemented in Genetic Profiler software (version 1.1). In addition, an independent manual review of all samples was also completed. Two independent readings of all genotypes were completed. When differences in the genotypes scored in the two readings were observed, the data were reverified to resolve inconsistencies. After raw data genotyping analyses were completed and the results were exported, allele rebinning was performed for each marker on the basis of the distribution of allele peak locations over all samples in this study. Once final allele bins were defined, allele codes were assigned for each individual or microsatellite.
Consistency of the data with Mendelian inheritance, for both the genome scan and the fine-mapping genotypes, was examined using PedCheck.33 Genotypes were reverified when inconsistencies were detected. When the inconsistency could not be resolved, the family was removed from the study.
The data were also examined to determine whether the removal of unlikely genotypes would alter the results of linkage analysis. Unlikely genotypes were identified using the error-checking algorithm implemented in MERLIN,34 which is based on the detection of double-recombinant events. The default parameter, which identifies erroneous genotypes as those with a likelihood ratio of P⩽.025, was used. The same procedure was used both for the original markers and for those used in fine mapping. As expected, given the framework map used in the linkage studies, there was little difference in the results whether unlikely genotypes were kept or removed; thus, results are reported for all genotype data.
Analyses were completed using two diagnostic classifications. The first set of analyses included as affected only those individuals with a diagnosis of TD (TD). The second set of analyses included as affected all individuals with a diagnosis of either TD or CT disorder (TD + CT). Both single-point and multipoint analyses were completed. Marker-allele frequencies were estimated using the program MENDEL,35 which uses full-pedigree data and accounts for the nonindependent relationship within the family. In the single-point analysis, the identity-by-descent distribution was estimated given the marker genotypes for each marker individually. In the multipoint analysis, NPL scores were computed for >4,000 different locations relative to the markers (average step size <1 cM). All analyses of the ASP families were conducted using MERLIN.34 For ASP families with more than two affected siblings, all possible pairs were evaluated, and the results were weighted by a factor equal to 2/n, where n is the number of affected children in the sibship. As noted above, the families included in the final analyses yielded a total of 304 pairwise independent sib pairs. Analyses of the multigenerational families were conducted with the computer program SIMWALK2.36–38 Finally, the two samples were combined and were analyzed using MERLIN and SIMWALK2. MERLIN was used to calculate the Z scores in all sib-pair families and in several multigenerational families, and those results were then imported, using MEGA2,39 into SIMWALK2, which was used to analyze the remaining multigenerational families. For the SIMWALK2 analyses, empirical P values were estimated by randomly sampling from the very large inheritance-vector space, to obtain an estimate of pointwise significance. This is in contrast to other linkage software programs that are constrained to smaller family units and that calculate an exact P value by calculating the value of the test statistic for all possible inheritance vectors. In much larger pedigrees, calculating exact P values is impossible, given the large number of possible inheritance vectors. However, SIMWALK2 can calculate the values for a large number of the inheritance vectors, thus estimating the pointwise P value. Analyses were completed using 10,000, 20,000, 40,000, and 100,000 simulations to estimate significance. The reason for conducting these analyses was that, for each of the first three sets of simulations, our most significant result was always equal to the minimum P value possible for that set of simulations. That is, for 10,000 simulations, our most significant results corresponded to P=1×10-4 (-logP=4.00); for 20,000 simulations, the most significant result observed corresponded to P=5×10-5 (-logP=4.30); for 40,000 simulations, the most significant P value observed was equal to 2.5×10-5 (-logP=4.60); for 100,000 simulations, the most significant P values observed were not at the bound of 1×10-6 (-logP=-5.00). In addition, the run time for the Markov chain–Monte Carlo analyses was extended more than three times the default value in SIMWALK2, to increase the likelihood of achieving the optimal solution in the sample of large families. Only the results of the analysis in which 100,000 simulations were completed are reported.
