Abstract
Recently, somatic recombination of human mitochondrial DNA (mtDNA) was discovered in skeletal muscle. To determine whether recombinant mtDNA molecules can be transmitted through the germ line, we investigated two families, each harboring two inherited heteroplasmic mtDNA mutations. Using allele-specific polymerase chain reaction and single-cell and single-molecule mutational analyses, we discovered, in both families, all four possible allelic combinations of the two heteroplasmic mutations (tetraplasmy), the hallmark of mtDNA recombination. We strongly suggest that these recombinant mtDNA molecules were inherited rather than de novo generated somatically, because they (1) are highly abundant and (2) are present in different tissues of maternally related family members, including young individuals. Moreover, the comparison of the complete mtDNA sequence of one of the families with database sequences revealed an irregular, nontreelike pattern of mutations, reminiscent of a reticulation. We therefore propose that certain reticulations of the human mtDNA phylogenetic tree might be explained by recombination of coexisting mtDNA molecules harboring multiple mutations.
Recombination of mtDNA has been described in plants, fungi, protists, mussels, and fish.1 In human mitochondrial genetics, however, the absence of mtDNA recombination is still a broadly accepted viewpoint.2 The presence of human mtDNA recombination was initially claimed to explain the large number of homoplasies in human mtDNA sequences3 and the results of a linkage-disequilibrium analysis,4 but the conclusions of the authors could not be confirmed on different data sets.5,6 More recently, we provided the first experimental evidence of mtDNA recombination in the skeletal muscle of an individual with biparental inheritance of the mitochondrial genome.7 Furthermore, in a set of skeletal-muscle samples from individuals with multiple heteroplasmy (who carried a mixture of wild-type and mutated alleles at more than one location in their mitochondrial genomes), we were able to show the frequent presence of recombinant mtDNA molecules.8 However, the relevance of these findings for mtDNA phylogeny depends on whether human mtDNA recombination is restricted to skeletal muscle or whether recombinant mtDNA molecules can enter and pass through the germ line.
Here, we present experimental data of two double-heteroplasmic families, which strongly suggests that recombinant mtDNA molecules can be inherited. Furthermore, one of the investigated families provides an example of how persisting heteroplasmy and recombination might explain certain reticulations of the human mtDNA phylogenetic tree.
Subjects and Methods
Subjects
The family with the A8344G/A16182C mutations includes a boy, aged 14 years, who has MERRF syndrome (myoclonus epilepsy with ragged red fibers [MIM 545000]) and his sister, aged 29 years, who has occasional seizures. Their parents (and the mother’s sister) were free of symptoms (fig. 1A). The family with the A3243G/G16428A mutations includes a man, aged 29 years, who received the diagnosis of systemic MELAS syndrome (myopathy, encephalopathy, lactic acidosis, and strokelike episodes [MIM 540000]); his maternal aunt, aged 44 years; and her daughter, aged 12 years (fig. 2A). The study was conducted following the guidelines of the University Bonn Ethical Commission, and informed consent was obtained from all studied subjects.
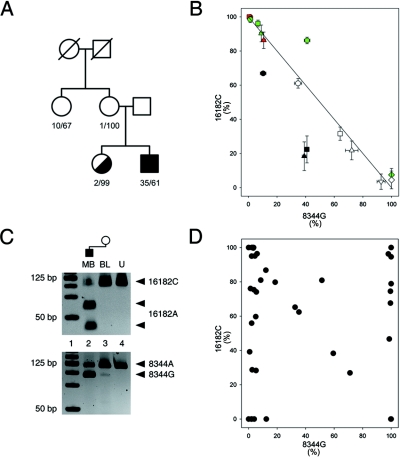
Figure 1. .
