Abstract
Alopecia areata (AA) is a genetically determined, immune-mediated disorder of the hair follicle that affects 1%–2% of the U.S. population. It is defined by a spectrum of severity that ranges from patchy localized hair loss on the scalp to the complete absence of hair everywhere on the body. In an effort to define the genetic basis of AA, we performed a genomewide search for linkage in 20 families with AA consisting of 102 affected and 118 unaffected individuals from the United States and Israel. Our analysis revealed evidence of at least four susceptibility loci on chromosomes 6, 10, 16 and 18, by use of several different statistical approaches. Fine-mapping analysis with additional families yielded a maximum multipoint LOD score of 3.93 on chromosome 18, a two-point affected sib pair (ASP) LOD score of 3.11 on chromosome 16, several ASP LOD scores >2.00 on chromosome 6q, and a haplotype-based relative risk LOD of 2.00 on chromosome 6p (in the major histocompatibility complex locus). Our findings confirm previous studies of association of the human leukocyte antigen locus with human AA, as well as the C3H-HeJ mouse model for AA. Interestingly, the major loci on chromosomes 16 and 18 coincide with loci for psoriasis reported elsewhere. These results suggest that these regions may harbor gene(s) involved in a number of different skin and hair disorders.
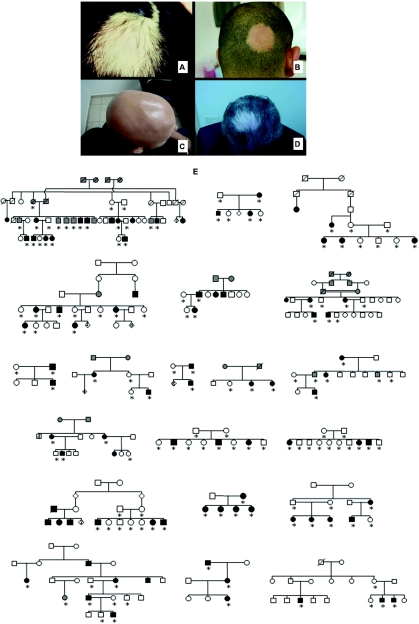
Alopecia areata (AA [MIM 104000]) is one of the most common human autoimmune diseases, with a lifetime risk of ∼2%.1–3 In the United States, >4.5 million people are affected with AA (National Alopecia Areata Foundation), which affects both sexes at all ages and in all ethnic groups. It is characterized by patchy hair loss on the scalp (fig. 1A and 1B), which can eventually involve the entire scalp, a condition known as “alopecia totalis,” or the entire body, a condition known as “alopecia universalis” (fig. 1C). The onset of the disease can be sudden, its progression is unpredictable, and it can be recurrent throughout life. Reports in the literature of overnight whitening of the hair represent the abrupt onset of AA, since it preferentially targets pigmented hairs, leaving only white hairs behind (fig. 1D).4–7 The pathology of alopecia extends far beyond the physical aspects of hair loss, and it can have a deeply disturbing psychological impact on affected individuals.8–10 Even among patients with minimal hair loss from AA, the loss carries significant emotional and psychological meaning that not only pertains to hair but also has a profound impact on an individual's quality of life, ability to function in society, and preservation of self-esteem, and it can lead to profound psychological disturbances.8,10
Figure 1. .
Clinical presentation of AA. A and B, AA appearing as well-circumscribed patches of hair loss on the scalp. C, Alopecia universalis, the complete form of AA, which leads to absence of all hair on the body. D, AA with selective loss of pigmented hair, with a patch of white hair left behind. E, Representative examples of pedigree structures of the families with AA. Participating family members are indicated by an asterisk (*).
Despite its high prevalence and the inherent visibility of the phenotype, the pathogenesis of AA is poorly understood, and there has been significant debate about whether the primary defect is in the hair follicle, the immune response, or both. Because of the presence of a peribulbar lymphocytic infiltrate in the scalp biopsy specimens of affected patients and the positive response of the disease to steroid treatment, an autoimmune mechanism has been postulated for many years, although an autoantigen has not yet been identified.11–16 More recently, attempts to arrive at a unified hypothesis have led to the description of AA as a tissue-specific autoimmune disease of the hair follicle.1 The hair follicle is an immune-privileged site, with low levels of major histocompatibility complex (MHC) expression, and the emerging view is that AA represents a breakdown in immune privilege and the subsequent destruction of the hair follicle by T-lymphocytes. Thus, AA can be considered a genetically determined, immune-mediated disorder, which therefore should be amenable to genetic linkage studies.
It is now generally accepted that AA fits the paradigm of a complex or multifactorial genetic trait, on the basis of several lines of evidence: (i) its prevalence in the population (∼2%),17,18 (ii) concordance in twins (55%),19 (iii) a Gaussian distribution of severity,2 (iv) a 10-fold increased risk for first-degree relatives of affected individuals,2,3,20 and (v) the aggregation of affected individuals in families, instead of a clear Mendelian pattern of inheritance.2,3 A systematic search for genetic factors underlying this disorder has not been previously undertaken. Additionally, linkage analysis in complex traits is not exempt from challenges, because of the characteristic genetic heterogeneity and the potential presence of numerous susceptibility alleles. These factors may be further complicated, in the case of AA, by the challenges inherent in defining the phenotype, exemplified by a range in ages of onset, the sometimes subtle nature of presenting signs, and the waxing-and-waning nature of the disease.
Until now, genetic studies in AA have been limited to association analyses, which suggest that a permissive human leukocyte antigen (HLA) status may potentiate the development of the disease.12 In early attempts to identify a genetic component of AA, a number of association studies with candidate genes have been conducted. Association between AA and particular HLA alleles (MIM *604305 and MIM *142860), interleukin-1 (IL1 [MIM *147760 and MIM *147720]) cluster genes, and the myxovirus resistance 1 gene (MX1 [MIM *147150]), on chromosome 21, have been suggested (for a review, see the work of Green and Sinclair2 and McDonagh and Tazi-Ahnini3 and references therein). Significant association has been reported between AA and DQB1*0301 (for severe AA) and DRB1*1104 alleles.12,21 A family-based study has revealed that 85% of patients with AA carry DQB1*03 alleles, compared with 46% of controls.22 Those authors also have shown linkage to HLA-DQB and HLA-DR. Recently, two new positive associations have been described. The MHC class I chain-related gene A (MICA [MIM *600169]) has been identified as both a potential candidate gene and a part of an extended HLA haplotype that may contribute to the susceptibility to and severity of AA.23 Additionally, the gene encoding the lymphoid protein tyrosine phosphatase (PTPN22 [MIM *600716]) has been shown to be associated with severe forms of AA.24
The presence of a perifollicular T-cell infiltrate suggests an important role for cytokine production in the pathogenesis of AA. Likewise, it is possible to transfer the disease with lesional human lymphocytes into an SCID mouse grafted with human scalp skin.25 Along these lines, several authors have suggested association between the more severe forms of AA and the IL-1 receptor antagonist gene (IL1RN [MIM *147679]), in particular, allele IL1RN*2, and the IL-1 receptor antagonist homologue (IL1F5 [MIM *605507]).26–28 Furthermore, there is an increased prevalence of AA among patients with Down syndrome (MIM #190685) (9%) compared with among control individuals (0.1%).29 The MX1 gene maps to chromosome 21 and encodes an interferon-inducible protein highly expressed in lesional anagen hair bulbs from patients with AA. Tazi-Ahnini et al.30 showed significant association between a SNP located within MX1, 9,959 bp from the transcription start site, and patchy AA. Finally, an association between autoimmune polyglandular syndrome type 1 (APS1 [MIM #240300]), caused by mutations in the AIRE gene (MIM *607358), on chromosome 21, and AA has also been reported,31 with AA observed in 37% of patients with APS1. Despite the genetic associations described above, it is likely that these alleles account for only part of the genetic susceptibility to AA. It is noteworthy that cosegregation with HLA was excluded in two Israeli families.32
A second line of evidence for the genetic basis of AA comes from the study of animal models. Particularly, the C3H/HeJ mouse is an inbred laboratory strain that spontaneously develops an adult-onset disease that resembles adult-onset AA in humans. Sundberg et al.33 have identified four genetic susceptibility loci on mouse chromosomes 8 (Alaa3), 9 (Alaa2), 15 (Alaa4), and 17 (Alaa1), wherein Alaa1 corresponds to HLA orthologs.
The lack of knowledge of the etiology of AA, the psychological impact on the quality of life of patients with AA, and the absence of an effective treatment underscore the importance of identifying the mechanisms underlying the disease. With this in mind, we initiated a comprehensive genetic analysis of families with multiple affected individuals, in search of genes that contribute to the development of AA.