Results
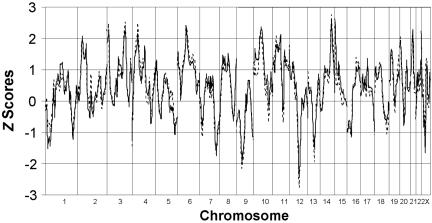
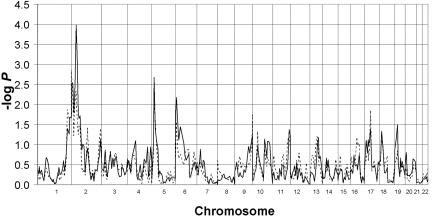
For both diagnostic classifications, the genome scans of the ASP sample and the multigenerational family sample were analyzed separately and together. For the analyses in the ASP sample in which only individuals with TD were included as affected, no Zpairs scores >3 were obtained, but scores ⩾2 (P <1×10−2) were observed for markers on chromosomes 2p, 3p, 3q, 4p, 6p, 10p, 15p, 21p, and Xp (fig. 1). The analysis of the multigenerational family sample, which included only individuals with TD as affected, yielded the highest peak on 2p (-logP=2.34, P=4.6×10-3) at marker D2S165. One other score >2 was observed on chromosome 5p (fig. 2). For the analyses in which individuals with either TD or CT disorder were included as affected, no NPLpairs scores >3 were obtained in the ASP sample. Scores >2 (P=2×10-2) were observed for markers on chromosomes 2p, 3p, 3q, 4p, 6p, 10p, 11p, 14q, 15p, 20p, 21p, and Xp (fig. 1). Analyses of the multigenerational families that used the same diagnostic grouping (TD + CT) yielded the highest peak on 2p (-logP=3.99, P=1×10-4) at marker D2S165. Additional scores >2 were observed on chromosomes 5p and 6p (fig. 2). It should be noted that the highest scores observed in the ASP sample on 2p for both sets of analyses (TD alone and TD + CT) were also observed for marker D2S165.
Figure 1. .
Zpairs scores for ASP families, with use of TD (broken line) and TD + CT (solid line) as diagnostic groupings (MERLIN)
Figure 2. .
Zpairs −logP scores for the multigenerational pedigrees, with use of TD (broken line) and TD + CT (solid line) as diagnostic groupings (SIMWALK2).
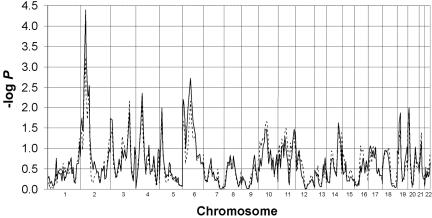
Analysis of the combined sample of ASP and multigenerational families slightly increased the linkage signal on 2p for the analyses that included individuals with TD or CT disorder as affected. For the analyses including only individuals with TD, the peak at D2S165 was -logP=3.23 (P=5.8×10-4), compared with 2.34 (P=4.6×10-3) when the data sets were not combined. When individuals with TD or CT disorder were included as affected, the peak at D2S165 was (-logP=4.40, P=4.0×10-5) (fig. 3), compared with 3.99 (P=1×10-4) when the analyses were completed with the large families alone.
Figure 3. .
Zpairs −logP scores for the combined sample of ASP and multigenerational families, with use of TD (broken line) and TD + CT (solid line) as diagnostic groupings (MERLIN and SIMWALK2).
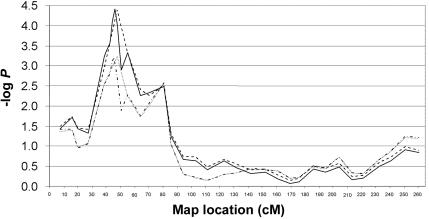
Given these findings, fine mapping on 2p in both ASP and large families was completed; five additional STR markers (D2S2233, D2S220, D2S2221, D2S144, and D2S352) were genotyped at an average spacing of 2 cM. Markers D2S2233, D2S220, D2S2221, and D2S144 are between D2S305 and D2S165 (the two markers giving the highest −logP score in the original genome scan data). Marker D2S352 is between D2S165 and D2S367. The analyses in the combined sample of ASP and multigenerational families yielded a maximum Zpairs -log(P)=4.42 (P=3.8×10-5) at marker D2S144 with the diagnostic classification of TD + CT, confirming the findings from the initial 10-cM genome scan (fig. 4). The 95% CI, calculated using BINOM,40 for this −logP score is 4.01–4.96. Furthermore, support at nearly the same level was maintained over an ∼5-cM region (sex averaged, encompassing ∼8 Mb). A larger region of ∼16 cM encompasses all markers with a drop of −logP−1.