Family with A8344G and A16182C heteroplasmic mutations. A, Pedigree. The first number indicates the percentage of 8344G; the second number indicates the percentage of 16182C in blood samples of the family members. B, Apparent cosegregation of the 16182A allele with the 8344G allele in tissue samples of double-heteroplasmic individuals. Mutation loads were determined by RFLP and were confirmed by sequencing. Circle = blood; square = buccal cells; triangle = urine sediment; diamond = skeletal-muscle biopsy sample; hexagon = fibroblasts; inverse triangle = brain biopsy sample. Black indicates the sister of the patient’s mother; red indicates the patient’s mother; white indicates the patient; green indicates the patient’s sister. The 8344G mutant allele primarily co-occurs with the 16182A wild-type allele (and the 8344A wild-type allele with the d-loop mutant 16182C). A third allelic combination, the putative “original” wild-type 8344A/16182A, is present in the tissue samples of the sister of the patient’s mother (the data points are below the line). Data points above the line in the tissue samples of the patient’s sister indicate that 8344G also occurs in combination with the 16182C allele (the fourth allelic combination, double mutant, and the potential recombinant). C, The recombinant 8344G/16182C allele, present in the blood of the patient’s mother. Note the faint band indicating the presence of 8344G in the blood of the patient’s mother, whereas 16182A is virtually absent in this sample (lane 3), which confirms the presence of the recombinant 8344G/16182C. Lane 1, DNA ladder. Lane 2, Patient’s muscle biopsy (MB) sample. Lane 3, Blood of the patient’s mother (BL). Lane 4, Undigested PCR product (U). D, Distribution of the two heteroplasmic mtDNA mutations in single fibroblasts of the patient’s sister. Data points populate all four corners of the graph, which indicates tetraplasmy.
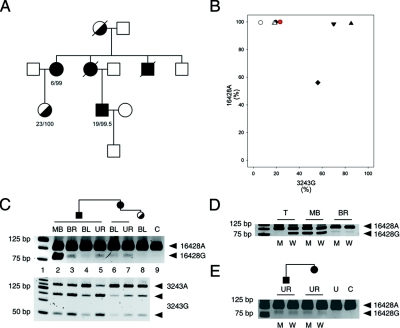
Figure 2. .
Family with A3243G and G16428A heteroplasmic mutations. A, Pedigree. The first number indicates the percentage of 3243G; the second number indicates the percentage of 16428A in blood samples of the family members. B, Apparent association of the 16428A allele with the 3243G allele in tissue samples of double-heteroplasmic individuals. The mutation loads were determined by RFLP and were confirmed by sequencing. Circle = blood; triangle = urine sediment; diamond = skeletal-muscle biopsy sample; inverse triangle = brain biopsy sample. Black = patient; red = daughter of the patient’s aunt; white = the patient’s aunt. C, Distribution of the heteroplasmic mutations in different tissue samples from the family members. Note the clear presence of 16428G in the samples of the patient’s maternal aunt (lane 7). Lane 1, DNA ladder. Lanes 2–5, Patient muscle biopsy (MB) sample, brain (BR), blood (BL), and urine sediment (UR). Lanes 6–7, Patient’s aunt's blood (BL) and urine sediment (UR). Lane 8, Patient’s cousin's blood (BL). Lane 9, Homoplamic 16428A control (C) (isolated PCR fragment). D, Tetraplasmic distribution of the two heteroplasmic mutations, A3243G and G16428A, in skeletal muscle (MB) and brain (BR) of the patient. Note the presence of both digested and undigested bands in the allele-specific reaction pairs. The potential recombinant 16428G/3243G is represented by the digested PCR product in the M lanes. M = mutant 3243G allele-specific PCR; W = wild-type 3243A allele-specific PCR. T = triplasmic control DNA mixture harboring wild-type, single-mutant 16428A and double-mutant 3243G/16428A mtDNA molecules. E, Tetraplasmy of the A3243G/G16428A alleles in urine sediments from the patient and his maternal aunt. Note the mtDNA tetraplasmy in both urine sediments, with the potential recombinant 16428G/3243G (digested PCR product in lanes M). U = undigested PCR product; C = control PCR product with the 16428A mutation.
