Abstract
A high level of cytogenetic expression of the rare folate-sensitive fragile site FRA12A is significantly associated with mental retardation. Here, we identify an elongated polymorphic CGG repeat as the molecular basis of FRA12A. This repeat is in the 5′ untranslated region of the gene DIP2B, which encodes a protein with a DMAP1-binding domain, which suggests a role in DNA methylation machinery. DIP2B mRNA levels were halved in two subjects with FRA12A with mental retardation in whom the repeat expansion was methylated. In two individuals without mental retardation but with an expanded and methylated repeat, DIP2B expression was reduced to approximately two-thirds of the values observed in controls. Interestingly, a carrier of an unmethylated CGG-repeat expansion showed increased levels of DIP2B mRNA, which suggests that the repeat elongation increases gene expression, as previously described for the fragile X–associated tremor/ataxia syndrome. These data suggest that deficiency of DIP2B, a brain-expressed gene, may mediate the neurocognitive problems associated with FRA12A.
The eukaryotic genome contains tissue-specific patterns of cytosines covalently modified by the addition of a methyl group.1 Of all CpG dinucleotides of the mammalian genome, 60%–90% are subject to this kind of epigenetic modification. Most of the unmethylated CpG sites are located in the promotor region of genes, where changes in their methylation status are linked to long-term changes in expression level of the underlying genes.2 Methylated cytosines may be targeted by methyl-binding domains of transcriptional repressors—which, on binding, recruit members of chromatin-remodeling complexes—resulting in the deacetylation of core histone proteins and transcriptional silencing of the underlying gene.3
Methylation abnormalities have been linked to a broad range of diseases.2 In tumor cells, the promotor regions with a CpG island are often hypermethylated. This epigenetic change is found in every type of tumor and is associated with a loss-of-gene transcription.3 Alterations of genomic imprinting or loss of imprinting has been shown to play a pivotal role in malignant tumorigenesis; for example, see the works of Robertson2 and Muller et al.4 Methylation defects of the DNA-methylation machinery itself can also give rise to different human diseases. For instance, mutations in the DNMT3B gene are known to cause human immunodeficiency syndrome (ICF) (immunodeficiency, centromeric heterochromatin, facial anomalies, mental retardation, prolonged respiratory infections, and infections of the skin and digestive system) because of hypomethylation of the pericentromeric satellite DNA, which causes centromeric decondensation and chromosomal rearrangements.5 In addition, faithful maintenance of the DNA-methylation pattern and the enzymes that bind methylated CpG residues are essential for proper functioning and survival of the CNS.6–8
Excessive DNA methylation may also be responsible for gene silencing at rare folate-sensitive fragile sites. Fragile sites are breakage-prone spots in the DNA that express as gaps and breaks in chromosomes when cells are grown in vitro under specific cell-culture conditions. Rare fragile sites are further subdivided according to specific culture conditions by which they are induced. Six folate-sensitive, rare fragile sites have been characterized at the molecular level—namely, FRAXA,9 FRAXE,10 FRAXF,11 FRA11B,12 FRA16A,13 and, more recently, FRA10A.14 All six were shown to be associated with an expanded CGG repeat. After repeat expansion, the CGG repeat and the surrounding CpG island located in the promotor become hypermethylated. Several of the rare folate-sensitive fragile sites have been associated with cognitive dysfunction; for example, fragile X syndrome, caused by expression of the fragile site FRAXA, is characterized by mental retardation and specific dysmorphic and behavioral features. Expansion of the FRAXE repeat is associated with a mild form of mental retardation. In vivo chromosome breakage at or near the FRA11B locus has been implicated in Jacobsen syndrome, characterized by mental retardation and specific associated abnormalities.
In the present study, we have cloned the fragile site FRA12A, another rare folate-sensitive fragile site that has been linked to mental retardation (for an overview, see the work of Berg et al.15), and we have identified the associated gene. The protein product of the FRA12A-associated gene DIP2B contains a DMAP1-binding domain and might thus be involved in DNA-methylation processes.
Subjects and Methods
Clinical Diagnosis and Chromosome Analysis
Family A
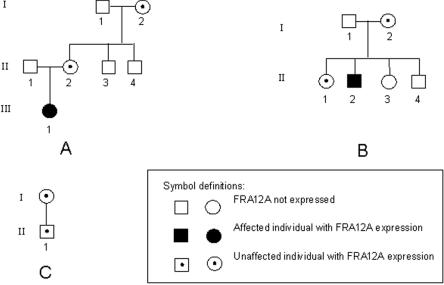
The proband (III.1; fig. 1A) was a 5-year-old girl who had global delay in her neurodevelopment during infancy, significant learning disability, subtle craniofacial dysmorphisms, and recurrent, inexplicable chest infections.15 She had bullous ichthyosiform erythroderma and areas of hyperkeratosis, particularly around the knees, elbows, wrists, and neck. Both the mother (II.2; fig. 1A) and the grandmother (I.2; fig. 1A) of the proband were phenotypically unaffected. Examination of metaphase chromosome spreads from the proband revealed a fragile site at 12q13 in 8 of the 20 metaphases examined. An identical fragile site was observed in metaphase chromosomes of the mother (4 of 20) and the grandmother (4 of 40). After receipt of informed consent, a peripheral-blood sample from the grandmother was obtained. Epstein-Barr virus (EBV)–transformed lymphoblastoid cell lines of the proband and her mother were obtained from the European Collection of Cell Cultures.
Figure 1. .
Family pedigree of FRA12A-expressing families A–C
Family B
The proband (subject II.2; fig. 1B) was a 15-year-old mentally retarded boy who had epileptic seizures and serious behavioral problems.16 A fragile site at 12q13 was seen in 20 of 50 of his metaphase chromosomes. His mother (subject I.2; fig. 1B) and sister (subject II.1; fig. 1B) showed the same fragility in 13 of 50 and 11 of 50 of their metaphase chromosomes, respectively. Yet they did not show any phenotypical abnormality. Peripheral-blood samples from the proband and his mother were obtained, with informed consent. From these peripheral-blood samples, EBV-transformed lymphoblastoid cell lines were established at our laboratory.
Family C
Subject II.1 (fig. 1C) of this family was a 35-year-old phenotypically unaffected man. The fragile site FRA12A was detected in peripheral-blood cells with a frequency of 2 of 63 metaphases in standard low-folate medium.17 When we added methotrexate and BrdU to the standard medium, frequency was increased to 11 of 50 metaphases. The addition of FudR also induced the FRA12A site in fibroblast cells, with a frequency of 8 of 77 metaphases. Peripheral-blood cells of his mother also displayed the site in standard medium (4 of 63 metaphases). Peripheral-blood samples from subject II.1 were obtained after informed consent, and we established an EBV-transformed lymphoblastoid cell line. None of the children of subject II.1 inherited the FRA12A fragile site.