Material and Methods
Ascertainment of Families
Families were recruited through a patient who received a diagnosis of AA. The inclusion criteria required that families have two or more affected relatives. With this requirement as a starting point, all family members willing to participate were recruited for the study. Pedigrees were enrolled from the United States, primarily through the National Alopecia Areata Registry, and from Israel. Those family members for whom clinical data were not available were classified as “unknown” for linkage purposes.
Clinical examiners diagnosed AA in the patients before the genetic studies. At the time of consultation, blood samples were drawn and written informed consent was obtained from all participants. The study was approved by the local institutional review boards. Overall, 38 pedigrees, consisting of 102 affected and 118 unaffected or unknown individuals who participated in the study, were collected (examples shown in fig. 1E). On average, each family contained three affected individuals. The largest pedigree, HAA01, originating from Israel, consisted of 22 participating family members, 10 of whom received a diagnosis of AA (fig. 1E).
Genotyping
Genomic DNA was extracted using the PureGene DNA Isolation Kit (Gentra Systems). Twenty pedigrees (fig. 1E) were initially genotyped using a semiautomated high-throughput genotyping approach with fluorescently labeled microsatellite markers.34–36 A panel of 342 microsatellite markers was used, with an average marker spacing of 10 cM and an average heterozygosity of 0.77. Most of the markers were chosen from version 8.0 of the Marshfield fluorescence-labeled genome screening set. DNA samples and PCR reagents were aliquoted with a TECAN Genesis RSP 150 robotic workstation. Multiplex PCR was performed in 384-well plates (Marsh) in PTC 225 thermocyclers (MJ Research). An average of 50 ng of genomic DNA was amplified in 10-μl PCRs containing 0.15–0.2 mM MgCl2, 0.2 mM dNTPs, and 0.5 units of Taq Platinum polymerase (Invitrogen). The primer concentration was adjusted (1–50 pmol) to achieve even amplification of each marker locus contained in the multiplex PCRs. As described elsewhere,37 and to improve allele-calling, the last nucleotide of the reverse, nonfluorescent primer was modified to a guanine to promote the nontemplated addition of adenine by Taq DNA polymerase onto the complementary, fluorescence-labeled strand. DNA from CEPH control individuals was used as a size standard for every marker locus. PCR products were electrophoresed on 377 DNA sequencers (PE Applied Biosystems). Raw data from the PCR products were collected by PRISM 377XL data-collection software (PE Applied Biosystems), and the products were sized by GENESCAN version 2.1 and GENOTYPER v.1.1.1. The genotypes were imported to LABMAN38 for allele binning, Mendelian checking, and generation of linkage files. The PEDCHECK program39 was used to check for genotype errors. The markers that showed genotype errors were recoded to unknown genotypes in each family in which a genotype error was observed. For all multipoint runs, the MEGA2 program40 was used to format pedigree data for computation of the likelihood-ratio Z (Zlr) statistics.
As follow-up to the genome scan, fine mapping with microsatellite markers was performed by deCODE Genetics for the entire cohort of 38 families. For each marker, the forward primer was fluorescently labeled. The primer pairs were extensively tested for optimizing the multiplex PCR reactions for cost benefits. PCR amplifications were set up on Zymark ALH 400, were run on MJR Tetrad, and were pooled on Gilson Cyberlab C200 robots. The reaction volume was 5 μl, and, for each PCR, 20 ng of genomic DNA was amplified in the presence of 2 pmol of each primer, 0.25 U AmpliTaq Gold, 0.2 mM dNTPs, and 2.5 mM MgCl2 (buffer was supplied by the manufacturer, Applera). Cycling conditions were as follows: at 95°C for 10 min, followed by 37 cycles at 94°C for 15 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min. The PCR products were supplemented with the internal size standard GS500-LIZ, and the pools were separated and detected on 3730 Sequencers. Alleles were automatically called using DAC, an allele-calling program developed at deCODE Genetics,41 and the program deCODE GT was used to fractionate called genotypes, according to quality, and to edit, when necessary.42
Six chromosomal regions on four chromosomes (6, 10, 16, and 18) were genotyped, corresponding to the six intervals: (i) the HLA region on chromosome 6, analyzed using a high-density panel of 60 microsatellite marker loci at an average marker spacing of 0.09 cM, which spanned the interval flanked by D6S2219 and D6S1560 (2.37 cM); (ii) chromosome 6 interval D6S1040–D6S1003 (16.72 cM; 12 marker loci); (iii) chromosome 10 interval D10S674–D10S2481 (10.26 cM; 11 marker loci); (iv) chromosome 10 interval D10S1239–D10S1230 (23.22 cM; 17 marker loci); (v) chromosome 16 interval D16S403–D16S3098 (60.13 cM; 34 marker loci); and (vi) chromosome 18 interval D18S59–D18S1157 (61.03 cM; 29 marker loci). Marker locations are reported in Haldane map units.
Statistical Analysis
Initial genome scan
A two-step linkage analysis was performed on the collection of 38 pedigrees (102 affected and 118 unaffected participating individuals). In the first stage, a genomewide scan was conducted for a collection of 20 pedigrees and a total of 131 DNA samples (69 affected and 62 unaffected family members; fig. 1E). The test statistics applied to the data set obtained in the genomewide scan were (i) the heterogeneity LOD score,43,44 maximized over four settings of penetrance parameters (hereafter, “MAXHLOD”) (see table 1 for the penetrance settings used); (ii) the mean test for affected sib pairs, as implemented in the ANALYZE program45 (hereafter, “ASP”); (iii) a test of allele sharing that uses all sibs45 (hereafter, “ALLSIBS”); and a likelihood version of the transmission/disequilibrium test,46 as developed by Terwilliger47 (TDT-like, hereafter referred to as “TDT”).
Table 1. .
Modes of Inheritance and Penetrance Values Used to Compute MAXHLOD
| Penetrancesb |
||||
| Genetic Modela |
Pr(aff/++) | Pr(aff/+d) | Pr(aff/dd) | Disease-Allele Frequency Pr(d)c |
| 1 | .0 | .5 | .5 | .02 |
| 2 | .0 | .8 | .8 | .01 |
| 3 | .0 | .0 | .5 | .20 |
| 4 | .0 | .0 | .8 | .16 |
Models correspond to autosomal dominant (models 1 and 2) and recessive (models 3 and 4) patterns of inheritance, with penetrance values of 50% (models 1 and 3) and 80% (models 2 and 4).
A plus sign (+) = wild-type allele at disease locus; d = disease allele at disease locus.
Pr(d) is altered in each model, to maintain a constant prevalence.
For the ASP test, when there are multiple sibs in a sibship, ANALYZE weights the sib pairs according to the sibship size, as follows two sibs equal one sib pair, three sibs equal two sib pairs, and four sibs equal three sib pairs. As noted by Terwilliger in the user notes for ANALYZE, “This weighting has been selected to conform with the information content of phase-unknown nuclear pedigrees in linkage analysis.”48(p9) A detailed discussion has been published.49,50
Our rationale for the use of these different tests was described elsewhere.51 Briefly, for the MAXHLOD calculations, we applied a model-based linkage analysis in which the LOD score was calculated under both autosomal dominant and autosomal recessive patterns of inheritance. For both models, two different values of penetrance were considered. ASP, ALLSIBS, and TDT tests were chosen because they are all genetic-model free,52,53 in the sense that they do not require a specification of the genetic model parameters (penetrance and disease-allele frequency). MAXHLOD was chosen because it has been shown that it has at least as much power to localize disease loci as do ASP and ALLSIBS,54 and, under certain circumstances, it is a more precise indicator of the location of a disease locus than are statistics such as ASP and ALLSIBS.55 Finally, the TDT was chosen because it has been shown that it may be more powerful than linkage tests (e.g., MAXHLOD, ASP, and ALLSIBS) when linkage and linkage disequilibrium exist between a disease and a marker locus.
All results are presented as LOD scores. The TDT P value was converted to a LOD score by use of the following approximation. Let P be the P value for the TDT statistic. Because TDT is asymptotically distributed as χ2 with 1 df, we convert P to a LOD score via the formula χ-1(P)/4.6, where χ-1 is the inverse of the one-tailed probability of the χ2 distribution. An identical conversion was used for the haplotype-based haplotype relative risk (HRR) minus 2 times n statistic (HRR-2×n; see the “Fine-mapping analyses” section).