mtDNA Mutation Analysis
Genomic DNA was extracted from 10-ml aliquots of EDTA-anticoagulated blood by use of a salting-out method9 or from frozen skeletal-muscle or brain-biopsy specimens, buccal swabs, urine sediment, or cultured fibroblasts with the QiaAmp DNA Mini Kit (Qiagen). Degrees of heteroplasmy were determined by mismatch PCR and RFLP analysis. To detect the A8344G MERRF mutation, primers 5′-GTATTTACCCTATAGCACCCCCTCTAC-3′ (nucleotide positions [np] 8255–8281 [numbered according to the work of Anderson et al.10]) and 5′-GGGGCATTTCACTGTAAAGAGGTGCCGG-3′ (np 8372–8345) (mismatched positions underlined) were used, and PCR products were digested with restriction endonuclease NaeI. For the MELAS mutation A3243G, primers 5′-GAAAGGACAAGAGAAATAAGGCCT-3′ (np 3127–3150) and 5′-GGAATTGAACCTCTGACTGTAAAG-3′ (np 3293–3270) and restriction endonuclease HaeIII were used; for mutation G16428A, primers 5′-CCTCCGTGAAATCAATAGCCC-3′ (np 16407–16427) and 5′-CAGATGTCGGATACAGTTCACTT-3′ (np 16503–16481) and restriction endonuclease Cac8I were used; for mutation A16182C, primers 5′-AAAAACCCAATCCACGTCAA-3′ (np 16162–16181) and 5′-GTTGGTATCCTAGTGGGTGAG-3′ (np 16282–16262) and restriction endonuclease HincII or primers 5′-AAAAACCCAATCCACATCCA-3′ (np 16162–16181) and 5′-AGGGGTGGCTTTGGAGTT-3′ (np 16263–16246) and restriction endonuclease MmeI were used. Amplification conditions were as follows: at 95°C for 5 min; 33 cycles at 95°C for 15 s, at 55°C or 60°C for 30 s, and at 72°C for 1 min; finally, at 72°C for 7 min. Restriction fragments were separated on 10% polyacrylamide gels and were visualized by SYBR Green I staining (Sigma-Aldrich). Proportions of wild-type and mutant mtDNA were estimated from band intensities through use of the ImageJ analysis software.
Allele-specific amplification of DNA fragments carrying either the wild-type or the mutant allele of np 3243 were performed with primer 5′-ACCCAATCCACATCAAAACC-3′ (np 16166–16185), together with either primer 5′-TTAAGTTTTATGCGATTACCGGCCT-3′ or primer 5′-TTAAGTTTTATGCGATTACCGGCCC-3′ (np 3267–3243) (mismatched nucleotide underlined; diverging nucleotides in italics). Amplification conditions were as follows: at 96°C for 30 s, 20 cycles at 94°C for 30 s and at 68°C for 8 min, and finally at 72°C for 7 min. One microliter of the amplified product was used as the template in a second-round mismatch PCR. Direct sequencing of PCR products was performed on an automatic sequence analyzer by a commercial sequencing service (MWG Biotech).
Single-Cell PCR Analysis
A suspension of trypsin-harvested fibroblasts was layered on slides with poly-l-lysine–treated polyethylene naphtalate membrane and was incubated at 37°C for 2 h, to allow cells to attach to the membrane. Slides were then immersed sequentially in 70% ethanol, 95% ethanol, absolute ethanol, and xylene. Single fibroblasts from air-dried membranes were cut using the PALM MicroBeam system operating with a nitrogen laser and were catapulted to a tube cap containing 20 μl magnesium-free PCR buffer, 10-fold diluted Tris-EDTA buffer, and 0.5% Tween 20. After a short centrifugation and incubation (for 1 h) at room temperature, samples were divided into two aliquots and were directly subjected to mismatch-PCR and RFLP analysis.
Single-Molecule PCR
Individual DNA molecules were amplified using nested PCR under limiting dilution conditions.11 The first 50 cycles of the nested PCR were performed with forward primer 5′-TAACTCCACCATTAGCACCC-3′ (np 15972–15991) and reverse primer 5′-GCTCGCAGTGCGCCGATCAGGGCGTAGTTTGAGTTT-3′ (np 3710–3675). A second round of 15 cycles was made using the inner forward primer 5′-ACCCAAAGCTAAGATTCTAA-3′ (np 15988–16007) and the same reverse primer as for the first round. Single-molecule PCR products were analyzed by HaeIII restriction digestion for the A3243G mutation. Before digestion, PCR products were converted into homoduplex form by twofold dilution in fresh PCR mixture and by performing one additional PCR cycle. The G16428A mutation was determined by direct sequencing of each single-molecule PCR product.