FISH Mapping
Slides for FISH analysis were made from fixed cell suspensions of subject I.1, by standard methods. BAC clone RP11-831J22 (Roswell Park Cancer Institute library) was obtained from the BACPAC Resource Center. The probe was labeled with digoxigenin-11-2-deoxyuridine 5-triphosphate (dUTP) or biotin-16-dUTP (Roche) by nick translation. DNA hybridization and antibody detection were performed in accordance with standard protocols. At least five metaphases were analyzed for each hybridization, with a Zeiss Axioplan 2 fluorescence microscope equipped with a triple band-pass filter (#83000 for 4,6-diamidino-2-phenylindole, fluorescein isothiocyanate, and Texas Red [Chroma Technology]). Images were collected using a cooled CCD camera (Princeton Instruments Pentamax [Roper Scientific]) and were analyzed using IPLab software (Scanalytics).
PCR Amplification and Hybridization of the FRA12A-Associated CGG Repeat
PCR amplification of the normal allele of the FRA12A repeat–containing region was performed with the aid of 2.5×PCR× Enhancer solution (Invitrogen) with use of primers derived from the sequences flanking the repeat (DIP2B-CGG-F primer, 5′-GTCTTCAGCCTGACTGGGCTGG-3′, and DIP2B-CGG-R, primer 5′-CCGGCGACGGCTCCAGGCCTCG-3′). To amplify the expanded allele, a PCR with use of deaza-dGTP (2-deoxyguanosine 5-triphosphate) instead of common dGTP was performed as described by Sarafidou et al.14 The expanded allele could not be detected on an agarose gel. Therefore, the PCR products were electrophoresed on a 1.5% agarose gel and, after denaturation and neutralization, were transferred to Hybond N+ membranes. Hybridization was performed at 65°C with use of a 255-bp PCR CGG probe, produced using primers DIP2B-CGG-F and DIP2B-CGG-R. Triplet-repeat primed PCR was performed according to the principles described by Warner et al.,18 to detect CAG-repeat expansion. We adapted their protocol by using 2.5×PCR× Enhancer solution (Invitrogen), to be able to amplify the CGG repeat. Three different primers were added to the PCR mixture: a single forward fluorescently labeled primer (DIP2B-CGG-F) and a combination of two reverse primers (P4, 5′-TACGCATCCCAGTTTGAGACGGCCGCCGCCGCCGCCGC-3′, and P3, 5′-TACGCATCCCAGTTTGAGACG-3′) in a 1:10 ratio. The reverse primer P4 anneals at different sites of the CGG repeat, which produces PCR products of different lengths that differ from each other by a multiple of three residues. After depletion of the P4 primer, the P3 primer takes over and amplifies the PCR products of different lengths. PCR products were size fractionated on a Prism ABI 3100 DNA sequencer (Applied Biosystems). The methylation status of the CGG repeat and the surrounding region was analyzed by bisulphite sequencing. Genomic DNA was bisulphite treated using the EZ DNA methylation kit (Baseclear). Bisulphite treatment converts all nonmethylated cytosines into thymines, whereas methylated cytosines remain unchanged. Primers specific for the methylated bisulphite–converted DNA were developed (DIP2B-FM, 5′-GATAGATATATATTGTTAAGTTTAG-3′, and DIP2B-RM, 5′-CCGCCATAAACAAAAATCACGTAAACG-3′). After PCR amplification, the CGG surrounding area was sequenced using primers DIP2B-FM and DIP2B-RM on a Prism ABI 3100 DNA sequencer (Applied Biosystems).
Gene-Expression Analysis
We used a multiple-tissue northern blot (FirstChoice Northern Human Blot I [Ambion]) to determine the size of the DIP2B transcript. The specific DIP2B probe was an 855-bp cDNA PCR product (forward primer, 5′-CCGCAGACACAAGAAACTGA-3′, located at the exon 2–exon 3 boundary; reverse primer, 5′-AGCAGGAGGCCAGTTACAGA-3′, located in exon 8) and was radiolabeled by the addition of [α-32P]dATP and [α-32P]dCTP (MP). Hybridization was performed according to the manufacturer’s instructions. Detailed information on the expression pattern of the DIP2B gene was obtained from a multiple tissue expression (MTE) array (BD Biosciences) that was hybridized to the radiolabeled 855-bp DIP2B cDNA probe. Hybridization was performed according to the manufacturer’s instructions.
The European Bioinformatics Institute (EBI) database was searched for coding SNPs (cSNPs) within the coding sequence of the DIP2B gene. The presence of cSNPs in the FRA12A carriers and patients was tested by sequencing at the genomic level. RNA was isolated for EBV-transformed lymphoblastoid cells of the FRA12A carriers through use of Trizol (Invitrogen) and was converted to cDNA through use of Superscript III reverse transcriptase (Invitrogen). cDNA was tested for the presence of cSNPs in FRA12A carriers and patients by PCR followed by sequencing. The following primer sets were used: for exon 16, 5′-GATGCACACAATCAGCGTAC-3′ (forward) and 5′-TCAGGCTTCAGTCCATGACT-3′ (reverse); for exon 20, 5′-AATCTGTGTTAGCTCCAGAACT-3′ (forward) and 5′-TCTACAGCCAATCCAGTAGC-3′ (reverse); for exon 24, 5′-CTCCACTAGGAGGAATCCAT-3′ (forward) and 5′-ACTGTAGGATCTCTGCCAGA-3′ (reverse); and, for exon 37, 5′-AGCGTCATGATGCATTGTATG-3′ (forward) and 5′-CCAACGATGAGGTAATGCTC-3′ (reverse). Big Dye Terminator sequencing was performed, and sequencing products were analyzed using an ABI 3100 (Applied Biosystems) automated sequencer.
Real-time quantitative PCR was performed using gene-specific Assays-on-Demand containing two PCR primers (forward and reverse) and a TaqMan MGB probe (FAM dye labeled), obtained from Applied Biosystems. Seven independent control individuals were included in the first experiment, and five control individuals in the second experiment. PCR products were synthesized from RNA samples first converted to cDNA, through use of the qPCR Mastermix Plus without UNG (Eurogentec). The reactions were run on an ABI Prism 7000 sequence-detection system. The thermal cycler conditions were 2 min at 50°C and 10 min at 95°C, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. The raw fluorescence data were analyzed using the Sequence Detection Software (SDS [Applied Biosystems]). The SDS results were subsequently exported as comma-separated value files and were imported into the relative quantification software qBase for further analysis. For normalization of the quantitative expression values, the geometric mean of three housekeeping genes (GAPD, UBC, and YWHAZ)19 were used. The final results were checked by the nonparametrical Mann-Whitney U test.
Sequencing the DIP2B Gene
All 38 exons of the DIP2B gene were PCR amplified at the genomic level. All primers used for mutation analysis of the DIP2B gene can be obtained from the authors on request. Big-Dye Sequencing products were analyzed on an ABI 3100 (Applied Biosystems) automated sequencer.