Fine-mapping analyses
For fine-mapping purposes, we focused on the marker loci that showed significant test scores (the top 10% of scores observed across all marker loci) for at least two of the test statistics listed above. At this stage, we were more concerned with power than with inflation of the false-positive rate.56
We performed fine-mapping analyses, using two-point and multipoint genetic model–free statistics. The two-point method was the ASP test. The multipoint method was the Kong and Cox57 Zlr statistic, which is implemented in the GENEHUNTER-PLUS software. Although we used affected and unaffected individuals in our initial genome scan (for the MAXHLOD and ALL-SIB statistics), we decided to focus on “affecteds-only” statistics in the fine-mapping analyses. Our reasoning was that the ASP method was generally more powerful (i.e., yielded higher LOD scores) than were the MAXHLOD and ALL-SIB statistics. We hypothesized that decrease in power for methods using unaffected individuals may stem from the fact that they are truly affected but did not present with symptoms at the time of diagnosis. The TDT statistic is also an affecteds-only statistic that takes advantage of any linkage disequilibrium that may occur between marker and disease locus. We added a family-based test of association, the HRR.58 The HRR statistic tests for association in the presence of linkage—unlike the TDT, which tests for linkage in the presence of association.59 More intuitively, the HRR tests whether the risk alleles are a single allele or a small set of alleles, whereas the TDT tests whether recombination between the trait and marker loci is small (close to 0). Our rationale for using a family-based test of association was based on the observation that, for several autoimmune diseases, associations with the MHC have been described, despite weak or no evidence of linkage. This phenomenon is thought to occur because of the high frequency of the associated allele, which creates multiple MHC haplotypes among affected members within families.
We use the HRR-2×n version of the HRR, as implemented in the ANALYZE program. As Terwilliger notes in the README notes on the program, “The test performed is that standard 2×n table χ2 test, which has n-1 df (where there are n alleles whose frequencies are compared in case and control samples). This test is less powerful by far when there is only one associated allele (i.e., linkage disequilibrium from a founder effect), but can be more sensitive when there are higher order associations with different alleles.”48(p7) Particularly for the MHC region, we expect that the latter situation will be true. The case and control samples for family-based data refer to the transmission of a particular allele to an affected child (case sample) and the nontransmission of a particular allele to an affected child (control sample).
Results
Initial Genome Scan
The top 15 scores resulting from each of the four statistical methods used are presented in table 2. A total of 10 marker loci resulted in LOD scores >2 (5 are shown in table 2). Among them, marker D6S1009, at 6q23.3, reached the highest linkage signal, with a MAXHLOD score of 3.554 and an ASP LOD score of 4.831. Even after correction for multiple testing, both scores are significant at the .05 level genomewide.60,61 Moreover, this locus showed the top score for three of the four statistical methods considered (table 2). It is interesting to note that several marker loci appear in the list of top scores for a few of the statistical methods. Although some of the methods may have correlated results,62 they are not identical. Finally, marker loci D6S1281 and D6S2427, which are located in the vicinity of the HLA region on chromosome 6, were among the markers with the top 15 MAXHLOD scores (Zmax=1.034 and 1.707, respectively).
Table 2. .
Top 15 Results for Four Genetic Statistical Methods Applied to the 20 Genotyped Pedigrees with AA
| Method and Result Rank |
Chromosome | Locus | LOD |
| MAXHLOD: | |||
| 1 | 6 | D6S1009 | 3.554 |
| 2 | 16 | D16S753 | 2.007 |
| 3 | 6 | D6S2427 | 1.707 |
| 4 | 10 | D10S1239 | 1.691 |
| 5 | 2 | D2S1776 | 1.545 |
| 6 | 9 | D9S301 | 1.518 |
| 7 | 1 | D1S1612 | 1.434 |
| 8 | 10 | D10S2481 | 1.384 |
| 9 | 17 | D17S1301 | 1.228 |
| 10 | 6 | D6S1270 | 1.173 |
| 11 | 12 | D12S1064 | 1.129 |
| 12 | 9 | D9S930 | 1.081 |
| 13 | 6 | D6S1003 | 1.068 |
| 14 | 18 | D18S976 | 1.055 |
| 15 | 6 | D6S1281 | 1.034 |
| ASP: | |||
| 1 | 6 | D6S1009 | 4.831 |
| 2 | 18 | D18S535 | 2.314 |
| 3 | 6 | D6S1003 | 1.660 |
| 4 | 17 | D17S1301 | 1.388 |
| 5 | 11 | D11S1390 | 1.213 |
| 6 | 17 | D17S1293 | 1.126 |
| 7 | 17 | D17S916 | .967 |
| 8 | 16 | D16S3253 | .964 |
| 9 | 6 | D6S1040 | .919 |
| 10 | 16 | D16S403 | .915 |
| 11 | 16 | D16S753 | .899 |
| 12 | 3 | D3S2398 | .881 |
| 13 | 10 | D10S1239 | .844 |
| 14 | 2 | D2S1776 | .820 |
| 15 | 2 | D2S1353 | .874 |
| ALLSIBS: | |||
| 1 | 6 | D6S1009 | 2.762 |
| 2 | 10 | D10S2481 | 1.354 |
| 3 | 2 | D2S338 | 1.340 |
| 4 | 2 | D2S1776 | 1.025 |
| 5 | 10 | D10S674 | .850 |
| 6 | 1 | D1S1612 | .845 |
| 7 | 10 | D10S1230 | .836 |
| 8 | 6 | D6S1270 | .809 |
| 9 | 6 | D6S1056 | .775 |
| 10 | 9 | D9S930 | .755 |
| 11 | 10 | D10S1239 | .747 |
| 12 | 8 | D8S1179 | .704 |
| 13 | 11 | ATA34E08 | .696 |
| 14 | 13 | D13S779 | .691 |
| 15 | 9 | D9S301 | .690 |
| TDT: | |||
| 1 | 17 | D17S1301 | 1.930 |
| 2 | 9 | D9S1122 | 1.659 |
| 3 | 17 | D17S1290 | 1.505 |
| 4 | 18 | D18S535 | 1.410 |
| 5 | 7 | D7S2846 | 1.389 |
| 6 | 1 | D1S1612 | 1.383 |
| 7 | 1 | D1S1595 | 1.312 |
| 8 | 18 | D18S1116 | 1.245 |
| 9 | 18 | D18S976 | 1.148 |
| 10 | 9 | D9S910 | 1.117 |
| 11 | 18 | D18S59 | 1.115 |
| 12 | 6 | D6S1003 | .942 |
| 13 | 5 | D5S1354 | .918 |
| 14 | 16 | D16S686 | .837 |
| 15 | 1 | D1S1597 | .820 |
Because certain chromosomes appear among the top scores for at least three of the four statistical methods applied (table 2), and given that these statistics test different hypotheses, these results point to regions on chromosomes 1, 2, 6, 9, 10, 16, 17, and 18 as potential loci harboring AA-susceptibility genes.
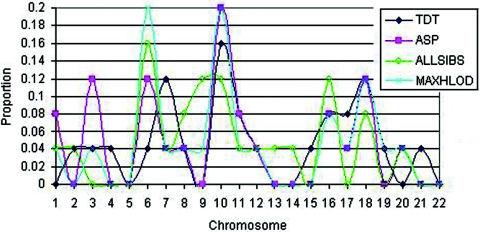
As an exploratory tool that might indicate which chromosomes harbor susceptibility loci for AA, in figure 2, we plotted the proportion of the top 25 scores for each test statistic that appear on a given chromosome. Importantly, for four chromosomes (6, 10, 16, and 18), at least three of the four test statistics have at least 8% of their top 25 scores on that chromosome. The 8% value is 4% higher than the value expected by random selection. Furthermore, if we consider a proportion of ⩾0.12, then, for three chromosomes (6, 10, and 18), three of the four tests have at least 12% of their top 25 scores on that chromosome. Thus, the results of the initial genome scan indicate several suggestive regions of linkage; the most significant LOD scores were 4.8 on chromosome 6 at marker D6S1009 and 2.3 (for a marker other than D6S1009) on chromosome 18 at marker D18S535.
Figure 2. .
Proportion of top 25 scores on a given chromosome for analysis of pedigrees with AA by use of each of the four test statistics.
We performed multipoint linkage analysis, using the MAXHLOD statistic with the initial genome scan data; however, none of the multipoint results were as significant as the two-point MAXHLOD scores. We postulate that this result stems from the fact that the marker density (10 cM) was insufficient to benefit from multipoint linkage analysis (full data not shown).