Figure 4. .
Zpairs −logP scores for fine mapping of chromosome 2p, with use of TD and TD + CT as diagnostic groupings (MERLIN and SIMWALK2). The dotted line represents TD-only genome-scan markers; the dotted and broken line represents TD-only fine-mapping markers; the broken line represents TD + CT genome-scan markers; and the solid line represents TD + CT fine-mapping markers.
Discussion
The current study represents the largest series of families with TD to have been included in a genome scan. The strongest evidence of linkage is at marker D2S144 on chromosome 2p23.2. Moreover, the evidence is strengthened by the fact that (1) both the sib-pair and the multigenerational pedigree samples support linkage to this region and (2) a more clearly defined linkage peak was observed when additional markers were typed. An additional piece of evidence that may support the existence of a TD locus in this region comes from the observation of a complex chromosomal rearrangement in which two children with TD, OCD, and mental retardation inherited an insertion of 2p21-23 (without any apparent disruption of any genes) into a region of chromosome 7q35-36.41 This segment of chromosome 2, which is trisomic in the affected offspring, contains the 15-Mb interval that lies beneath the linkage peak (between D2S305 and D2S367). This region has the highest priority for follow-up studies.
Other regions may also be important to follow up. When examined separately, several regions emerged as potentially interesting in the sib-pair sample, with segments on 3p, 3q, and 14q all displaying NPL scores >2.5. However, none of these regions were supported substantially by the total multigenerational sample. Similarly, two regions (5p and 6p) yielded −logP scores >2.0 in the pedigree sample. Both regions had positive NPL scores in the ASP sample (5p, -logP=1.11; 6p, -logP=2.42). Of note is that, in the analyses of the combined samples, −logP=2.73 in the 6p region. Although none of these regions achieve statistical significance, they all warrant additional follow-up, since they may represent the effect of gene variants that are contributing to TD susceptibility in only a subset of the families. Given the size of this sample, it was not computationally feasible to complete simulations necessary to provide −logP scores that define the thresholds for suggestive and significant linkage. For example, one complete genome analysis of the combined data set on a Linux cluster of 30 processors requires >3 mo to complete.
In a previous linkage study, using a subset of the sib pairs employed in the current genome scan, the TSAICG18 reported moderate evidence of linkage on chromosomes 4q and 8p (MLS >2). A third region on chromosome 4p showed weak evidence of linkage (MLS >1.0) in that initial study.18 Of note, in the current study, the evidence of linkage to 4p is strengthened in both the ASP (Zpairs score >2.0) and the large family (−logP >2.0) samples, as well as in the combined sample (−logP >2.0); however, neither the region on 4q nor the region on 8p are supported, even in the ASP sample alone. The diminution of evidence in successive linkage studies of complex traits has been observed for several other disorders.42,43 In addition, theoretical studies have predicted that this would be the case,44 on the basis of the inherent etiologic heterogeneity of such traits, especially where there is variability in either the ascertainment or the assessment of the subjects. In our study, we attempted to minimize such variability (e.g., through the use of our best-estimate assessment procedures); nevertheless, we ascertained the families for this study in multiple sites in Europe and North America.