Results
Cytogenetic Expression of FRA12A and Likelihood of Mental Retardation
The percentage of cells expressing FRA12A on cytogenetic analysis was available for 41 individuals from 16 published families (table 1). In seven of the individuals with significant neurocognitive problems, the average level of expression was 43.7% (95% CI 33.6%–54.8%). In two of these individuals, the FRA12A expression was de novo; for three individuals, information about the parents was not available; and the remaining two individuals with mental retardation inherited the fragile site maternally, and approximately half of the cells of the unaffected mother expressed FRA12A. The average level of FRA12A expression in the individuals with FRA12A but without mental retardation was 16.6% (95% CI 14.3%–19.0%). Thus, the percentage of cytogenetic expression is significantly associated with the clinical phenotype of the patients.
Table 1. .
Overview of FRA12A Expression of 41 Individuals from 16 Published Families
| Family and Subject |
Sex | MR/Delaya | Percentage of Cells with FRA12A Expression |
Phenotype and Remarks | Reference (Year) |
| 1: | Giraud et al.20 (1976) | ||||
| 1 | F | N | Mother of baby with lethal arthrogryposis, baby had no evidence of FRA12A (subject 9) | ||
| 2: | Giraud et al.20 (1976) | ||||
| 2 | M | Y | Male with MR and multiple congenital abnormalities (not detailed) (subject 10) | ||
| 3: | Stetten et al.21 (1988) | ||||
| 3 | F | N | Woman, aged 38 years, with two miscarriages | ||
| 4: | Smeets et al.16 (1985) | ||||
| 4 | M | Y | 40 | Male with MR (40% of cells), unaffected sister (22% of cells), and unaffected mother (26% cells) | |
| 5 | F | N | 22 | Unaffected sister of subject 4 | |
| 6 | F | N | 26 | Unaffected mother of subject 4 | |
| 5: | Moric-Petrovic and Laca22 (1984) | ||||
| 7 | M | Y | Male with MR, hypotonia, cryptorchidism, and micropenis; neither parent had fragile site | ||
| 6: | Donti et al.23 (1979) | ||||
| 8 | F | Y | 30 | Female with MR, hypotonia, facial dysmorphisms, arachnodactyly, intracranial calcification; neither parent had fragile site | |
| 7: | Mules et al.17 (1983) | ||||
| 9 | M | N | Incidental finding in the father of child with ring chromosome 13 | ||
| 10 | F | N | 8 | Mother of subject 9; low-folate culture failed, 4 of 63 FRA12A seen on standard-culture cultured fibroblasts 2 of 63 in subject 9 | |
| 8: | Sutherland and Hecht24 (1985) | ||||
| 11 | M | N | 13 | Phenotypically unaffected male neonate | |
| 12 | F | N | 14 | Phenotypically unaffected mother of subject 11 | |
| 13 | F | N | 24 | Phenotypically unaffected maternal grandmother of subject 11 | |
| 14 | M | N | 12 | Phenotypically unaffected maternal great-grandfather of subject 11 | |
| 9: | Sutherland and Hinton25 (1981) | ||||
| 15 | M | N | 16 | Found in family with autosomal dominant epidermylosis bullosa simplex (EBS), unaffected | |
| 16 | M | N | 16 | Brother of subject 15; unaffected with EBS | |
| 17 | F | N | 16 | Sister of subject 15; affected with EBS | |
| 18 | F | N | 28 | Mother of subject 15; affected with EBS | |
| 19 | F | N | 12 | Maternal aunt of subject 15 | |
| 20 | F | N | 18 | Maternal first cousin of subject 15 | |
| 21 | M | N | 30 | Maternal first cousin of subject 15 | |
| 22 | F | N | 14 | Maternal aunt of subject 15; unaffected with EBS | |
| 23 | F | N | 11 | Maternal first cousin of subject 15 | |
| 24 | F | N | 12 | Maternal grandmother of subject 15; affected with EBS | |
| 25 | M | N | 8 | Maternal great-uncle of subject 15 | |
| 10: | Kumar et al.26 (1986) | ||||
| 26 | F | N | 30 | Probably incidental finding in a woman with craniofrontonasal dysplasia | |
| 27 | F | N | 27 | Unaffected sister of subject 26 | |
| 28 | F | N | 20 | Mother of subject 26; also affected with craniofrontonasal dysplasia | |
| 11: | Webb et al.27 (1987) | ||||
| 29 | F | Y | Female with mild nondysmorphic MR; parents not available | ||
| 12: | Guichaoua et al.28 (1982) | ||||
| 30 | F | N | Mother of an infant with multiple malformations | ||
| 13: | Guichaoua et al.28 (1982) | ||||
| 31 | M | Y | Male, aged 7 years, with dysmorphic features | ||
| 14: | Giardino29 (1990) | ||||
| 32 | M | N | 9 | Male whose wife had multiple miscarriages | |
| 33 | F | N | 14 | Unaffected daughter of subject 32 | |
| 34 | F | N | 16 | Unaffected daughter of subject 32 | |
| 15: | Kahkonen30 (1989) | ||||
| 35 | F | N | 25 | Unaffected female; coincidental finding on investigation of familial t(X;5) | |
| 36 | F | N | 12 | Unaffected mother of subject 35 | |
| 37 | F | N | 18 | Unaffected maternal aunt of subject 35 | |
| 38 | M | N | 10 | Unaffected maternal uncle of subject 35 | |
| 16: | Berg et al.15 (2000) | ||||
| 39 | F | Y | 40 | Girl, aged 11 years, with MR, pulmonary stenosis, and bullus ichthyosiform erythroderma | |
| 40 | F | N | 20 | Mother of subject 39; unaffected | |
| 41 | F | N | 2 | Maternal grandmother of subject 39; unaffected |
Mental retardation (MR) or developmental delay.
FISH Mapping of FRA12A
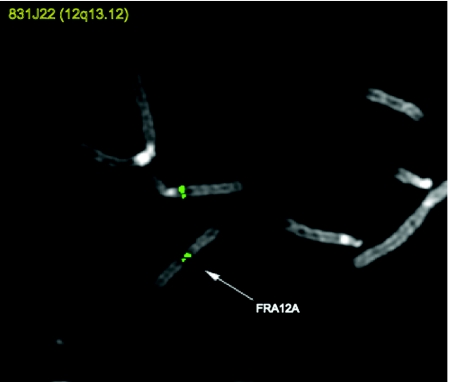
Through use of FISH experiments on metaphase chromosome spreads obtained from lymphoblastoid cells of proband III.1 of family A (fig. 1A), a single BAC clone RP11-831J22 (49080K–49260K) (National Center for Biotechnology Information [NCBI]) spanning the fragile site FRA12A was identified (fig. 2). Since all hitherto cloned folate-sensitive rare fragile sites are caused by a CGG-repeat expansion, the identified BAC clone was searched for CGG repeats. A single (CGG)7 repeat was identified, located in the CpG island at the 5′ end of the Disco-interacting protein 2 homologue DIP2B gene (alternate names KIAA1463, FLJ34278, and MGC104005). The DIP2B gene maps to chromosome 12q13.12.
Figure 2. .
FISH mapping of BAC clone RP11-831J22, spanning the fragile site FRA12A in patient I.1. The fragile site FRA12A is indicated by an arrow.