Fine Mapping
Several authors have defined criteria for declaring suggestive and significant linkage for complex traits, but there is still controversy about what thresholds should be applied and how to extend the theoretical situations on which they are based to real data sets.63–65 Considering this, we applied the following criteria to the results obtained in the genome scan, to prioritize the follow-up regions for the second stage of our study: (i) regions yielding LOD scores among the top 15 scores for two or more statistical tests, (ii) marker loci showing LOD scores values >2; and (iii) chromosomal regions “overrepresented” by having consecutive or nearly consecutive marker loci among the top 15 scores for different statistical tests. On the basis of this algorithm, and to optimize the use of DNA samples, we chose to first genotype additional marker loci in six regions on chromosomes 6, 10, 16, and 18 (see the “Material and Methods” section for details on the intervals). The HLA region was included in this second stage on the basis of the reports of genetic association of AA with different HLA alleles. Regions on chromosomes 1, 2, 9, and 17 will be analyzed in subsequent studies.
The follow-up marker density was increased from 1 marker every 10 cM (in the genome scan) to 1 every 1.82 cM, on average. For the analysis of the HLA region, we used a high-density panel of microsatellites designed by deCODE Genetics with marker loci spaced at an average distance of 0.09 cM (see the “Material and Methods” section). Results obtained after genotyping additional microsatellite marker loci are plotted in figures 3–6.
Figure 3. .
Results of the fine-mapping study for chromosome 6. A, Region 52.33–54.71 cM. B, Region 134.2–150.93 cM.
Figure 4. .
Results of the fine-mapping study for chromosome 10. A, Region 41.19–51.42 cM. B, Region 124.64–147.86 cM.
Figure 5. .
Results of the fine-mapping study for chromosome 16
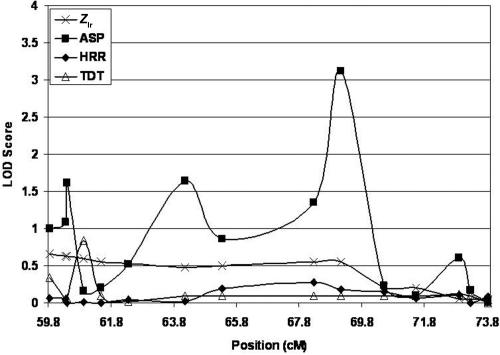
Figure 6. .
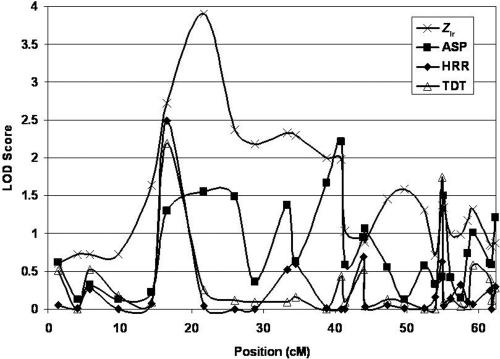
Results of the fine-mapping study for chromosome 18
Chromosome 6
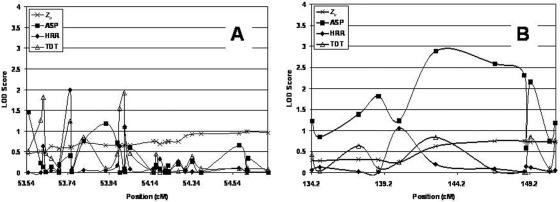
On chromosome 6, we fine mapped two regions: 52.33–54.71 cM and 134.2–150.93 cM. In figure 3A and 3B, we plotted LOD scores for both regions. The largest ASP LOD score observed on chromosome 6 in the fine-mapping data occurred at position 142.7 cM (marker D6S1009). The ASP LOD score at that marker is 2.89. This marker locus also showed the most significant ASP LOD score in the original genome scan (table 2). The decrease in significance is because of the addition of new families in the fine mapping, some of which were unlinked to this locus. It is important to note that the next most significant ASP LOD scores all occur for marker loci within ∼6 cM of D6S1009 (fig. 3B). For example, the second-most significant ASP LOD score is 2.59, for position 146.8 cM (marker D6S1569), and the third-most significant ASP LOD score is 2.31, for position 148.8 cM (marker D6S1684).
The most significant TDT LOD score of 1.9 (P=.0015) occurred at position 54.01 cM (marker D6S2889). As with the ASP LOD scores, the next five most significant TDT LOD scores all occurred within 0.5 cM of this position (fig. 3A). Finally, the most significant Zlr LOD score of 1.49 (P=.004) occurred at position 54.6 cM (marker D6S2727), within 0.6 cM of the most significant TDT LOD score. Another important observation is that the Zlr multipoint LOD scores are almost always more significant than are the ASP two-point LOD scores in the region from 52 cM to 56 cM, whereas the converse is true in the region from 134.2 cM to 150.93 cM (fig. 3A and 3B).
Consideration of the HRR statistic for these data provided additional information. In particular, marker D6S273 (position 53.75 cM) showed an HRR LOD score of 1.99 (P=.001). This score was the second largest among HRR LOD scores for all fine-mapping data. It is interesting to note that this marker is in the MHC locus, for which previous associations with AA have been reported.19,22,23,30,66
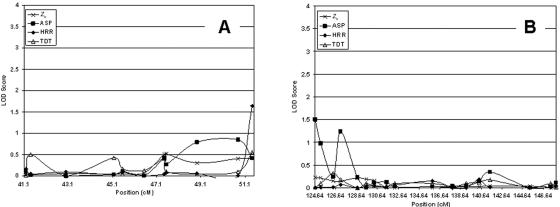
Chromosome 10
On chromosome 10, we fine mapped two regions: 41.19–51.42 cM and 124.64–147.86 cM. In figure 4A and 4B, we plotted the LOD scores for the three statistics for these two regions. Overall, the highest LOD scores occurred for three marker loci by use of the ASP statistic. Marker D10S1239, at position 124.64 cM, showed an ASP LOD score of 1.50 (P=.004). Marker D10S254, at position 127.08 cM, showed an ASP LOD score of 1.23 (P=.009). Finally, marker D10S1738, at position 125.15 cM, showed an ASP LOD score of 0.975 (P=.017). The largest Zlr LOD score (0.093; P=.064) occurred at position 47.53 cM (marker D10S1734). The largest LOD score for the TDT method was at position 51.42 cM (marker D10S2481). The largest HRR LOD at position 51.42 cM (marker D10S2481) was 1.63 (P=.003). This score was the third largest among HRR LOD scores for all fine-mapping data.
Chromosome 16
On chromosome 16, we fine mapped the region from 47 cM to 73.82 cM. The most significant result occurred for marker D16S415, at position 69.14 cM, where we observed an ASP LOD score of 3.11. The multipoint Zlr LOD score for the same marker was 0.08, indicating that the two-point and multipoint results show substantially different evidence of linkage. The TDT and HRR LOD scores were generally nonsignficant (P>.10), with the exception of that for marker D16S261 (60.925 cM), which displayed a TDT LOD score of 0.84 (P=.02).
Chromosome 18
For chromosome 18, we fine mapped the region from 1 cM to 62 cM. The most significant result occurred for marker D18S967, at position 21.74 cM, where we observed an Zlr LOD of 3.93 (fig. 6). Relative peaks for the ASP, TDT, and HRR methods also occurred near this position (fig. 6). In fact, the Zlr, TDT, and HRR LOD scores for marker D18S976 (position 16.55 cM) are in the top 10% of the distribution of LOD scores for each statistic’s distribution of fine-mapping LOD scores. It is important to note that, for chromosome 18, the multipoint LOD scores (Zlr values) increased over the two-point ASP LODs in the region from ∼10 cM to 30 cM.
Stratified Fine-Mapping Analyses
In table 3, we present results of our stratified fine-mapping analyses for only U.S. pedigrees, only Israeli pedigrees, and all pedigrees combined. As has been shown in other linkage studies,67–70 stratification of data by a certain criteria (e.g., disease diagnosis or ethnic origin) often will increase linkage information in at least one of the substrata. The results for all pedigrees combined are included in table 3 for comparison.
Table 3. .
Summary of Fine-Mapping Results for U.S., Israeli, and Combined Pedigrees
| Chromosome and Sample |
Maximum LOD | Position (cM) |
Method | Marker |
| 6: | ||||
| U.S. | 3.03 | 138.78 | ASP | D6S270 |
| Israeli | 1.42 | 142.71 | ASP | D6S1009 |
| Combined | 2.88 | 142.71 | ASP | D6S1009 |
| 10: | ||||
| U.S. | 1.88 | 41.4 | TDT | D10S1661 |
| Israeli | 1.63 | 51.42 | HRR | D10S2481 |
| Combined | 1.63 | 51.42 | HRR | D10S2481 |
| 16: | ||||
| U.S. | 1.28 | 69.14 | ASP | D16S415 |
| Israeli | 2.14 | 65.35 | ASP | D16S2623 |
| Combined | 3.11 | 69.14 | ASP | D16S415 |
| 18: | ||||
| U.S. | 3.73 | 21.74 | Zlr | D18S967 |
| Israeli | 1.38 | 26.03 | HRR | D18S1163 |
| Combined | 3.93 | 21.74 | Zlr | D18S967 |
The combined results for chromosome 6 are relatively consistent across all sets of pedigrees. That is, the position of the maximum LOD score is between 138.78 cM and 142.71 cM, and the maximum LOD score was achieved using the ASP method. The maximum LOD score increases to 3.03 when only the U.S. pedigrees are considered, compared with the maximum LOD of 2.88 for all pedigrees (see also fig. 3B).