This study also does not provide support for other regions implicated in prior smaller-scale linkage studies of TD, some of which focused on families that were included in the current study. Mérette and colleagues45 reported moderate evidence of linkage to chromosome 11q23 in one large French Canadian kindred. In a genomewide linkage study of seven multigenerational families, Barr et al.46 observed a linkage signal for TD in two regions (19p13.3 and 5p13-q11.2). Whereas neither of these regions showed significant linkage in our combined sib-pair and pedigree analysis, the positive linkage results on chromosome 5 in the multigenerational families in the current sample are consistent with the Barr et al. findings.46 However, this may be due largely to the inclusion of four of the families from the Barr et al.46 study in the present study. Paschou and colleagues47 identified suggestive linkage results in 17q (spanning the interval from D17S784 to D17S928) in three multigenerational families. Although the families included in the Paschou et al. study47 are also included in the current sample, the findings of the combined sample of ASP and pedigree families and the total pedigree sample do not support the results of Paschou et al.47
Other approaches have also been employed in the search for TD susceptibility loci. A recent finding reported a possible association between the gene “slit and Trk-like 1” (SLITRK1) and TD48; SLITRK1 was examined as a candidate gene because of its proximity to a de novo chromosomal inversion on chromosome 13q31.1 in a child with TD and on the basis of the finding of a frameshift mutation and two independent occurrences of the identical variant in the binding site for microRNA hsa-miR-189 among 174 unrelated probands (but not in >3,600 control chromosomes). Unfortunately, there was no support for a locus on chromosome 13 in the current study, suggesting that, if SLITRK1 is a susceptibility gene for TD, it is not one with major effect in the population studied and reported in this article.
The fact that we obtained our strongest linkage finding when we included as affected those individuals with CT disorder, as well as those given diagnoses of TD, underscores the continuing uncertainty regarding the optimal phenotypic definition for linkage and association studies of these disorders. Since we have obtained, in the families studied, information on other qualitative phenotypes (e.g., OCD and ADHD) that may be related to TD, as well as a wide range of quantitative phenotypic features, it will be possible to conduct further analyses of the genome-scan data. These analyses may suggest additional genome regions that warrant follow-up investigation in these families, as well as in the trios with TD that we have sampled.
In conclusion, this sample of 238 ASP families and 18 multigenerational families provides significant evidence of linkage to marker D2S144 on chromosome 2p32.2. Other chromosomal regions—including 3p, 3q, and 14q—had NPL scores >2.5 in the sib-pair sample but not in the multigenerational pedigrees. These results are consistent with a complex inheritance model that includes locus heterogeneity and a gene of major effect on 2p32.2. Since it appears likely that other regions may harbor additional genes that contribute to susceptibility for TD, it is important that analyses be undertaken to examine the extent of linkage heterogeneity and phenotypic variation in this sample and to attempt to determine if genes in these regions may be linked in a subset of individuals with TD.
Sib-pair and pedigree samples are most suitable for identifying relatively infrequent variants of moderate-to-large effect.49 Given that our collection of such samples has involved an exhaustive international effort, our efforts so far represent an effective screen for most such variants that contribute to TD. However, it remains important to employ different strategies, such as whole-genome association studies, that will facilitate the identification of variants that may have a lesser effect in any given individual with TD but that will make a relatively larger contribution to the manifestation of TD in the population as a whole.