Analysis of the DIP2B CGG Candidate Repeat
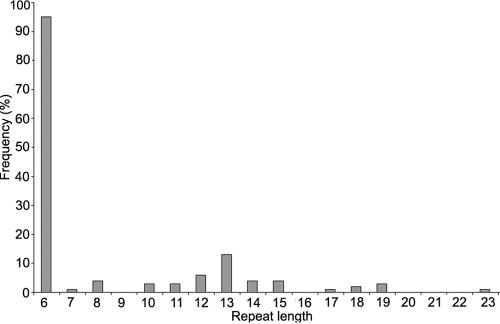
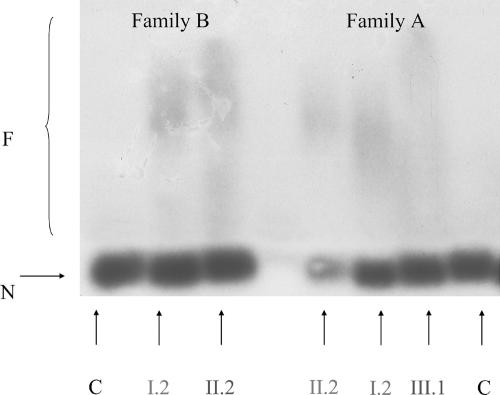
PCR-based amplification of the DIP2B candidate CGG repeat in 70 controls—a total of 140 chromosomes—revealed 13 different alleles, with a range of 6–23 CGG-repeat units; the most common allele contained 6 repeats (fig. 3). We analyzed six individuals with cytogenetic expression of the fragile site FRA12A (subjects I.2, II.2, and III.1 of family A; subjects I.2 and II.2 of family B; and subject II.1 of family C). A single CGG allele in the normal-size range was identified in all FRA12A carriers through use of primers flanking the repeat and standard PCR techniques. Triplet-repeat primed–PCR analysis18 did show a characteristic ladder on the fluorescence trace, with a periodicity of 3 bp, for all FRA12A-expressing individuals but not for controls, which indicates repeat expansion (not shown). Subsequent PCR amplification incorporating deaza-dGTP, followed by Southern blotting and hybridization with use of a CGG-repeat probe, confirmed the presence of an expanded allele in all patients and carriers with FRA12A (fig. 4). This experiment was repeated several times, with comparable outcomes. All individuals expressing FRA12A showed much larger PCR fragments, indicating CGG-repeat expansion. The length of the expanded allele in the unaffected FRA12A carriers (subjects I.2 and II.2 of family A and subject I.2 of family B) exceeded that in control individuals (250 bp) by at least 400–600 bp. In the two patients with FRA12A (subject III.1 of family A and subject II.2 of family B), the length of the expanded allele exceeded the normal length by ∼800–900 bp.
Figure 3. .
Allele frequencies of the FRA12A-associated 5′ CGG repeat in a population of 70 control individuals.
Figure 4. .
Analysis of the DIP2B CGG repeat in controls (“C”) and individuals expressing the FRA12A fragile site (in family B, subjects I.2 and II.2; in family A, subjects I.2, II.2, and III.1). In addition to a normal allele (“N”; 250 bp), individuals who cytogenetically express the FRA12A site show additional PCR fragments of larger size (“F”), indicating repeat expansion. The length of the expanded allele in the unaffected FRA12A carriers (subjects I.2 and II.2 of family A and subject I.2 of family B) is 650–850 bp. In the two patients with FRA12A (subject III.1 of family A and subject II.2 of family B), the length of the expanded allele is >1,050–1,150 bp.
To assess the methylation status of the expanded CGG allele and the surrounding CpG island, a 304-bp region of the promotor CpG island was subjected to bisulphite sequencing. The Gene2Promotor program (Genomatix) identified a 601-bp promotor region starting at −656 bp before the start codon of the DIP2B gene. The FRA12A-associated CGG repeat and the surrounding CpG island lie within the predicted promotor. The bisulphite sequencing method allowed a detailed analysis of methylation in a particular region by converting all nonmethylated cytosines into thymines; methylated cytosines remained unchanged. In control individuals, the promotor region was unmethylated. In family A, this promotor region was fully methylated in FRA12A-affected patient III.1 but was unmethylated in the patient's unaffected FRA12A-expressing mother (subject II.2) and grandmother (subject I.2). In family B, methylation of the 5′ promotor region of the DIP2B gene was detected in FRA12A-affected patient II.2 and his FRA12A-carrier mother (subject I.2). Also in the unaffected FRA12A carrier (subject II.1) of family C, the promotor region was methylated (table 2). All cytosines at CpG dinucleotides in the DIP2B promotor in the patients and carriers were methylated as indicated in figure 5.
Table 2. .
Overview of Methylation and Disease Status of FRA12A-Expressing Individuals
| Family and Subject | Methylation | Status |
| A: | ||
| II.2 | No | Unaffected |
| III.1 | Yes | Affected |
| B: | ||
| I.2 | Yes | Unaffected |
| II.2 | Yes | Affected |
| C: | ||
| II.1 | Yes | Unaffected |
Figure 5. .
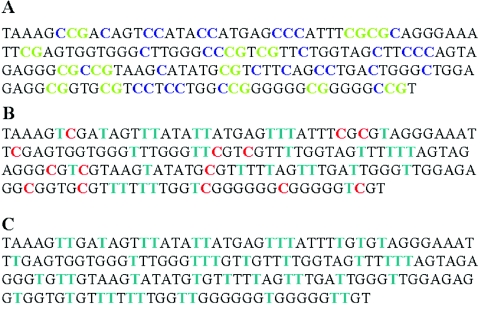
A, Sequence of the DIP2B promotor region. All non-CpG cytosines are indicated in dark blue; CpG cytosines are indicated in green. B, Sequence of the DIP2B promotor after bisulphite treatment in FRA12A-expressing individuals with a methylated CGG-repeat expansion. All nonmethylated cytosines are converted into thymines (blue), whereas methylated cytosines remain unchanged (red). C, Sequence of the DIP2B promotor after bisulphite treatment in FRA12A-expressing individuals with an unmethylated repeat expansion. All cytosines are converted into thymines (blue).
DIP2B Protein
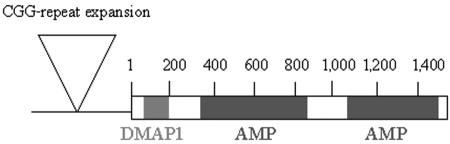
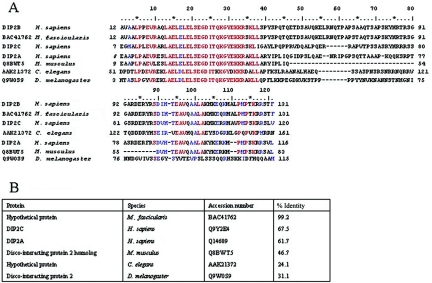
The DIP2B gene contains 38 exons, which encode a putative 1,576-aa protein. The DIP2B gene is a member of the Disco-interacting 2 family (Ensembl family ENSF00000000904), which contains two additional members: DIP2A on chromosome 21q22.3 and DIP2C on chromosome 10p15.3. Percentage similarities, as calculated by the program Align (EBI), are listed in table 3. The DIP2B protein contains three putative functional domains: one DMAP1-binding domain (location 14–131) and two AMP-binding domains (locations 370–822 and 1025–1495) (fig. 6). The conserved domain database (NCBI) points out that the DMAP1-binding domain is highly conserved throughout the eukaryotic kingdom (fig. 7), an indication of its important function.