We observed similar results for the chromosome 10 analyses. The maximum LOD score increases slightly from 1.63 for the combined set of pedigrees to 1.88 for only the U.S. pedigrees, and the position of the maximum LOD shifts from 51.42 cM (combined set) to 41.40 cM (U.S. pedigrees).
On chromosome 16, the results for the stratified sets (U.S. and Israeli pedigrees) appear additive. That is, the combined maximum LOD score of 3.11 is approximately the sum of the maximum LOD score for the U.S. set (1.28) and the maximum LOD score for Israeli set (2.14), although the position for the maximum LOD differs by ∼4 cM for the U.S. and Israeli sets.
Finally, for chromosome 18, results of the stratification suggest that the maximum LOD score of 3.93 for the combined set of pedigrees is largely from the U.S. pedigrees. The maximum LOD for the only the U.S. pedigrees is 3.73 at the same position (21.74 cM).
Conclusions from the Fine-Mapping Results
Taken together, the fine-mapping results indicated the strongest evidence of linkage on chromosome 18 in the region near 21 cM. It is worth noting that the fine-mapping analyses of chromosome 18 with additional families produced significantly larger LOD scores than those from the original genome scan, and the maximum multipoint LOD score of 3.9 is highly significant.
The next strongest evidence of linkage occurred for chromosome 6 in the HLA region. We see multiple marker loci with TDT and/or HRR LOD scores >1.5. These results are not surprising, given that population-based association methods have implicated HLA marker loci as being associated with AA. Although there are some two-point (ASP) LOD scores >3 for marker loci on chromosome 16, when the multipoint (Zlr) method is applied, these LOD scores decrease substantially.
Stratification of pedigrees by ethnic origin resulted in an increased linkage signal on chromosomes 6 and 10 for the U.S. pedigrees, no increase in the linkage signal on chromosome 16, and the observation that the maximum LOD score for chromosome 18 appears to be from the U.S. pedigrees. As a methodologic note, the TDT and HRR methods provided highly correlated results, and, in fact, the correlation among the TDT and HRR P values (−log transformed) was 0.32 (P<.001 for test of correlation).
Discussion
The mapping of complex disorders has only recently begun to yield successes in gene identification. The identification of susceptibility genes for psoriasis and atopic dermatitis are among the most significant results obtained to date for complex diseases in dermatology.71–76 One of the first significant findings in complex-disease mapping was the identification of alleles predisposing to Crohn disease (MIM #266600) in the NOD2 gene (MIM *605956).77,78 More recently, an allele of the complement factor H gene (CFH [MIM *134370]) was significantly associated with age-related macular degeneration (MIM #153800).79–82 In all examples, the genetic location of the disease susceptibility locus was first identified by performing genome scans on families.71,75,83 Subsequent to the linkage analysis, association studies were used to determine the particular alleles that confer disease susceptibility. Although many studies of complex diseases have focused on the collection of individual cases or sib-pair samples because of the difficulty of finding a significant number of pedigrees segregating the disease, these types of studies are more vulnerable to the effects of genetic heterogeneity and the polygenic nature of such diseases. Therefore, the identification of novel disease loci has traditionally been accomplished with family-based linkage studies. For this reason, we chose to approach this first genomewide susceptibility search of AA by using a collection of pedigrees with multiple affected individuals. Furthermore, 14 of the pedigrees originate from Israel, increasing the probability that they share underlying genetic factors.
The statistical tests used in this study were chosen because they all are genetic model-free tests,52,53 in the sense that (with the exception of MAXHLOD) they do not require a specification of the genetic-model parameters (penetrance and disease-allele frequency). MAXHLOD is a parametric linkage analysis in which the LOD score is calculated under autosomal dominant and recessive patterns of inheritance and penetrance values of 50% and 80% (table 1). It has been shown that it is at least as powerful in localizing disease loci as tests like ASP and ALLSIBS54 and is, under certain circumstances, a more precise indicator of the location of a disease locus than are statistics like ASP and ALLSIBS.55 Several authors have used parametric linkage analyses to look for susceptibility loci contributing to complex traits, since, when calculated under both dominant and recessive models even though they do not describe the pattern of inheritance of such diseases, these methods can be a powerful way to localize susceptibility loci.84 Furthermore, these methods can be useful in a sample like ours, where many of the affected pairs are not siblings.
TDT was chosen because it has been shown that it may be more powerful than linkage tests (MAXHLOD, ASP, ALLSIBS), when there is linkage and linkage disequilibrium between a disease and marker locus. Some of the families with AA collected in Israel are of Ashkenazi and Sephardic Jewish extraction. These are considered genetically isolated populations, and the extent of linkage disequilibrium is therefore thought to extend over larger regions of the human genome.85,86 Overall, the strategy used to identify chromosomes of interest for follow-up is similar to exploratory methods employed by other researchers analyzing genome-scan data for complex traits.87 Previous authors subdivided the genome into chromosomal bins, whereas, in this work, we examine whole chromosomes at a time. We consider the whole chromosome as the unit of measure because it has been observed, in simulated data sets, that the methods we employ have better power to determine the correct chromosome than to determine a particular subregion of a chromosome harboring a disease-susceptibility locus.56
In this study, we did not specifically model sporadic cases, where the probability of being affected given two copies of the wild-type allele at the disease locus, Pr(aff/++), could carry one or two copies of mutated allele; that is, Pr(aff/++) is perhaps nonzero. However, our previous research55 suggests that the effect of sporadic cases can be modeled by locus heterogeneity. Therefore, we used the MAXHLOD statistic rather than the MAXLOD statistic.
It has been suspected for many years that an autoimmune pathogenesis underlies AA, yet firm evidence of an autoantigen is lacking.11–16 The results of association studies with HLA in AA suggest a role,12 although perhaps it explains only a part of the genetic susceptibility to AA. When compared with the four AA-susceptibility loci identified in the C3H-HeJ mouse model,33 our results overlap at the HLA locus. In our data sets, the two marker loci in the vicinity of the HLA locus yield MAXHLOD values of 1.034 (D6S1281) and 1.707 (D6S2427). The other three loci in the mouse (on mouse chromosomes 8, 9, and 15) correspond to regions on 10 different human chromosomes (4, 5, 8, 11, 12, 13, 16, 17, 19, and 22). Although the majority of these loci do not correspond to our findings, the region on chromosome 8 does correspond to part of the region that we fine mapped on human chromosome 16. Since this study represents a first attempt to identify genetic factors contributing to AA in humans, our results should be replicated in an independent population. It is possible that new loci will be identified once the sample is extended and further suggestive chromosomal regions are tested, such as those on chromosomes 1, 2, 9, and 17.
Several studies published elsewhere have reported the association of AA with various genes, including MX1 and AIRE on chromosome 21 and PTPN22 on chromosome 1. None of these regions were implicated in our study.
We noted two significant peaks of linkage on chromosome 6, one on each arm. The main peak was on 6q (ASP LOD score 2.89), outside the HLA region. The HLA region on 6p also showed suggestive linkage (TDT and HRR LOD score of 1.9), which is consistent with previous reports.19,22,23,30,66
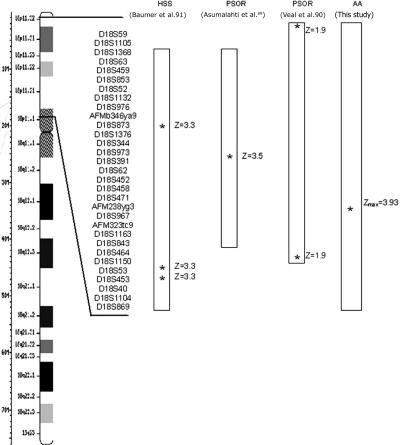
Two of the regions identified in this study have previously been implicated in conferring susceptibility to psoriasis. The region on chromosome 16 was identified in a genome scan performed on patients with psoriasis (Zmax=2.31)88 and also overlaps with a region near a Crohn disease–susceptibility locus.83 On chromosome 18, we observed consistent linkage peaks >3.0 (Zmax=3.93). This same region of chromosome 18p also contains a psoriasis-susceptibility region that was identified independently in families from the United Kingdom (Z=1.97) and Finland (Z=3.58).89,90 There has also been a reported association between deletion of chromosome 18p and psoriasis vulgaris (fig. 7).66 Interestingly, this same area of chromosome 18 has also been implicated by linkage in autosomal dominant hereditary hypotrichosis simplex (Z=3.31), although no causative gene has been identified yet.91 Taken together, these lines of evidence suggest that a gene(s) on chromosome 18p is linked to AA and may also be involved in other inherited skin and hair disorders.