Acknowledgments
The members of TSAICG are as follows, listed alphabetically by city: D. Cath and P. Heutink, Departments of Psychiatry and Human Genetics, Free University Medical Center Amsterdam, Amsterdam, The Netherlands; M. Grados, H. S. Singer, and J. T. Walkup, Departments of Psychiatry and Neurology, Johns Hopkins University School of Medicine, Baltimore; C. Illmann, J. M. Scharf, S. Santangelo, S. E. Stewart, J. Platko, and D. L. Pauls, Psychiatric and Neurodevelopmental Genetics Unit, Center for Human Genetic Research, Massachusetts General Hospital, Harvard Medical School, Boston; N. J. Cox, Departments of Medicine and Human Genetics, University of Chicago, Chicago; M. M. Robertson, Department of Mental Health Sciences, University College London Institute of Neurology, National Hospital for Neurology and Neurosurgery, Queen Square, London; S. Service, D. Keen-Kim, C. Sabatti, and N. Freimer, Departments of Psychiatry, Human Genetics and Statistics, University of California–Los Angeles Medical School, Los Angeles; G. A. Rouleau, J.-B. Riviere, S. Chouinard, F. Richer, P. Lesperance, and Y. Dion, University of Montreal, Montreal; R. A. King, J. R. Kidd, A. J. Pakstis, J. F. Leckman, and K. K. Kidd, Department of Genetics and the Child Study Center, Yale University School of Medicine, New Haven, CT; G. Gericke, Department of Biomedical Sciences, Tshwane University of Technology, Pretoria, South Africa; R. Kurlan, P. Como, and D. Palumbo, Department of Neurology, University of Rochester School of Medicine, Rochester, NY; A. Verkerk and B. A. Oostra, Department of Clinical Genetics, Erasmus University, Rotterdam, The Netherlands; W. McMahon, M. Leppert, and H. Coon, Departments of Psychiatry and Human Genetics, University of Utah School of Medicine, Salt Lake City; C. Mathews, Department of Psychiatry, University of California–San Francisco, San Francisco; and P. Sandor and C. L. Barr, Department of Psychiatry, The Toronto Hospital and University of Toronto, Toronto. C. Betard and D. Zelenika, Centre National de Génotypage, Evry, France, also contributed to this work. The TSAICG is grateful to all the families with TD, in all of the participating centers, who generously agreed to be part of this study. Furthermore, the members of the Consortium are deeply indebted to the Tourette Syndrome Association (TSA)—in particular, to Ms. Judit Ungar, TSA president, and Ms. Sue Levi-Pearl, TSA director of medical and scientific programs. Both have dedicated their professional lives to the understanding and treatment of TD. Their tireless efforts to help move the research forward were critical to the success of the project. Without their support, guidance, and prodding, this study would not have been possible. The Consortium also sincerely thanks the members of the TSA Board of Directors for their continuing support. Finally, the members of the Consortium thank the advisors to the collaborative group, who include the following volunteer members of the TSA Scientific Advisory Board’s Subcommittee for Genetics: P. Michael Conneally, Francis McMahon, John Rice, Neal Swerdlow, Peter Hollenbeck, and Jonathan Mink. The TSAICG acknowledges J. Hebebrand, B. Klug, and H. Remschmidt, Department of Child and Adolescent Psychiatry, Phipps University, Marburg, Germany; J. L. Weber, B. C. Hiner, and M. Spindler, Center for Medical Genetics, Marshfield Medical Foundation, Marshfield, WI; and J. Jankovic, Department of Neurology, Baylor College of Medicine, Houston, for their assistance in collecting data from sib-pair families that were included in this genome scan. This work was supported by funds from the TSA and by National Institutes of Health grant NS 40024. The final manuscript was prepared by Drs. Pauls, Cox, Freimer, Oostra, McMahon, and Walkup, with input from members of the Consortium.