Results
Family with the A8344G/A16182C Mutations
In skeletal muscle of a patient harboring the pathogenic A8344G MERRF mutation, we identified an additional heteroplasmic mutation in the noncoding d-loop region at np 16182. The mutation loads of both heteroplasmic mutations were determined in different available tissue samples from the patient and family members (fig. 1A) by RFLP analysis. Additionally, regions of the mitochondrial genome flanking each mutation were sequenced in each sample to confirm heteroplasmy and to exclude contamination. Figure 1B shows a combined view of the mutation loads of both heteroplasmic mutations in all analyzed tissue samples. Data points accumulate mainly in the upper left corner and in the lower right corner, as well as along the axis between these two corners. This indicates an abundance of two allelic combinations in the family, 8344A/16182C (upper left) and 8344G/16182A (lower right). In all family members, blood samples harbored the most 8344A/16182C alleles, whereas buccal mucosa, urine sediment, fibroblasts, brain, and skeletal muscle contained increasing amounts of the other alleles, 8344G and 16182A. However, certain deviations from this approximate distribution pattern were observed. First, all data points for the tissue samples from the sister of the patient’s mother (black symbols in fig. 1B) are located below the theoretical axis. Such a pattern cannot be interpreted without the assumption of the presence of the wild-type allelic combination 8344A/16182A. Second, the data points representing fibroblasts and skeletal muscle from the patient’s sister (green symbols in fig. 1B) are located above the theoretical axis. This can be accomplished only if these samples harbor the fourth, double-mutant allelic combination 8344G/16182C. Third, when the blood sample from the patient’s mother was examined in detail, evidence of the presence of the double-mutant 8344G/16182C combination was also obtained (fig. 1C; lane 3), since this sample is apparently homoplasmic for the 16182C allele, whereas it contains both 8344A and 8344G alleles. In addition to estimations of bulk tissue-mutation loads, we examined single fibroblasts of the patient’s sister to find out which allelic combinations are present in individual cells. Figure 1D shows the distribution of the two heteroplasmic mutations in single fibroblasts. Data points populate all four corners of the graph, which indicates the presence of all four possible allelic combinations. Notably, the double-mutant allelic combination 8344G/16182C (upper right corner of the graph in fig. 1D) is present in ∼25% of the cells.
In summary, we observed, in different tissue samples from the A8344G/A16182C double-heteroplasmic family, that (1) all four allelic combinations of the two heteroplasmic mutations were present in the family, although, as expected, the distribution showed significant differences between individuals; (2) two family members carried the double-mutant (and possibly recombinant) allelic combination; and (3) high amounts of the possibly recombinant genotype, along with all other possible allelic combinations, were present in the fibroblast sample from one individual.
Family with the A3243G/G16428A Mutations
In a second family, we identified the rare G16428A d-loop mutation in heteroplasmic state in addition to the pathogenic A3243G MELAS mutation (fig. 2A). A wide range of A3243G mutation loads was observed in various tissues of family members (fig. 2B). In contrast, the mutant 16428A allele was in most samples close to homoplasmy; only the patient’s muscle harbored a high degree of the wild-type 16428G allele. However, except for the blood from the daughter of the patient’s aunt, which was apparently homoplasmic 16428A, the presence of the 16428G allele was confirmed in all other samples (fig. 2C; upper panel). We examined brain and urine sediment and skeletal muscle from the patient and urine sediment from the patient’s maternal aunt, by a two-round allele-specific/mismatch PCR approach, to determine which allelic combinations were present in these samples. In all investigated samples, we found that both alleles of G16428A were present in combination with both alleles of A3243G, at comparable proportions (tetraplasmy) (fig. 2D and 2E). These findings were confirmed by single-molecule PCR of the bulk skeletal-muscle sample from the patient. Of 36 molecules analyzed, 20 were 3243G/16428G, 8 were 3243A/16428A, 4 were 3243G/16428A, and 4 were 3243A/16428G.
Findings in the family with the A3243G/G16428A mutations, therefore, showed that (1) all four allelic combinations of the two heteroplasmic mutations were present in two family members and (2) the proportions of the rarer 16428G allele were balanced between both alleles 3243A and 3243G.
Comparison with Database Sequences
To estimate mutation frequencies and to examine our families in the phylogenetic context, we sequenced the complete mitochondrial genomes of one family member from each family (GenBank accession numbers DQ862536 [for the patient with the A3243G/G16428A mutations] and DQ862537 [for the sister of the patient with the A8344G/A16182C mutations]) and compared the obtained sequences with >1,600 publicly available complete human mitochondrial genome sequences (GenBank,12; mtDB–Human Mitochondrial Genome Database,13).