Figure 7. .
Comparison of chromosome 18 results with published studies.90,91 Boxes represent regions that showed linkage. Asterisks (*) indicate marker loci with the highest linkage scores. HSS = hereditary hypotrichosis simplex; PSOR = psoriasis.
Understanding the genetic factors underlying AA may help to elucidate the mechanisms responsible for the disease, as well as to define at-risk individuals, to improve the disease prognosis, and to determine its response to environmental triggers; eventually, it could lead to the design of new treatment strategies. Initially, the genes in AA could provide targets for transgenic and knockout mice, which would be essential for in vivo testing of new therapies. Ultimately, it is anticipated that discovery and modulation of the genes in AA will provide novel therapeutic targets for this psychologically devastating disorder.
Acknowledgments
We thank all participating families for their contribution to this study. We are deeply appreciative of the constant support and encouragement of the National Alopecia Areata Foundation (NAAF), and particularly Vicki Kalabokes, NAAF Executive Director. We are also grateful to the National Alopecia Areata Registry for its invaluable contributions and HaMut Lam for her expert technical assistance. This work was supported in part by grants from the NAAF (to A.M.C. and A.Z.), Israeli Alopecia Areata Fund (to A.Z.), Columbia University Clinical Trials Office Pilot Award (to A.M.-M.), the North American Hair Research Society Mentorship Award (to A.M.-M.), and the National Institutes of Health: National Institute of Arthritis and Musculoskeletal and Skin Diseases grants N01AR02249 (AA Registry [to M.D.]), R03AR050158 (to A.M-M. and J.M.), and R01AR52579 (to A.M.C.) and National Institute of Mental Health grant R01MH44292 (to J.O.).
Web Resources
The URLs for data presented herein are as follows:
- National Alopecia Areata Foundation, http://www.naaf.org/
- National Alopecia Areata Registry, http://www.mdanderson.org/departments/alopecia/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for AA, HLA alleles, IL1, MX1, MICA, PTPN22, IL1RN, IL1F5, Down syndrome, ASP1, AIRE, Crohn disease, NOD2, CFH, and age-related macular degeneration)
References
- 1.Gilhar A, Kalish RS (2006) Alopecia areata: a tissue specific autoimmune disease of the hair follicle. Autoimmun Rev 5:64–69 10.1016/j.autrev.2005.07.001 [DOI] [PubMed] [Google Scholar]
- 2.Green J, Sinclair RD (2000) Genetics of alopecia areata. Australas J Dermatol 41:213–218 10.1046/j.1440-0960.2000.00439.x [DOI] [PubMed] [Google Scholar]
- 3.McDonagh AJ, Tazi-Ahnini R (2002) Epidemiology and genetics of alopecia areata. Clin Exp Dermatol 27:405–409 10.1046/j.1365-2230.2002.01077.x [DOI] [PubMed] [Google Scholar]
- 4.Bahmer FA (2002) The phenomen of sudden graying of the hair in the poem “Die Fusse im Feuer” by Conrad Ferdinand Meyer [in German]. Hautarzt 53:492–494 10.1007/s00105-002-0375-3 [DOI] [PubMed] [Google Scholar]
- 5.Barlag K, Ruzicka T (1995) Sudden graying or whitening of the scalp hair [in German]. Dtsch Med Wochenschr 120:158–159 [PubMed] [Google Scholar]
- 6.Goldenhersh MA (1992) Rapid whitening of the hair first reported in the Talmud: possible mechanisms of this intriguing phenomenon. Am J Dermatopathol 14:367–368 [DOI] [PubMed] [Google Scholar]
- 7.Plinck EP, Peereboom-Wynia JD, Vuzevski VD, Westerhof W, Stolz E (1993) Turning white overnight, is it possible? [in Dutch]. Ned Tijdschr Geneeskd 137:1207–1210 [PubMed] [Google Scholar]
- 8.Cash TF (2001) The psychology of hair loss and its implications for patient care. Clin Dermatol 19:161–166 10.1016/S0738-081X(00)00127-9 [DOI] [PubMed] [Google Scholar]
- 9.Gulec AT, Tanriverdi N, Duru C, Saray Y, Akcali C (2004) The role of psychological factors in alopecia areata and the impact of the disease on the quality of life. Int J Dermatol 43:352–356 10.1111/j.1365-4632.2004.02028.x [DOI] [PubMed] [Google Scholar]
- 10.Maffei C, Fossati A, Rinaldi F, Riva E (1994) Personality disorders and psychopathologic symptoms in patients with androgenetic alopecia. Arch Dermatol 130:868–872 10.1001/archderm.130.7.868 [DOI] [PubMed] [Google Scholar]
- 11.Khoury EL, Price VH, Greenspan JS (1988) HLA-DR expression by hair follicle keratinocytes in alopecia areata: evidence that it is secondary to the lymphoid infiltration. J Invest Dermatol 90:193–200 10.1111/1523-1747.ep12462213 [DOI] [PubMed] [Google Scholar]
- 12.Welsh EA, Clark HH, Epstein SZ, Reveille JD, Duvic M (1994) Human leukocyte antigen-DQB1*03 alleles are associated with alopecia areata. J Invest Dermatol 103:758–763 10.1111/1523-1747.ep12412584 [DOI] [PubMed] [Google Scholar]
- 13.Sundberg JP, Oliver RF, McElwee KJ, King LE Jr (1995) Alopecia areata in humans and other mammalian species. J Invest Dermatol 104:32S–33S 10.1111/1523-1747.ep12613464 [DOI] [PubMed] [Google Scholar]
- 14.Tobin DJ, Hann SK, Song MS, Bystryn JC (1997) Hair follicle structures targeted by antibodies in patients with alopecia areata. Arch Dermatol 133:57–61 10.1001/archderm.133.1.57 [DOI] [PubMed] [Google Scholar]
- 15.Tobin DJ, Sundberg JP, King LE Jr, Boggess D, Bystryn JC (1997) Autoantibodies to hair follicles in C3H/HeJ mice with alopecia areata-like hair loss. J Invest Dermatol 109:329–333 10.1111/1523-1747.ep12335848 [DOI] [PubMed] [Google Scholar]
- 16.Gilhar A, Ullmann Y, Berkutzki T, Assy B, Kalish RS (1998) Autoimmune hair loss (alopecia areata) transferred by T lymphocytes to human scalp explants on SCID mice. J Clin Invest 101:62–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sawaya ME, Hordinsky MK (1992) Advances in alopecia areata and androgenetic alopecia. Adv Dermatol 7:211–226 [PubMed] [Google Scholar]
- 18.Schwartz RA, Janniger CK (1997) Alopecia areata. Cutis 59:238–241 [PubMed] [Google Scholar]
- 19.Jackow C, Puffer N, Hordinsky M, Nelson J, Tarrand J, Duvic M (1998) Alopecia areata and cytomegalovirus infection in twins: genes versus environment? J Am Acad Dermatol 38:418–425 10.1016/S0190-9622(98)70499-2 [DOI] [PubMed] [Google Scholar]
- 20.van der Steen P, Traupe H, Happle R, Boezeman J, Strater R, Hamm H (1992) The genetic risk for alopecia areata in first degree relatives of severely affected patients: an estimate. Acta Derm Venereol 72:373–375 [PubMed] [Google Scholar]
- 21.Morling N, Frentz G, Fugger L, Georgsen J, Jakobsen B, Odum N, Svejgaard A (1991) DNA polymorphism of HLA class II genes in alopecia areata. Dis Markers 9:35–42 [PubMed] [Google Scholar]
- 22.de Andrade M, Jackow CM, Dahm N, Hordinsky M, Reveille JD, Duvic M (1999) Alopecia areata in families: association with the HLA locus. J Investig Dermatol Symp Proc 4:220–223 [DOI] [PubMed] [Google Scholar]
- 23.Barahmani N, de Andrade M, Slusser JP, Zhang Q, Duvic M (2006) Major histocompatibility complex class I chain-related gene A polymorphisms and extended haplotypes are associated with familial alopecia areata. J Invest Dermatol 126:74–78 10.1038/sj.jid.5700009 [DOI] [PubMed] [Google Scholar]
- 24.Kemp EH, McDonagh AJ, Wengraf DA, Messenger AG, Gawkrodger DJ, Cork MJ, Tazi-Ahnini R (2006) The non-synonymous C1858T substitution in the PTPN22 gene is associated with susceptibility to the severe forms of alopecia areata. Hum Immunol 67:535–539 10.1016/j.humimm.2006.04.006 [DOI] [PubMed] [Google Scholar]