Web Resources
The URLs for data presented herein are as follows:
- BINOM version 20, http://www.genemapping.cn/linkutil.htm
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for TD) [PubMed]
References
- 1.American Psychiatric Association (2000) Diagnostic and statistical manual, 4th edition–text revision (DSM-IV-TR). American Psychiatric Press, Washington, DC [Google Scholar]
- 2.Apter A, Pauls DL, Bleich A, Zohar AH, Kron S, Ratzoni G, Dycian A, Kotler M, Weizman A, Gadot N, et al (1993) An epidemiologic study of Gilles de la Tourette’s syndrome in Israel. Arch Gen Psychiatry 50:734–738 [DOI] [PubMed] [Google Scholar]
- 3.Robertson MM, Stern JS (1998) Tic disorders: new developments in Tourette syndrome and related disorders. Curr Opin Neurol 11:373–380 10.1097/00019052-199808000-00014 [DOI] [PubMed] [Google Scholar]
- 4.Pauls DL, Raymond CL, Stevenson JM, Leckman JF (1991) A family study of Gilles de la Tourette syndrome. Am J Hum Genet 48:154–163 [PMC free article] [PubMed] [Google Scholar]
- 5.Walkup JT, LaBuda MC, Singer HS, Brown J, Riddle MA, Hurko O (1996) Family study and segregation analysis of Tourette syndrome: evidence for a mixed model of inheritance. Am J Hum Genet 59:684–693 [PMC free article] [PubMed] [Google Scholar]
- 6.Hebebrand J, Klug B, Fimmers R, Seuchter SA, Wettke-Schafer R, Deget F, Camps A, Lisch S, Hebebrand K, von Gontard A, et al (1997) Rates for tic disorders and obsessive compulsive symptomatology in families of children and adolescents with Gilles de la Tourette syndrome. J Psychiatr Res 31:519–530 10.1016/S0022-3956(97)00028-9 [DOI] [PubMed] [Google Scholar]
- 7.Hyde TM, Aaronson BA, Randolph C, Rickler KC, Weinberger DR (1992) Relationship of birth weight to the phenotypic expression of Gilles de la Tourette’s syndrome in monozygotic twins. Neurology 42:652–658 [DOI] [PubMed] [Google Scholar]
- 8.Price RA, Kidd KK, Cohen DJ, Pauls DL, Leckman JF (1985) A twin study of Tourette syndrome. Arch Gen Psychiatry 42:815–820 [DOI] [PubMed] [Google Scholar]
- 9.Pauls DL, Leckman JF (1986) The inheritance of Gilles de la Tourette’s syndrome and associated behaviors: evidence for autosomal dominant transmission. N Engl J Med 315:993–997 [DOI] [PubMed] [Google Scholar]
- 10.Eapen V, Pauls DL, Robertson MM (1993) Evidence for autosomal dominant transmission in Tourette’s syndrome: United Kingdom cohort study. Br J Psychiatry 162:593–596 [DOI] [PubMed] [Google Scholar]
- 11.Pauls DL, Alsobrook J II, Almasy L, Leckman JF, Cohen DJ (1991) Genetic and epidemiological analyses of the Yale Tourette’s Syndrome Family Study data. Psychiatr Genet 2:28 [Google Scholar]
- 12.Hasstedt SJ, Leppert M, Filloux F, van de Wetering BJM, McMahon WM (1995) Intermediate inheritance of Tourette syndrome, assuming assortative mating. Am J Hum Genet 57:682–689 [PMC free article] [PubMed] [Google Scholar]
- 13.Seuchter SA, Hebebrand J, Klug B, Knapp M, Lehmkuhl G, Poustka F, Schmidt M, Remschmidt H, Baur MP (2000) Complex segregation analysis of families ascertained through Gilles de la Tourette syndrome. Genet Epidemiol 18:33–47 [DOI] [PubMed] [Google Scholar]
- 14.Scharf JM, Pauls DL. Genetics of tic disorders. In: Rimoin DL, Connor JM, Pyeritz RE, Korf BR (eds) Emery and Rimoin’s principles and practices of medical genetics, 5th ed. Elsevier, New York (in press) [Google Scholar]
- 15.McMahon WM, Leppert M, Filloux F, van de Wetering BJM, Hasstedt S (1992) Tourette syndrome in 171 related family members. Adv Neurol 58:159–165 [PubMed] [Google Scholar]