Table 3. .
Percentage Identity among the Human DIP2B Protein and Other Homologous Protein Sequences
| Protein and Species | Accession Number | Percentage Identity |
| Protein similar to DIP2B: | ||
| Rattus norvegicus | XP_235656 (GenBank) | 97.5 |
| LOC239667: | ||
| Mus musculus | NP_766407 (GenBank) | 96.1 |
| Protein similar to DIP2B, partial: | ||
| Bos taurus | XP_580943.1 (GenBank) | 93.3 |
| Macaca fascicularis | BAC41762 (GenBank) | 83.7 |
| DIP2C: | ||
| H. sapiens | NP_055789 (GenBank) | 72.7 |
| Disco-interacting protein 2 (DIP2) homologue: | ||
| M. musculus | Q8BWT5 (UniProtKB/Swiss-Prot) | 70.7 |
| DIP2A: | ||
| H. sapiens | NP_055966 (GenBank) | 71.2 |
Figure 6. .
Position of the three conserved domains in the DIP2B protein.
Figure 7. .
A, Alignment of the six most similar homologues of the DMAP1-binding domain of the DIP2B protein. Amino acid–sequence positions are indicated at the beginning and end of each line. Identical amino acids are indicated in red, and the similar ones in blue. The source species and the accession number of the sequence are indicated at the left of each line. B, Percentage similarity among the DIP2B DMAP1-binding domain and its homologues.
DIP2B Gene Expression
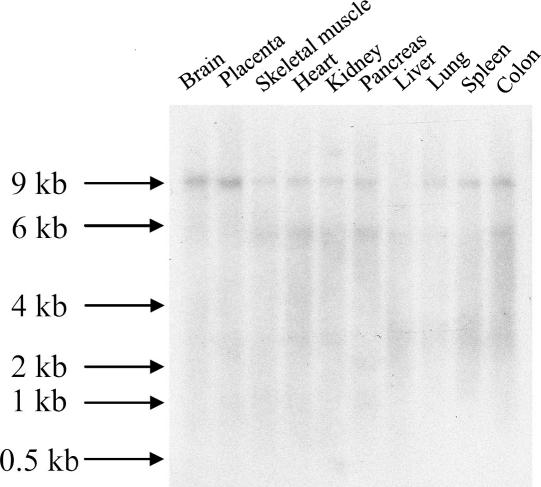
For northern-blot analysis, we used a specific DIP2B probe that contained exons 3–7. A single transcript of ∼9 kb, which corresponds well to the predicted cDNA size, was detected in several human tissues (fig. 8). DIP2B expression was found in brain, placenta, skeletal muscle, heart, kidney, pancreas, lung, spleen, and colon. Hybridization of an MTE array (data not shown) indicated that the DIP2B gene is expressed in several brain regions: the cerebral cortex; the frontal, parietal, occipital, and temporal lobes; the paracentral gyrus; the pons; the corpus callosum; and the hippocampus. Highest expression levels in the brain were found in the cerebral cortex and the frontal and parietal lobes.
Figure 8. .
Northern-blot analysis of the DIP2B gene. A specific cDNA probe was hybridized to a human multiple-tissue northern blot. A single transcript of ∼9 kb was detected in various human tissues.
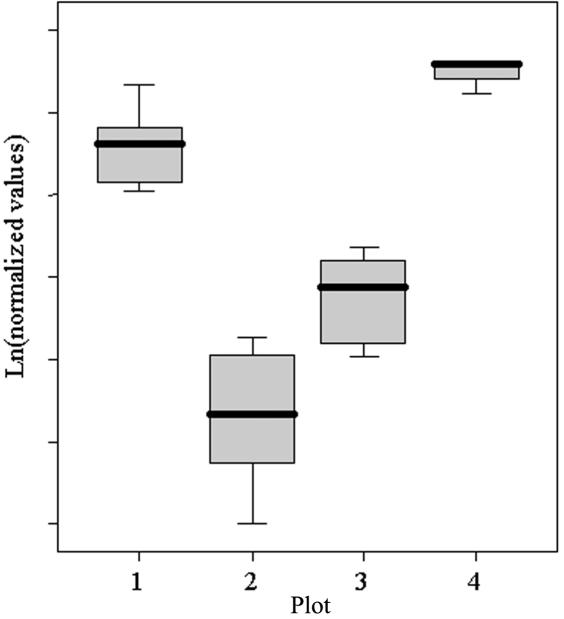
To detect possible differences in the expression levels of DIP2B among FRA12A-affected patients, FRA12A carriers, and control individuals, real-time PCR was performed. To have a representative sampling, cDNA of each FRA12A carrier and patient was prepared three times independently, and each preparation was analyzed in duplicate (fig. 9). This procedure was repeated, with comparable results (table 4). As expected, for both patients with FRA12A, DIP2B expression was reduced to about half the amount of that in controls (55% [P=.002] and 45% [P=.001] in the first and second experiment, respectively). For subject II.2 of family A, a carrier of an unmethylated FRA12A expansion, real-time PCR results revealed enhanced expression values (200% [P=.009) and 142% [P=.013]). Subjects I.2 of family B and II.1 of family C, both carriers of a methylated FRA12A expansion, showed a reduction of DIP2B expression, approximately two-thirds of that of controls (67% [P=.056] and 63% [P=.001]).
Figure 9. .
Box plot showing the significant difference in the normalized amount of RNA for DIP2B in function of the methylation status and the disease status. Plot 1 represents control individuals, plot 2 represents patients with a methylated repeat expansion, plot 3 represents healthy individuals with a methylated repeat expansion, and plot 4 represents a healthy individual with an unmethylated repeat expansion. The significance was tested using one-way ANOVA (analysis of variance).
Table 4. .
Relative DIP2B Expression for Families A, B, and C in Comparison with That for Control Individuals for Two Independent Real-Time PCR Analyses
| Experiment and Subject(s) | Relative Expression (%) |
P |
| 1: | ||
| Healthy individual with unmethylated repeat expansion | 200 | ≪.01 |
| Affected individuals with methylated repeat expansion | 55 | ≪.01 |
| Healthy individuals with methylated repeat expansion | 67 | ∼.05 |
| 2: | ||
| Healthy individual with unmethylated repeat expansion | 142 | ⩽.01 |
| Affected individuals with methylated repeat expansion | 45 | ≪.01 |
| Healthy individuals with methylated repeat expansion | 63 | ≪.01 |
To verify whether the reduced gene expression in patients with FRA12A is due to the silencing of the expanded allele, cSNP analysis was performed by sequencing. Four cSNPs were found in the DIP2B cDNA sequence (GenBank accession number NM_173602): one at position 2073 in exon 16 (c.2073A/T), one at position 2530 in exon 20 (c.2530A/G), one at position 3102 in exon 24 (c.3102T/C), and one at position 4587 in exon 37 (c.4587C/T). All FRA12A carriers and patients were tested for these cSNPs. The cSNP in exon 16 appeared informative in DNA of carrier I.2 of family B (genotype g.2073A/T). By sequencing the cDNA of subject I.2, only the c.2073A allele could be detected, indicating the transcriptional silencing of the T allele. Determination of the inheritance in family B demonstrated that it is the FRA12A allele that is silenced: descendant II.2 of carrier I.2 has the genotype g.2073T/T. The fact that he expresses FRA12A indicates that the c.2073T allele is linked to the CGG-repeat expansion.