- 25.Gilhar A, Shalaginov R, Assy B, Serafimovich S, Kalish RS (1999) Alopecia areata is a T-lymphocyte mediated autoimmune disease: lesional human T-lymphocytes transfer alopecia areata to human skin grafts on SCID mice. J Investig Dermatol Symp Proc 4:207–210 [DOI] [PubMed] [Google Scholar]
- 26.Tarlow JK, Clay FE, Cork MJ, Blakemore AI, McDonagh AJ, Messenger AG, Duff GW (1994) Severity of alopecia areata is associated with a polymorphism in the interleukin-1 receptor antagonist gene. J Invest Dermatol 103:387–390 10.1111/1523-1747.ep12395398 [DOI] [PubMed] [Google Scholar]
- 27.Barahmani N, de Andrade M, Slusser J, Zhang Q, Duvic M (2002) Interleukin-1 receptor antagonist allele 2 and familial alopecia areata. J Invest Dermatol 118:335–337 10.1046/j.0022-202x.2001.01676.x [DOI] [PubMed] [Google Scholar]
- 28.Tazi-Ahnini R, Cox A, McDonagh AJ, Nicklin MJ, di Giovine FS, Timms JM, Messenger AG, Dimitropoulou P, Duff GW, Cork MJ (2002) Genetic analysis of the interleukin-1 receptor antagonist and its homologue IL-1L1 in alopecia areata: strong severity association and possible gene interaction. Eur J Immunogenet 29:25–30 10.1046/j.1365-2370.2002.00271.x [DOI] [PubMed] [Google Scholar]
- 29.Du Vivier A, Munro DD (1975) Alopecia areata, autoimmunity, and Down’s syndrome. Br Med J 1:191–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tazi-Ahnini R, di Giovine FS, McDonagh AJ, Messenger AG, Amadou C, Cox A, Duff GW, Cork MJ (2000) Structure and polymorphism of the human gene for the interferon-induced p78 protein (MX1): evidence of association with alopecia areata in the Down syndrome region. Hum Genet 106:639–645 10.1007/s004390050037 [DOI] [PubMed] [Google Scholar]
- 31.Betterle C, Greggio NA, Volpato M (1998) Clinical review 93: autoimmune polyglandular syndrome type 1. J Clin Endocrinol Metab 83:1049–1055 10.1210/jc.83.4.1049 [DOI] [PubMed] [Google Scholar]
- 32.Zlotogorski A, Weinrauch L, Brautbar C (1990) Familial alopecia areata: no linkage with HLA. Tissue Antigens 36:40–41 [DOI] [PubMed] [Google Scholar]
- 33.Sundberg JP, Silva KA, Li R, Cox GA, King LE (2004) Adult-onset alopecia areata is a complex polygenic trait in the C3H/HeJ mouse model. J Invest Dermatol 123:294–297 10.1111/j.0022-202X.2004.23222.x [DOI] [PubMed] [Google Scholar]
- 34.Aita VM, Liu J, Knowles JA, Terwilliger JD, Baltazar R, Grunn A, Loth JE, Kanyas K, Lerer B, Endicott J, et al (1999) A comprehensive linkage analysis of chromosome 21q22 supports prior evidence for a putative bipolar affective disorder locus. Am J Hum Genet 64:210–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J, Juo SH, Holopainen P, Terwilliger J, Tong X, Grunn A, Brito M, Green P, Mustalahti K, Maki M, et al (2002) Genomewide linkage analysis of celiac disease in Finnish families. Am J Hum Genet 70:51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Nyholt DR, Magnussen P, Parano E, Pavone P, Geschwind D, Lord C, Iversen P, Hoh J, Ott J, et al (2001) A genomewide screen for autism susceptibility loci. Am J Hum Genet 69:327–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Magnuson VL, Ally DS, Nylund SJ, Karanjawala ZE, Rayman JB, Knapp JI, Lowe AL, Ghosh S, Collins FS (1996) Substrate nucleotide-determined non-templated addition of adenine by Taq DNA polymerase: implications for PCR-based genotyping and cloning. Biotechniques 21:700–709 [DOI] [PubMed] [Google Scholar]
- 38.Adams P (1994) LABMAN and LINKMAN: a data management system specifically designed for genome searches of complex diseases. Genet Epidemiol 11:87–98 10.1002/gepi.1370110109 [DOI] [PubMed] [Google Scholar]
- 39.O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukhopadhyay N, Almasy L, Schroeder M, Mulvihill WP, Weeks DE (2005) Mega2: data-handling for facilitating genetic linkage and association analyses. Bioinformatics 21:2556–2557 10.1093/bioinformatics/bti364 [DOI] [PubMed] [Google Scholar]
- 41.Fjalldal JB, Sigurdsson J, Benediktsson K, Ellingssen LM (2001) Automated genotyping: combining neural networks and decision trees to perform robust allele calling. Proc Int Joint Conf Neural Netw A1–A6 [Google Scholar]
- 42.Palsson B, Palsson F, Perlin M, Gudbjartsson H, Stefansson K, Gulcher J (1999) Using quality measures to facilitate allele calling in high-throughput genotyping. Genome Res 9:1002–1012 10.1101/gr.9.10.1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith CA (1963) Testing for heterogeneity of recombination fraction values in human genetics. Ann Hum Genet 27:175–182 [DOI] [PubMed] [Google Scholar]
- 44.Ott J (1999) Analysis of human genetic linkage. The Johns Hopkins University Press, Baltimore, pp 225–226 [Google Scholar]
- 45.Terwilliger JD, Ott J (1994) Handbook of human genetic linkage. The Johns Hopkins University Press, Baltimore, pp 227–232 [Google Scholar]
- 46.Spielman RS, McGinnis RE, Ewens WJ (1993) Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet 52:506–516 [PMC free article] [PubMed] [Google Scholar]
- 47.Terwilliger JD (1995) A powerful likelihood method for the analysis of linkage disequilibrium between trait loci and one or more polymorphic marker loci. Am J Hum Genet 56:777–787 [PMC free article] [PubMed] [Google Scholar]
- 48.Documentation for ANALYZE program (http://bioweb.pasteur.fr/docs/doc-gensoft/analyze/analyze.doc)
- 49.Blackwelder WC, Elston RC (1985) A comparison of sib-pair linkage tests for disease susceptibility loci. Genet Epidemiol 2:85–97 10.1002/gepi.1370020109 [DOI] [PubMed] [Google Scholar]
- 50.Suarez BK, Van Eerdewegh P (1984) A comparison of three affected-sib-pair scoring methods to detect HLA-linked disease susceptibility genes. Am J Med Genet 18:135–146 10.1002/ajmg.1320180117 [DOI] [PubMed] [Google Scholar]
- 51.Martinez-Mir A, Zlotogorski A, Ott J, Gordon D, Christiano AM (2003) Genetic linkage studies in alopecia areata. J Investig Dermatol Symp Proc 8:199–203 10.1046/j.1087-0024.2003.00809.x [DOI] [PubMed] [Google Scholar]
- 52.Elston RC (1989) Man bites dog? The validity of maximizing lod scores to determine mode of inheritance. Am J Med Genet 34:487–488 10.1002/ajmg.1320340407 [DOI] [PubMed] [Google Scholar]
- 53.Hodge SE, Elston RC (1994) Lods, wrods, and mods: the interpretation of lod scores calculated under different models. Genet Epidemiol 11:329–342 10.1002/gepi.1370110403 [DOI] [PubMed] [Google Scholar]
- 54.Abreu PC, Greenberg DA, Hodge SE (1999) Direct power comparisons between simple LOD scores and NPL scores for linkage analysis in complex diseases. Am J Hum Genet 65:847–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Finch SJ, Chen CH, Gordon D, Mendell NR (2001) A study comparing precision of the maximum multipoint heterogeneity LOD statistic to three model-free multipoint linkage methods. Genet Epidemiol 21:315–325 10.1002/gepi.1037 [DOI] [PubMed] [Google Scholar]
- 56.Gordon D, Hoh J, Finch SJ, Levenstien MA, Edington J, Li W, Majewski J, Ott J (2001) Two approaches for consolidating results from genome scans of complex traits: selection methods and scan statistics. Genet Epidemiol Suppl 1 21:S396–S402 [DOI] [PubMed] [Google Scholar]
- 57.Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Falk CT, Rubinstein P (1987) Haplotype relative risks: an easy reliable way to construct a proper control sample for risk calculations. Ann Hum Genet 51:227–233 [DOI] [PubMed] [Google Scholar]
- 59.Ewens WJ, Spielman RS (2005) What is the significance of a significant TDT? Hum Hered 60:206–210 10.1159/000090544 [DOI] [PubMed] [Google Scholar]