- 16.McMahon WM, van de Wetering BJM, Filloux F, Betit K, Coon H, Leppert M (1996) Bilineal transmission and phenotypic variation of Tourette’s disorder in a large pedigree. J Am Acad Child Adolesc Psychiatry 35:672–680 10.1097/00004583-199605000-00024 [DOI] [PubMed] [Google Scholar]
- 17.Kurlan R, Eapen V, Stern J, McDermott MP, Robertson MM (1994) Bilineal transmission in Tourette’s syndrome families. Neurology 44:2336–2342 [DOI] [PubMed] [Google Scholar]
- 18.The Tourette Syndrome Association International Consortium for Genetics (1999) A complete genome screen in sib pairs affected by Gilles de la Tourette syndrome. Am J Hum Genet 65:1428–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leckman JF, Riddle MA, Hardin MT, Ort SI, Swartz KL, Stevenson J, Cohen DJ (1989) The Yale Global Tic Severity Scale: initial testing of a clinician-rated scale of tic severity. J Am Acad Child Adolesc Psychiatry 28:566–573 10.1097/00004583-198907000-00015 [DOI] [PubMed] [Google Scholar]
- 20.Robertson MM, Banerjee S, Kurlan R, Cohen DJ, Leckman JF, McMahon W, Pauls DL, Sandor P, van de Wetering BJ (1999) The Tourette syndrome diagnostic confidence index: development and clinical associations. Neurology 53:2108–2112 [DOI] [PubMed] [Google Scholar]
- 21.Goodman WK, Price LH, Rasmussen SA, Mazure C, Fleischmann RL, Hill CL, Heninger GR, Charney DS (1989) The Yale-Brown Obsessive-Compulsive Scale: development, use, and reliability. Arch Gen Psychiat 46:1006–1011 [DOI] [PubMed] [Google Scholar]
- 22.Scahill L, Riddle MA, McSwiggin-Hardin M, Ort SI, King RA, Goodman WK, Cicchetti D, Leckman JF (1997) Children’s Yale-Brown Obsessive Compulsive Scale: reliability and validity. J Am Acad Child Adolesc Psychiatry 36:844–852 10.1097/00004583-199706000-00023 [DOI] [PubMed] [Google Scholar]
- 23.Conners CK, Sitarenios G, Parker JD, Epstein JN (1998) The revised Conners’ Parent Rating Scale (CPRS-R): factor structure, reliability, and criterion validity. J Abnorm Child Psychol 26:257–268 10.1023/A:1022602400621 [DOI] [PubMed] [Google Scholar]
- 24.Conners CK, Erhardt D, Sparrow E (1999) Conners Adult ADHD Rating Scales (CAARS) technical manual. Multi-Health Systems, North Tonawanda, NY [Google Scholar]
- 25.Kaufman J, Birmaher B, Brent D, Rao U, Ryan N (1995) Kiddie-SADS-Lifetime Version (K-SADS-PL). University of Pittsburgh, Pittsburgh [Google Scholar]
- 26.Spitzer R, Williams J, Gibbon M, First M (1992) The Structured Clinical Interview for DSM-III-R (SCID). I. History, rationale, and description. Arch Gen Psychiat 49:624–629 [DOI] [PubMed] [Google Scholar]
- 27.Leckman JF, Walker DE, Cohen DJ (1993) Premonitory urges in Tourette’s syndrome. Am J Psychiatry 150:98–102 [DOI] [PubMed] [Google Scholar]
- 28.Leckman JF, Walker WK, Goodman WK, Pauls DL, Cohen DJ (1994) “Just right” perceptions associated with compulsive behaviors in Tourette’s syndrome. Am J Psychiat 151:675–680 [DOI] [PubMed] [Google Scholar]
- 29.Leckman JF, Grice DE, Boardman J, Zhang H, Vitale A, Bondi C, Alsobrook J, Peterson BS, Cohen DJ, Rasmussen SA, et al (1997) Symptoms of obsessive compulsive disorder. Am J Psychiatry 154:911–917 [DOI] [PubMed] [Google Scholar]
- 30.Rosenfeld R, Dar R, Anderson D, Kobak KA, Griest JH (1992) A computer administered version of the Yale-Brown Obsessive-Compulsive Scale. Psychol Assess 4:329–332 10.1037/1040-3590.4.3.329 [DOI] [Google Scholar]
- 31.Pauls DL, Alsobrook JP II, Goodman W, Rasmussen S, Leckman JF (1995) A family study of obsessive-compulsive disorder. Am J Psychiatry 152:76–84 [DOI] [PubMed] [Google Scholar]
- 32.Leckman J, Sholomskas D, Thompson W, Belanger A, Weissman MM (1982) Best estimate of lifetime psychiatric diagnoses: a methodological study. Arch Gen Psychiatry 39:879–883 [DOI] [PubMed] [Google Scholar]