The A16182C mutation is present in 7.7% of all publicly available sequences; however, in most cases, it occurs along with other mutations as a haplogroup-defining or a subgroup-defining mutation. In such cases, A16182C is likely to be acquired once in the respective haplogroup and to be transferred through maternal lineages rather than to be generated de novo in each individual. This assumption is also supported by the study of Meyer et al.,14 who suggested that the mutation rate at position 16182 is lower than the average HVRI (hypervariable region I) mutation rate. Furthermore, the analysis of database sequences reveals that the generation of the A16182C mutation is strictly context dependent. The available data strongly suggest a unidirectional sequence of mutational events: T16189C→A16183C→A16182C. This chain of mutation occurs several times in the phylogenetic tree scattered among different haplogroups. In certain cases, A16183C and A16182C occur together as haplogroup-defining mutations (e.g., haplogroup T2), whereas, in other haplogroups, such as haplogroup H, A16183C is present without the additional appearance of A16182C. Since our A8344G/A16182C family belongs to haplogroup H (subgroup H7), the probability of repeated 16182A→C mutational events within this family has to be considered low.
The G16428A mutation occurs twice in the database (GenBank accession numbers AY339529 and AY339472). These two database sequences seem to be unrelated, since they carry only one more common mutation, A16399G, and 15 and 25 different mutations, respectively. One of the two database sequences, AY339529, is closely related to the sequence of our patient, since these two sequences harbor 20 common mutations and are different at only two sites.
We strongly suggest that, taken together, neither G16428A nor A16182C is a mutational hotspot of the human mitochondrial genome. Since most human mitochondrial sequences in databases were obtained from blood DNA, we also performed an RFLP screening of our collection of muscle samples, to exclude the possibility that nucleotides 16428 and 16182 are tissue-specific mutational hotspots. With this experimental approach, we were able to detect low levels of heteroplasmy that might be missed by regular sequencing. The determined allele frequencies shown in table 1 prove that heteroplasmy at these particular d-loop sites is very rare.
Table 1. .
Frequency of Investigated Heteroplasmic Mutations in a Collection of Skeletal-Muscle Biopsies
| Mutation | Frequency of Heteroplasmy in Skeletal Muscle (%) |
No. of RFLP-Screened Unrelated Muscle Samples |
| A3243G | 4.1 | 266 |
| A8344G | 1.1 | 266 |
| A16182C | .4 | 266 |
| G16428A | .5 | 198 |
Discussion
To address the question of whether recombinant mtDNA molecules can be inherited, we examined families harboring two heteroplasmic mutations in several family members. In each investigated family, one heteroplasmic mutation was a well-characterized pathogenic mutation in the coding region, whereas the other heteroplasmic mutation was located in the noncoding d-loop region. The two heteroplasmic mutations were separated by several thousand nucleotides in both families, leaving enough distance for possible recombination events.8 To conclude inheritance of recombinant mtDNA molecules, the following questions have to be addressed. (1) Are all four possible combinations of the two heteroplasmic alleles present in the family? Two sequentially occurring independent mutation events would create a mixture of only three genotypes (triplasmy). For the creation of all four genotypes (tetraplasmy), either recurrent mutation events or recombination is required. (2) How high is the probability that a specific mutational event occurs repeatedly in an individual or in a family? Such recurrent mutations would also have a chance to create tetraplasmy, a pattern that is similar to the outcome of recombination. (3) Can recombinant mtDNA molecules be observed in different family members? If yes, are they likely to be results of independent recombination events or, rather, transmitted through the germ line?
Elsewhere, we addressed the first two questions in the study of skeletal muscle of double-heteroplasmic individuals,8 and we showed that tetraplasmy was present in cases where the probability of recurrent mutations was negligible. We interpreted this finding as a sign of recombination of the human mitochondrial genome. Apart from addressing the same questions in new cases and other tissues, here, we extend our investigations to the family level to evaluate the potential impact of mtDNA recombination on mtDNA phylogeny.