Finally, all exons of the DIP2B gene were sequenced at the DNA level, and intron/exon boundaries were checked for both FRA12A-affected patients. No sequence alterations could be detected in the DIP2B coding sequences.
Discussion
Our study provides evidence that the molecular basis of the rare folate-sensitive fragile site FRA12A is a CGG-repeat expansion at the 5′ end of the DIP2B gene. The DIP2B CGG repeat was shown to be expanded in individuals expressing the FRA12A site. An expanded CGG repeat has also been found in previously cloned folate-sensitive rare fragile sites. The FRA12A CGG repeat, located in the CpG island in the promotor region of DIP2B, is polymorphic in the general population and is expanded on transmission to the next generation in the FRA12A-affected families we analyzed.
The putative DIP2B protein contains a highly conserved DMAP1-binding domain and two AMP-binding domains. The AMP-binding domains are rich in serine, threonine, and glycine and possess a catalytic activity. The DNMT1-associated protein (DMAP1) was discovered recently and has a transcription-repressor activity.31 The chief enzyme that maintains DNA methylation of the mammalian genome during S phase is DNA (cytosine-5)–methyltransferase 1 (DNMT1).32,33 The carboxy-terminal catalytic domain of DNMT1 has a strong affinity for hemimethylated DNA and transfers methyl groups from S-adenosylmethionine to cytosines in CpG regions, whereas the N-terminal portion directs the enzyme to the replication foci. More recently, it has been shown that the noncatalytic N-terminal region of DNMT1 also forms a repressive transcription complex consisting of DNMT1, the histone deacetylase HDAC2, and the DNMT1-associated protein DMAP1. The DMAP1-DIP2B interaction may modify the DNMT1 enzymatic activity to perform an effective repression by methylation. If this is the case, the methylation of the DIP2B promotor is able to directly influence the DNMT1 methylation machinery itself.
The DIP2B protein is a member of the DIP2 protein family. We demonstrated expression of DIP2B in the brain. In situ hybridization of the mouse dip2 homologue suggested that mdip2 may participate in defining the segmental borders in the CNS and may have a role in providing positional cues for axon pathfinding and patterning.34 A Drosophila homologue of DIP2B, discovered by the yeast-2-hybrid interaction–trap technique, is a protein that interacts with the Drosophila disconnected (disco) gene shown to be required for proper neuronal connections in both the larval and adult visual system of Drosophila.35,36
For FRAXA, the most extensively studied folate-sensitive rare fragile site, a clear-cut relationship exists in males between fragile-site expression and disease. Patients with fragile X express the FRAXA fragile site in vitro because of large CGG-repeat expansions, which cause hypermethylation of the promotor region of the underlying FMR1 gene. These so-called full mutations originate from smaller expansions of the FMR1 CGG repeat (premutations), which are unmethylated and do not cause fragile X syndrome. A comparable situation is described for family A, where a repeat grows stepwise in three generations to a methylated CGG-repeat expansion in the youngest generation. It is tempting to speculate that the de novo gene silencing of the DIP2B gene is causative of the mental retardation in this patient. A relationship between fragile-site expression and mental retardation was predicted, since patients with cytogenetic FRA12A expression and mental retardation expressed the fragile site in a significantly higher percentage of cells than did unaffected FRA12A carriers. However, in family B, the unaffected mother of the patient (subject I.2) has a methylated repeat expansion as well, which could imply that the coexistence of DIP2B methylation and the mental retardation in patient II.2 is purely coincidental. However, it can also be speculated that disease penetrance is not 100%. This phenomenon is also seen in FRAXE mental retardation, where expansion and methylation of the CGG repeat in front of the FMR2 gene is not always associated with mental retardation.37 Some evidence of nonpenetrance is provided by investigation of the DIP2B expression in our FRA12A-affected families; it seems to suggest that gene expression in unaffected carriers is higher than in patients.
Other proteins involved in methylation metabolism have been implicated in disease as well. Mutations in the methyl-CpG binding protein 2 gene (MECP2) cause Rett syndrome, a form of severe mental retardation in females. MECP2 is a transcriptional repressor that binds to methylated CpG residues and forms chromatin remodeling complexes with various transcriptional repressors.38–40 Absence of the MECP2 protein in mice results in neurological abnormalities and premature death.41,42 Additional evidence of the importance of the DNA methylation machinery for the brain comes from studies on Dnmt1-knockout mice. Normal mice contain surprisingly high levels of Dnmt1 in neurons,43–46 whereas absence of this enzyme in the brain results in death shortly after birth.47 Moreover, in brains containing both normal and Dnmt1-negative cells, the Dnmt1-negative cells are rapidly lost and mutant neurons are quickly eliminated.47 Mutations in one of the methyltransferases, the DNMT3B gene, causes ICF, in which neurological abnormalities are also present.48 Finally, additional suggestions for a link between DNA methylation defects and brain disorders come from the ATR-X syndrome (α-thalassemia/mental retardation syndrome, X linked), characterized by severe psychomotoric retardation, characteristic facial features, genital abnormalities, and α-thalassemia. It was shown that the syndrome is caused by mutations in the ATR-X gene, resulting in changes in the methylation pattern of several highly repeated sequences, including the rDNA arrays, a Y-specific satellite, and subtelomeric repeats.49 More recently, the ATR-X gene has also been shown to be involved in other X-linked mental retardation syndromes.50,51
A second interesting finding, although based on a single case, is the observation that the elongated, unmethylated CGG repeat of carrier II.2 for family A has a 1.4–2-fold overexpression of the DIP2B gene. This implies two-to-threefold overexpression of the expanded allele, with the assumption of unaffected expression of the unexpanded allele. A similar situation is found in fragile X premutation carriers, who have an enhanced expression of the FMR1 mRNA of unmethylated elongated repeats. Our data thus suggest that CGG-repeat expansion in general may act as a transcriptional enhancer. Males with premutation fragile X alleles are at risk for a neurodegenerative disorder called “FXTAS” (fragile X–associated tremor/ataxia syndrome).52 It would therefore be of interest to determine the phenotype of aged FRA12A carriers.
In conclusion, we report the identification of the molecular cause of the fragile site FRA12A. The associated DIP2B gene possesses a DMAP1-binding domain and might therefore be involved in the methylation machinery. Our finding warrants further investigations of the possible role of the DIP2B gene in mental retardation.