- 33.O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abecasis GR, Cherny SS, Cookson WOC, Cardon LR (2002) MERLIN—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- 35.Lange K, Cantor RM, Horvath S, Perola M, Sabatti C, Sinsheimer J, Sobel E (2001) MENDEL version 4.0: a complete package for the exact genetic analysis of discrete traits in pedigree and population data sets. Am J Hum Genet Suppl 69:A1886 [Google Scholar]
- 36.Sobel E , Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- 37.Sobel E, Sengul H, Weeks DE (2001) Multipoint estimation of identity-by-descent probabilities at arbitrary positions among marker loci on general pedigrees. Hum Hered 52:121–131 10.1159/000053366 [DOI] [PubMed] [Google Scholar]
- 38.Sobel E, Papp JC, Lange K (2002) Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet 70:496–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mukhopadhyay N, Almasy L, Schroeder M, Mulvihill WP, Weeks DE (2005) Mega2: data-handling for facilitating genetic linkage and association analyses. Bioinformatics 21:2556–2557 10.1093/bioinformatics/bti364 [DOI] [PubMed] [Google Scholar]
- 40.Ott J (2002) Program BINOM version 20. Rockefeller University, New York [Google Scholar]
- 41.Verkerk AJ, Matthews CA, Joosse M, Eussen BH, Heutink P, Oostra BA, Tourette Syndrome Association International Consortium for Genetics (2003) CNTNAP2 is disrupted in a family with Gilles de la Tourette syndrome and obsessive compulsive disorder. Genomics 82:1–9 10.1016/S0888-7543(03)00097-1 [DOI] [PubMed] [Google Scholar]
- 42.Kerner B, Brugman DL, Freimer NB (2006) Evidence of linkage to psychosis on chromosome 5q33-34 in pedigrees ascertained for bipolar disorder. Am J Med Genet B Neuropsychiatr Genet (http://www3.interscience.wiley.com/cgi-bin/fulltext/112776793/HTMLSTART) (electronically published September 6, 2006; accessed December 12, 2006) [DOI] [PubMed]
- 43.Suarez BK, Lin J, Witte JS, Conti DV, Resnick MI, Klein EA, Burmester JK, Vaske DA, Banerjee TK, Catalona WJ (2000) Replication linkage study for prostate cancer susceptibility genes. Prostate 45:106–114 [DOI] [PubMed] [Google Scholar]
- 44.Suarez BK, Hampe CL, Van Eersewegh P (1994) Problems of replicating linkage claims in psychiatry. In: Gershon ES, Cloniniger CR (eds) Genetic approaches to mental disorders. American Psychiatric Press, Washington, DC, pp 23–46 [Google Scholar]
- 45.Merette C, Brassard A, Potvin A, Bouvier H, Rousseau F, Emond C, Bissonnette L, Roy MA, Maziade M, Ott J, et al (2000) Significant linkage for Tourette syndrome in a large French Canadian family. Am J Hum Genet 67:1008–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barr CL, Wigg KG, Pakstis AJ, Kurlan R, Pauls DL, Kidd KK, Tsuai L-C, Sandor P (1999) Genome scan for linkage to Gilles de la Tourette syndrome. Am J Med Genet 88:437–445 [DOI] [PubMed] [Google Scholar]
- 47.Paschou P, Feng Y, Pakstis AJ, Speed WC, DeMille MM, Kidd JR, Jaghori B, Kurlan R, Pauls DL, Sandor P, et al (2004) Indications of linkage and association of Gilles de la Tourette syndrome in two independent family samples: 17q25 is a putative susceptibility region. Am J Hum Genet 75:545–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abelson JF, Kwan KY, O’Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, et al (2005) Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science 310:317–320 10.1126/science.1116502 [DOI] [PubMed] [Google Scholar]
- 49.Freimer N, Sabatti C (2004) The use of pedigree, sib-pair and association studies of common diseases for genetic mapping and epidemiology. Nat Genet 36:1045–1051 10.1038/ng1433 [DOI] [PubMed] [Google Scholar]