We demonstrate in this study that individuals of both investigated families harbor all four possible allelic combinations of the two heteroplasmic mutations. This approach is analogous to the “four-gamete” test found in the classic literature about DNA recombination.15 The two pathogenic heteroplasmic mutations investigated here, A3243G (MELAS) and A8344G (MERRF), are rare (see table 1), and they have been recognized as having the potential to be inherited through several generations. Recent reports provided evidence that long-persisting heteroplasmy appears to not be an exclusive feature of certain pathogenic mutations; rather, it is also a feature of d-loop polymorphisms.16,17 Whether two heteroplasmic instances of a specific mtDNA mutation occurred independently or are related (i.e., originate from a single mutation event and have been inherited in the heteroplasmic state) may be difficult to distinguish. Since members of the published families16,17 carry heteroplasmy at the same site, inheritance of heteroplasmy in these families is more plausible, unless one assumes a family-specific and site-specific instability of the mitochondrial genome. In respect to the both d-loop sites—A16182C and G16428A—investigated in our report, we conclude from comparison with database sequences and a screening of a large collection of brain and muscle sample that heteroplasmy at these sites is rare. Therefore, similar to the pathogenic mutations A3243G and A8344G, the d-loop mutations A16182C and G16428A are likely to originate from single mutation events within the respective families. Since recurrence of mutational events has such a low probability, we conclude that the tetraplasmy observed in the study individuals indicates recombination of the mitochondrial genome.
The limited number of published cases in which recombination might contribute to the allelic distribution of heteroplasmic mitochondrial mutations makes it impossible to estimate the frequency of recombination events in humans. The reported proportions of putative recombinant genotypes highly diverge between individuals and have probably only a limited conclusive value concerning frequency of recombination. Similar to genomes with newly obtained mutations, recombinant mtDNA molecules are likely to be subjects of major segregational propagation or extinction. When segregational drifts are taken into account, it is more plausible to assume that recombinant mtDNA molecules were transmitted through the germ line rather than repeatedly generated in somatic cells (1) if recombinant molecules represent a large portion of all mitochondrial genomes in tissues and especially if this can be observed in young individuals, (2) if recombinant molecules not only appear in isolated tissues but are present in the whole body, and (3) if recombinant genotypes are detectable in different members of a family. Both families investigated here fulfill all three criteria. In all samples in which genotype frequencies were determined experimentally, all four allelic combinations were present at considerable levels. Since earlier observations indicate a rather low frequency of somatic recombination,7 our data strongly suggest the inheritance of the recombinant mitochondrial genotypes, even in the absence of direct evidence of recombinant mtDNA in female germ cells.
Interestingly, the mitochondrial genome of the patient with the A3243G/G16428A mutation appears to be closely related to a database sequence that also carries the G16428A mutation (AY339529).18 These two sequences are identical, with the exceptions that the mutation A249G, which is present in the database sequence, is missing from our patient, whereas the C150T mutation can be found in our patient and not in AY339529. Furthermore, an additional sequence (AY339528)18 is present in the database that harbors all mutations common between our patient and AY339529 but does not carry G16428A and harbors A249G. Thus, the two mutations A249G and G16428A create a remarkable pattern in this small subgroup of three individuals: each mutation is missing from one of the three individuals. Their phylogenetic relationship can be described only by a network. Similar deviations from the standard tree structure—called “reticulations”—are relatively common in mtDNA phylogenetic trees19,20 and always represent a serious problem for tree reconstruction and interpretation. Traditional explanations for reticulation include recurrent mutations at group-specific hotspots and reverse mutations, but recombination has also been suggested to create reticulations.3,21
A schematic view of possible mutation events that lead to a reticulation is shown in figure 3. Normally, two sequential mutational events create three types of genomes (fig. 3A). In the event of a reticulation (fig. 3B), four genome types are present. One possible explanation is that one of the reticulating mutations happens more than once in the specific subgroup, which creates all possible allelic combinations (fig. 3C). However, it is difficult to explain why a specific site mutates repeatedly in a very small subgroup when the same site shows very high stability throughout the entire human population. Haplogroup-specific changes in mutation rates of specific mtDNA sites have been proposed,16,22 but the the smaller the reticulating group is and the rarer the reticulating mutations are in the general population, the more dramatic the changes of mutation rates have to be assumed. Another explanation of reticulations postulates that a second mutation makes a previously mutated site unstable, so that it “mutates back” to wild type (fig. 3D). This scenario would, however, create a mutational drift toward the wild type, thus eliminating the mutant at the phylogenic scale, which appears to contradict statistical observations.21 Furthermore, for both reverse and recurrent mutations (with the exception of simple repeats23), no molecular mechanism is known at present that could explain the large variations of mutation rates.2
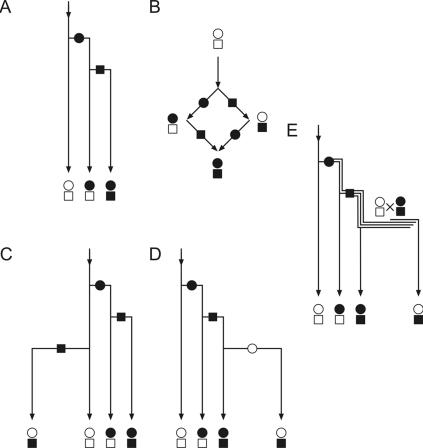
Figure 3. .