Acknowledgments
Financial support for this study was obtained through grants from the Belgian National Fund for Scientific Research–Flanders and an Interuniversity Attraction Pole program. Special thanks go to Olivia Beck, Gabriella Munteanu, and Lieve Wiericx for the establishment of the EBV-transformed lymphoblastoid cell lines.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- EBI, http://www.ebi.ac.uk/ (for the Align program)
- Ensembl, http://www.ensembl.org/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for DIP2B cDNA sequence [accession number NM_173602], R. norvegicus protein similar to DIP2B [accession number XP_235656], M. musculus LOC239667 [accession number NP_766407], B. taurus protein similar to DIP2B, partial [accession number XP_580943.1], M. fascicularis protein similar to DIP2B, partial [accession number BAC41762], H. sapiens DIP2C [accession number NP_055789], and H. sapiens DIP2A [accession number NP_055966])
- Genomatix, http://www.genomatix.de/ (for the Gene2Promotor program)
- NCBI, http://www.ncbi.nlm.nih.gov/
- qBase, http://medgen.ugent.be/qbase/
- UniProtKB/Swiss-Prot, http://au.expasy.org/uniprot/ (for Q8BWT5)
References
- 1.Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16:6–21 10.1101/gad.947102 [DOI] [PubMed] [Google Scholar]
- 2.Robertson KD (2005) DNA methylation and human disease. Nat Rev Genet 6:597–610 10.1038/nrm1228 [DOI] [PubMed] [Google Scholar]
- 3.Jones PA, Baylin SB (2002) The fundamental role of epigenetic events in cancer. Nat Rev Genet 3:415–428 10.1038/nrg962 [DOI] [PubMed] [Google Scholar]
- 4.Muller S, Zirkel D, Westphal M, Zumkeller W (2000) Genomic imprinting of IGF2 and H19 in human meningiomas. Eur J Cancer 36:651–655 10.1016/S0959-8049(99)00328-7 [DOI] [PubMed] [Google Scholar]
- 5.Hassan KM, Norwood T, Gimelli G, Gartler SM, Hansen RS (2001) Satellite 2 methylation patterns in normal and ICF syndrome cells and association of hypomethylation with advanced replication. Hum Genet 109:452–462 10.1007/s004390100590 [DOI] [PubMed] [Google Scholar]
- 6.Robertson KD, Wolffe AP (2000) DNA methylation in health and disease. Nat Rev Genet 1:11–19 10.1038/35049533 [DOI] [PubMed] [Google Scholar]
- 7.Tucker KL (2001) Methylated cytosine and the brain: a new base for neuroscience. Neuron 30:649–652 10.1016/S0896-6273(01)00325-7 [DOI] [PubMed] [Google Scholar]
- 8.Fan G, Hutnick L (2005) Methyl-CpG binding proteins in the nervous system. Cell Res 15:255–261 10.1038/sj.cr.7290294 [DOI] [PubMed] [Google Scholar]
- 9.Verkerk AJMH, Pieretti M, Sutcliffe JS, Fu Y-H, Kuhl DPA, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang F, et al (1991) Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65:905–914 10.1016/0092-8674(91)90397-H [DOI] [PubMed] [Google Scholar]
- 10.Knight SJL, Flannery AV, Hirst MC, Campbell L, Christodoulou Z, Phelps SR, Pointon J, Middleton-Price HR, Barnicoat A, Pembrey ME, et al (1993) Trinucleotide repeat amplification and hypermethylation of a CpG island in FRAXE mental retardation. Cell 74:127–134 10.1016/0092-8674(93)90300-F [DOI] [PubMed] [Google Scholar]
- 11.Parrish JE, Oostra BA, Verkerk AJMH, Richards CS, Reynolds J, Spikes AS, Shaffer LG, Nelson DL (1994) Isolation of a GCC repeat showing expansion in FRAXF, a fragile site distal to FRAXA and FRAXE. Nat Genet 8:229–235 10.1038/ng1194-229 [DOI] [PubMed] [Google Scholar]
- 12.Jones C, Penny L, Mattina T, Yu S, Baker E, Voullaire L, Langdon WY, Sutherland GR, Richards RI, Tunnacliffe A (1995) Association of a chromosome deletion syndrome with a fragile site within the proto-oncogene CBL2. Nature 376:145–149 10.1038/376145a0 [DOI] [PubMed] [Google Scholar]
- 13.Nancarrow JK, Kremer E, Holman K, Eyre H, Doggett NA, Le Paslier D, Callen DF, Sutherland GR, Richards RI (1994) Implications of FRA16A structure for the mechanism of chromosomal fragile site genesis. Science 264:1938–1941 10.1126/science.8009225 [DOI] [PubMed] [Google Scholar]
- 14.Sarafidou T, Kahl C, Martinez-Garay I, Mangelsdorf M, Gesk S, Baker E, Kokkinaki M, Talley P, Maltby EL, French L, et al (2004) Folate-sensitive fragile site FRA10A is due to an expansion of a CGG repeat in a novel gene, FRA10AC1, encoding a nuclear protein. Genomics 84:69–81 10.1016/j.ygeno.2003.12.017 [DOI] [PubMed] [Google Scholar]
- 15.Berg J, Grace E, Teik KW, Hammond H, Tidman M, FitzPatrick D (2000) Bullous ichthyosiform erythroderma, developmental delay, aortic and pulmonary stenosis in association with a FRA12A. Clin Dysmorphol 9:213–219 [DOI] [PubMed] [Google Scholar]
- 16.Smeets DFCM, Scheres JMJC, Hustinx TWJ (1985) Heritable fragility at 11q13 and 12q13. Clin Genet 28:145–150 [DOI] [PubMed] [Google Scholar]
- 17.Mules EH, Stamberg J, Jabs EW, Leonard CO (1983) Two different structural abnormalities of chromosome 13 in offspring of chromosomally normal parents with two fragile sites. Clin Genet 23:380–385 [DOI] [PubMed] [Google Scholar]
- 18.Warner JP, Barron LH, Goudie D, Kelly K, Dow D, Fitzpatrick DR, Brock DJ (1996) A general method for the detection of large CAG repeat expansions by fluorescent PCR. J Med Genet 33:1022–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:34.1–34.11 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giraud F, Ayme S, Mattei JF, Mattei MG (1976) Constitutional chromosomal breakage. Hum Genet 34:125–136 10.1007/BF00278880 [DOI] [PubMed] [Google Scholar]
- 21.Stetten G, Sroka B, Norbury-Glaser M, Corson VL (1988) Fragile site in chromosome 12 in a patient with two miscarriages. Am J Med Genet 31:521–525 10.1002/ajmg.1320310306 [DOI] [PubMed] [Google Scholar]
- 22.Moric-Petrovic S, Laca Z (1984) Fragile site at 12q13 associated with phenotypic abnormalities. J Med Genet 21:216–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donti E, Venti G, Bocchini V, Rosi G, Armellini R, Trabalza N (1979) The constitutional fragility of chromosome 12 in a case of 46,XX,var(12)(qh’,RHG,GAG,CBG). Hum Genet 48:53–59 10.1007/BF00273274 [DOI] [PubMed] [Google Scholar]
- 24.Sutherland GR, Hecht F (1985) Fragile sites on human chromosomes. Oxford University Press, New York [Google Scholar]