- 60.Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 10.1038/ng1195-241 [DOI] [PubMed] [Google Scholar]
- 61.Hodge SE, Abreu PC, Greenberg DA (1997) Magnitude of type I error when single-locus linkage analysis is maximized over models: a simulation study. Am J Hum Genet 60:217–227 [PMC free article] [PubMed] [Google Scholar]
- 62.Hodge SE (2001) Model-free vs model-based linkage analysis: a false dichotomy? Am J Med Genet 105:62–64 [DOI] [PubMed] [Google Scholar]
- 63.Kruglyak L, Daly MJ (1998) Linkage thresholds for two-stage genome scans. Am J Hum Genet 62:994–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Altmuller J, Palmer LJ, Fischer G, Scherb H, Wjst M (2001) Genomewide scans of complex human diseases: true linkage is hard to find. Am J Hum Genet 69:936–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Page GP, George V, Go RC, Page PZ, Allison DB (2003) “Are we there yet?” Deciding when one has demonstrated specific genetic causation in complex diseases and quantitative traits. Am J Hum Genet 73:711–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mikelsaar RV, Muru K, Kulla A, Suvari A (2002) Psoriasis vulgaris in a male with partial deletion 18p. Am J Med Genet 108:252–253 10.1002/ajmg.10259 [DOI] [PubMed] [Google Scholar]
- 67.Bartlett CW, Goedken R, Vieland VJ (2005) Effects of updating linkage evidence across subsets of data: reanalysis of the autism genetic resource exchange data set. Am J Hum Genet 76:688–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brant SR, Shugart YY (2004) Inflammatory bowel disease gene hunting by linkage analysis: rationale, methodology, and present status of the field. Inflamm Bowel Dis 10:300–311 10.1097/00054725-200405000-00019 [DOI] [PubMed] [Google Scholar]
- 69.Gordon D, Haynes C, Finch SJ, Brown AM (2006) Increase in linkage information by stratification of pedigree data into gold-standard and standard diagnoses: application to the NIMH Alzheimer Disease Genetics Initiative Dataset. Hum Hered 61:97–103 10.1159/000093303 [DOI] [PubMed] [Google Scholar]
- 70.Leal SM, Ott J (2000) Effects of stratification in the analysis of affected-sib-pair data: benefits and costs. Am J Hum Genet 66:567–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bhalerao J, Bowcock AM (1998) The genetics of psoriasis: a complex disorder of the skin and immune system. Hum Mol Genet 7:1537–1545 10.1093/hmg/7.10.1537 [DOI] [PubMed] [Google Scholar]
- 72.Bradley M, Soderhall C, Luthman H, Wahlgren CF, Kockum I, Nordenskjold M (2002) Susceptibility loci for atopic dermatitis on chromosomes 3, 13, 15, 17, and 18 in a Swedish population. Hum Mol Genet 11:1539–1548 10.1093/hmg/11.13.1539 [DOI] [PubMed] [Google Scholar]
- 73.Helms C, Cao L, Krueger JG, Wijsman EM, Chamian F, Gordon D, Heffernan M, Daw JA, Robarge J, Ott J, et al (2003) A putative RUNX1 binding site variant between SLC9A3R1 and NAT9 is associated with susceptibility to psoriasis. Nat Genet 35:349–356 10.1038/ng1268 [DOI] [PubMed] [Google Scholar]
- 74.Helms C, Saccone NL, Cao L, Daw JA, Cao K, Hsu TM, Taillon-Miller P, Duan S, Gordon D, Pierce B, et al (2005) Localization of PSORS1 to a haplotype block harboring HLA-C and distinct from corneodesmosin and HCR. Hum Genet 118:466–476 10.1007/s00439-005-0048-2 [DOI] [PubMed] [Google Scholar]
- 75.Lee YA, Wahn U, Kehrt R, Tarani L, Businco L, Gustafsson D, Andersson F, Oranje AP, Wolkertstorfer A, v Berg A, et al (2000) A major susceptibility locus for atopic dermatitis maps to chromosome 3q21. Nat Genet 26:470–473 10.1038/82625 [DOI] [PubMed] [Google Scholar]
- 76.Soderhall C, Bradley M, Kockum I, Wahlgren CF, Luthman H, Nordenskjold M (2001) Linkage and association to candidate regions in Swedish atopic dermatitis families. Hum Genet 109:129–135 10.1007/s004390100556 [DOI] [PubMed] [Google Scholar]
- 77.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, et al (2001) Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411:599–603 10.1038/35079107 [DOI] [PubMed] [Google Scholar]
- 78.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, et al (2001) A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 411:603–606 10.1038/35079114 [DOI] [PubMed] [Google Scholar]
- 79.Edwards AO, Ritter R 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA (2005) Complement factor H polymorphism and age-related macular degeneration. Science 308:421–424 10.1126/science.1110189 [DOI] [PubMed] [Google Scholar]
- 80.Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, et al (2005) Complement factor H variant increases the risk of age-related macular degeneration. Science 308:419–421 10.1126/science.1110359 [DOI] [PubMed] [Google Scholar]
- 81.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, et al (2005) Complement factor H polymorphism in age-related macular degeneration. Science 308:385–389 10.1126/science.1109557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wiggs JL (2006) Complement factor H and macular degeneration: the genome yields an important clue. Arch Ophthalmol 124:577–578 10.1001/archopht.124.4.577 [DOI] [PubMed] [Google Scholar]
- 83.Hugot JP, Laurent-Puig P, Gower-Rousseau C, Olson JM, Lee JC, Beaugerie L, Naom I, Dupas JL, Van Gossum A, Orholm M, et al (1996) Mapping of a susceptibility locus for Crohn’s disease on chromosome 16. Nature 379:821–823 10.1038/379821a0 [DOI] [PubMed] [Google Scholar]
- 84.Brzustowicz LM, Hodgkinson KA, Chow EW, Honer WG, Bassett AS (2000) Location of a major susceptibility locus for familial schizophrenia on chromosome 1q21-q22. Science 288:678–682 10.1126/science.288.5466.678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gordon D, Simonic I, Ott J (2000) Significant evidence for linkage disequilibrium over a 5-cM region among Afrikaners. Genomics 66:87–92 10.1006/geno.2000.6190 [DOI] [PubMed] [Google Scholar]
- 86.Ostrer H (2001) A genetic profile of contemporary Jewish populations. Nat Rev Genet 2:891–898 10.1038/35098506 [DOI] [PubMed] [Google Scholar]
- 87.Wise LH, Lewis CM (1999) A method for meta-analysis of genome searches: application to simulated data. Genet Epidemiol Suppl 1 17:S767–S771 [DOI] [PubMed] [Google Scholar]
- 88.Nair RP, Henseler T, Jenisch S, Stuart P, Bichakjian CK, Lenk W, Westphal E, Guo SW, Christophers E, Voorhees JJ, et al (1997) Evidence for two psoriasis susceptibility loci (HLA and 17q) and two novel candidate regions (16q and 20p) by genome-wide scan. Hum Mol Genet 6:1349–1356 10.1093/hmg/6.8.1349 [DOI] [PubMed] [Google Scholar]
- 89.Asumalahti K, Laitinen T, Lahermo P, Suomela S, Itkonen-Vatjus R, Jansen C, Karvonen J, Karvonen SL, Reunala T, Snellman E, et al (2003) Psoriasis susceptibility locus on 18p revealed by genome scan in Finnish families not associated with PSORS1. J Invest Dermatol 121:735–740 10.1046/j.1523-1747.2003.12483.x [DOI] [PubMed] [Google Scholar]
- 90.Veal CD, Clough RL, Barber RC, Mason S, Tillman D, Ferry B, Jones AB, Ameen M, Balendran N, Powis SH, et al (2001) Identification of a novel psoriasis susceptibility locus at 1p and evidence of epistasis between PSORS1 and candidate loci. J Med Genet 38:7–13 10.1136/jmg.38.1.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baumer A, Belli S, Trueb RM, Schinzel A (2000) An autosomal dominant form of hereditary hypotrichosis simplex maps to 18p11.32-p11.23 in an Italian family. Eur J Hum Genet 8:443–448 10.1038/sj.ejhg.5200506 [DOI] [PubMed] [Google Scholar]