Schematic representation of different theoretical scenarios that result in reticulation. A, Standard phylogenetic tree. Two sequential mutations create three different types of genomes (no reticulation). In some cases, all four possible combinations of the two mutations are present in a population (B). Then they have to be formally related by a nontree network structure (reticulation). The square-shaped reticulations can be easily recognized within typical mtDNA phylogenetic trees.19,20 Reticulations are usually “resolved” into tree structures by the assumption of either recurrent mutations (C) or reverse mutations (D). We propose that, alternatively, reticulations can result from recombination of mtDNA in double-heteroplasmic individuals (E). The persistent heteroplasmy (double line) is switched to a triplasmic double-heteroplasmic state (triple line) by a second mutational event. Recombination can then create the fourth allelic combination that leads to reticulation.
We propose an alternative mechanism that does not require site-specific or group-specific molecular mechanisms to explain a reticulation. As shown in figure 3E, the recombination of coexisting heteroplasmic mutations (indicated by double or triple lines) creates a reticulating pattern. This scenario is possible only if a newly generated mtDNA mutation persists in the heteroplasmic state until a second mutation occurs, so that the two heteroplasmic mutations coexist within an individual, which permits productive recombination. Coexistence of heteroplasmic mutations has been already demonstrated in several published examples.8,16,17,24–26
To summarize, our findings strongly suggest that recombination of the human mtDNA is not restricted to somatic tissues; rather, recombinant mtDNA molecules can be transmitted through the germ line. This feature could explain the presence of certain allelic combinations in maternal lineages that cannot be inferred from a plain sequential order of mutational events. Thus, our hypothesis—that recombination of coexisting heteroplasmic mutations creates reticulations—eliminates the need to search specific molecular mechanisms for an explanation of individual reticulations in the human mtDNA phylogenetic tree.
Acknowledgments
This study was supported by Deutsche Forschungsgemeinschaft grants KU 911/15-1 and SCHR 562/4-2 (to W.S.K.) and by National Institutes of Health grants AG19787 and ES11343 (to K.K.). The technical assistance of Ulrike Strube is gratefully acknowledged.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for the complete mitochondrial sequences of the patient with the A3243G/G16428A mutations [accession number DQ862536] and of the sister of the patient with the A8344G/A16182C mutations [accession number DQ862537] and Homo sapiens isolates F128 [accession numbers AY339529] and F71 [accession number AY339472])
- ImageJ, http://rsb.info.nih.gov/ij/ (for Image Analysis Software)
- mtDB–Human Mitochondrial Genome Database, http://www.genpat.uu.se/mtDB/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for MERRF syndrome and MELAS syndrome)
References
- 1.Rokas A, Ladoukakis E, Zouros E (2003) Animal mitochondrial DNA recombination revisited. Trends Ecol Evol 18:411–417 10.1016/S0169-5347(03)00125-3 [DOI] [Google Scholar]
- 2.Pakendorf B, Stoneking M (2005) Mitochondrial DNA and human evolution. Annu Rev Genomics Hum Genet 6:165–183 10.1146/annurev.genom.6.080604.162249 [DOI] [PubMed] [Google Scholar]
- 3.Eyre-Walker A, Smith NH, Smith JM (1999) How clonal are mitochondria? Proc Biol Sci 266:477–483 10.1098/rspb.1999.0662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Awadalla P, Eyre-Walker A, Smith JM (1999) Linkage equilibrium and recombination in hominid mitochondrial DNA. Science 286:2524–2525 10.1126/science.286.5449.2524 [DOI] [PubMed] [Google Scholar]
- 5.Elson JL, Andrews RM, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (2001) Analysis of European mtDNAs for recombination. Am J Hum Genet 68:145–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Innan H, Nordborg M (2002) Recombination or mutational hot spots in human mtDNA? Mol Biol Evol 19:1122–1127 [DOI] [PubMed] [Google Scholar]