- 25.Sutherland GR, Hinton L (1981) Heritable fragile sites on human chromosomes. Hum Genet 57:217–219 10.1007/BF00282028 [DOI] [PubMed] [Google Scholar]
- 26.Kumar D, Clark JW, Blank CE, Patton MA (1986) A family with craniofrontonasal dysplasia, and fragile site 12q13 segregating independently. Clin Genet 29:530–537 [DOI] [PubMed] [Google Scholar]
- 27.Webb TP, Thake AI, Bundey SE, Todd J (1987) A cytogenetic survey of a mentally retarded school-age population with special reference to fragile sites. J Ment Defic Res 31:61–71 [DOI] [PubMed] [Google Scholar]
- 28.Guichaoua M, Matteï MG, Matteï JF, Giraud F (1982) Aspects genetiques des sites fragiles autosomiques: apropos de 40 cas. J Genet Hum 30:183–197 [PubMed] [Google Scholar]
- 29.Giardino D, Bettio D, Simoni G (1990) 12q13 Fragility in a family with recurrent spontaneous abortions: expression of the fragile site under different culture conditions. Ann Genet 33:88–91 [PubMed] [Google Scholar]
- 30.Kähkönen M, Tengstrom C, Alitalo T, Matilainen R, Kaski M, Airaksinen E (1989) Population cytogenetics of folate-sensitive fragile sites. II. Autosomal rare fragile sites. Hum Genet 82:3–8 10.1007/BF00288261 [DOI] [PubMed] [Google Scholar]
- 31.Rountree MR, Bachman KE, Baylin SB (2000) DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet 25:269–277 10.1038/77023 [DOI] [PubMed] [Google Scholar]
- 32.Bestor T, Laudano A, Mattaliano R, Ingram V (1988) Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells: the carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol 203:971–983 10.1016/0022-2836(88)90122-2 [DOI] [PubMed] [Google Scholar]
- 33.Yen RW, Vertino PM, Nelkin BD, Yu JJ, el-Deiry W, Cumaraswamy A, Lennon GG, Trask BJ, Celano P, Baylin SB (1992) Isolation and characterization of the cDNA encoding human DNA methyltransferase. Nucleic Acids Res 20:2287–2291 10.1093/nar/20.9.2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukhopadhyay M, Pelka P, DeSousa D, Kablar B, Schindler A, Rudnicki MA, Campos AR (2002) Cloning, genomic organization and expression pattern of a novel Drosophila gene, the disco-interacting protein 2 (dip2), and its murine homolog. Gene 293:59–65 10.1016/S0378-1119(02)00694-7 [DOI] [PubMed] [Google Scholar]
- 35.Steller H, Fischbach KF, Rubin GM (1987) Disconnected: a locus required for neuronal pathway formation in the visual system of Drosophila. Cell 50:1139–1153 10.1016/0092-8674(87)90180-2 [DOI] [PubMed] [Google Scholar]
- 36.Campos AR, Lee KJ, Steller H (1995) Establishment of neuronal connectivity during development of the Drosophila larval visual system. J Neurobiol 28:313–329 10.1002/neu.480280305 [DOI] [PubMed] [Google Scholar]
- 37.Gécz J, Oostra BA, Hockey A, Carbonell P, Turner G, Haan EA, Sutherland GR, Mulley JC (1997) FMR2 expression in families with FRAXE mental retardation. Hum Mol Genet 6:435–441 10.1093/hmg/6.3.435 [DOI] [PubMed] [Google Scholar]
- 38.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP (1998) Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet 19:187–191 10.1038/561 [DOI] [PubMed] [Google Scholar]
- 39.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A (1998) Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386–389 10.1038/30764 [DOI] [PubMed] [Google Scholar]
- 40.Kokura K, Kaul SC, Wadhwa R, Nomura T, Khan MM, Shinagawa T, Yasukawa T, Colmenares C, Ishii S (2001) The Ski protein family is required for MeCP2-mediated transcriptional repression. J Biol Chem 276:34115–34121 10.1074/jbc.M105747200 [DOI] [PubMed] [Google Scholar]
- 41.Chen RZ, Akbarian S, Tudor M, Jaenisch R (2001) Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet 27:327–331 10.1038/85906 [DOI] [PubMed] [Google Scholar]
- 42.Guy J, Hendrich B, Holmes M, Martin JE, Bird A (2001) A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet 27:322–326 10.1038/85899 [DOI] [PubMed] [Google Scholar]
- 43.Goto K, Numata M, Komura JI, Ono T, Bestor TH, Kondo H (1994) Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation 56:39–44 [DOI] [PubMed] [Google Scholar]
- 44.Brooks PJ, Marietta C, Goldman D (1996) DNA mismatch repair and DNA methylation in adult brain neurons. J Neurosci 16:939–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inano K, Suetake I, Ueda T, Miyake Y, Nakamura M, Okada M, Tajima S (2000) Maintenance-type DNA methyltransferase is highly expressed in post-mitotic neurons and localized in the cytoplasmic compartment. J Biochem (Tokyo) 128:315–321 [DOI] [PubMed] [Google Scholar]
- 46.Feng J, Chang H, Li E, Fan G (2005) Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res 79:734–746 10.1002/jnr.20404 [DOI] [PubMed] [Google Scholar]
- 47.Fan G, Beard C, Chen RZ, Csankovszki G, Sun Y, Siniaia M, Biniszkiewicz D, Bates B, Lee PP, Kuhn R, et al (2001) DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J Neurosci 21:788–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu GL, Bestor TH, Bourc’his D, Hsieh CL, Tommerup N, Bugge M, Hulten M, Qu X, Russo JJ, Viegas-Pequignot E (1999) Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402:187–191 10.1038/46052 [DOI] [PubMed] [Google Scholar]
- 49.Gibbons RJ, Picketts DJ, Villard L, Higgs DR (1995) Mutations in a putative global transcriptional regulator cause X-linked mental retardation with α-thalassemia (ATR-X syndrome). Cell 80:837–845 10.1016/0092-8674(95)90287-2 [DOI] [PubMed] [Google Scholar]
- 50.Gibbons RJ, Higgs DR (2000) Molecular-clinical spectrum of the ATR-X syndrome. Am J Med Genet 97:204–212 [DOI] [PubMed] [Google Scholar]
- 51.Wieland I, Sabathil J, Ostendorf A, Rittinger O, Ropke A, Winnepenninckx B, Kooy F, Holinski-Feder E, Wieacker P (2005) A missense mutation in the coiled-coil motif of the HP1-interacting domain of ATR-X in a family with X-linked mental retardation. Neurogenetics 6:45–47 10.1007/s10048-004-0190-3 [DOI] [PubMed] [Google Scholar]
- 52.Hagerman PJ, Hagerman RJ (2004) Fragile X-associated tremor/ataxia syndrome (FXTAS). Ment Retard Dev Disabil Res Rev 10:25–30 10.1002/mrdd.20005 [DOI] [PubMed] [Google Scholar]