- 7.Kraytsberg Y, Schwartz M, Brown TA, Ebralidse K, Kunz WS, Clayton DA, Vissing J, Khrapko K (2004) Recombination of human mitochondrial DNA. Science 304:981 10.1126/science.1096342 [DOI] [PubMed] [Google Scholar]
- 8.Zsurka G, Kraytsberg Y, Kudina T, Kornblum C, Elger CE, Khrapko K, Kunz WS (2005) Recombination of mitochondrial DNA in skeletal muscle of individuals with multiple mitochondrial DNA heteroplasmy. Nat Genet 37:873–877 10.1038/ng1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 10.1093/nar/16.3.1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465 10.1038/290457a0 [DOI] [PubMed] [Google Scholar]
- 11.Kraytsberg Y, Khrapko K (2005) Single-molecule PCR: an artifact-free PCR approach for the analysis of somatic mutations. Expert Rev Mol Diagn 5:809–815 10.1586/14737159.5.5.809 [DOI] [PubMed] [Google Scholar]
- 12.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL (2005) GenBank. Nucleic Acids Res 33:D34–D38 10.1093/nar/gki063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingman M, Gyllensten U (2006) mtDB: Human Mitochondrial Genome Database, a resource for population genetics and medical sciences. Nucleic Acids Res 34:D749–D751 10.1093/nar/gkj010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer S, Weiss G, von Haeseler A (1999) Pattern of nucleotide substitution and rate heterogeneity in the hypervariable regions I and II of human mtDNA. Genetics 152:1103–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hudson RR, Kaplan NL (1985) Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111:147–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carelli V, Achilli A, Valentino ML, Rengo C, Semino O, Pala M, Olivieri A, Mattiazzi M, Pallotti F, Carrara F, et al (2006) Haplogroup effects and recombination of mitochondrial DNA: novel clues from the analysis of Leber hereditary optic neuropathy pedigrees. Am J Hum Genet 78:564–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howell N, Kubacka I, Keers SM, Turnbull DM, Chinnery PF (2005) Co-segregation and heteroplasmy of two coding-region mtDNA mutations within a matrilineal pedigree. Hum Genet 116:28–32 10.1007/s00439-004-1203-x [DOI] [PubMed] [Google Scholar]
- 18.Moilanen JS, Finnilä S, Majamaa K (2003) Lineage-specific selection in human mtDNA: lack of polymorphisms in a segment of MTND5 gene in haplogroup J. Mol Biol Evol 20:2132–2142 10.1093/molbev/msg230 [DOI] [PubMed] [Google Scholar]
- 19.Finnilä S, Lehtonen MS, Majamaa K (2001) Phylogenetic network for European mtDNA. Am J Hum Genet 68:1475–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrnstadt C, Elson JL, Fahy E, Preston G, Turnbull DM, Anderson C, Ghosh SS, Olefsky JM, Beal MF, Davis RE, et al (2002) Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet 70:1152–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eyre-Walker A (2000) Do mitochondria recombine in humans? Philos Trans R Soc Lond B Biol Sci 355:1573–1580 10.1098/rstb.2000.0718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galtier N, Enard D, Radondy Y, Bazin E, Belkhir K (2006) Mutation hot spots in mammalian mitochondrial DNA. Genome Res 16:215–222 10.1101/gr.4305906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howell N, Smejkal CB (2000) Persistent heteroplasmy of a mutation in the human mtDNA control region: hypermutation as an apparent consequence of simple-repeat expansion/contraction. Am J Hum Genet 66:1589–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohno K, Yamamoto M, Engel AG, Harper CM, Roberts LR, Tan GH, Fatourechi V (1996) MELAS- and Kearns-Sayre-type co-mutation with myopathy and autoimmune polyendocrinopathy. Ann Neurol 39:761–766 10.1002/ana.410390612 [DOI] [PubMed] [Google Scholar]
- 25.Bidooki SK, Johnson MA, Chrzanowska-Lightowlers Z, Bindoff LA, Lightowlers RN (1997) Intracellular mitochondrial triplasmy in a patient with two heteroplasmic base changes. Am J Hum Genet 60:1430–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zsurka G, Schroder R, Kornblum C, Rudolph J, Wiesner RJ, Elger CE, Kunz WS (2004) Tissue dependent co-segregation of the novel pathogenic G12276A mitochondrial tRNALeu(CUN) mutation with the A185G D-loop polymorphism. J Med Genet 41:E124 10.1136/jmg.2004.022566 [DOI] [PMC free article] [PubMed] [Google Scholar]