Abstract
During development of plant seeds, embryos import nutrients and store massive amounts of reserves. Seed reserves are rapidly degraded and mobilized to support seedling development after germination. HIGH-LEVEL EXPRESSION OF SUGAR-INDUCIBLE GENE 2 (HSI2) of Arabidopsis thaliana is a B3 DNA-binding domain protein that represses the transcription of sugar-inducible reporter gene. Although disruption of HSI2 or HSI2-Like 1 (HSL1) did not affect growth, seeds with disruption of both HSI2 and HSL1 (KK mutant) developed abortive seedlings that stopped growing 7–9 days after imbibition. KK seedlings developed swollen hypocotyls that accumulated seed storage proteins and oil on medium containing sucrose or other metabolizable sugars, and calluses developed from KK seedlings also accumulated seed storage reserves. The expression of seed maturation genes, which include LEAFY COTYLEDON-type master regulators, in KK seedlings depended on the concentration of sucrose, suggesting that sugar controls the expression of seed maturation genes. Our results suggest that HSI2 and HSL1 repress the sugar-inducible expression of the seed maturation program in seedlings and play an essential role in regulating the transition from seed maturation to seedling growth.
Keywords: plant oil, seed germination, sugar signal
Plant seeds develop through embryogenesis and seed maturation phases. During the maturation phase, seeds accumulate massive storage reserves, after which the seeds undergo programmed dehydration, and the embryos transition to dormancy. Upon seed germination, the embryos are released from dormancy and initiate vegetative development as seedlings. Seed maturation depends on the import of sugar and other nutrients, and during seedling development, the seed reserves are rapidly degraded, and the nutrients are remobilized. The seed plants evolved by incorporating the seed maturation phase into the continuous mode of embryogenesis and subsequent morphogenetic changes that are characteristic of many lower plants that do not make seeds. Therefore, it is likely that unique mechanisms control the initiation of the seed maturation program and its termination and transition to active seedling growth (1).
LEAFY COTYLEDON 1 (LEC1), which is the HapIII subunit of CAAT-binding factor, controls various aspects of seed development from early embryogenesis to late seed maturation (2, 3). Three transcriptional regulators with plant-specific B3 DNA-binding domains, ABSCISIC ACID-INSENSITIVE 3 (ABI3), LEC2, and FUSCA 3 (FUS3), also play key roles in the control of seed maturation in cooperation with LEC1 and abscisic acid (ABA; ref. 4). Enforced expression of LEC1 (2) and LEC2 (5) is sufficient to confer embryonic traits to seedlings. On the other hand, PICKLE (PKL) is known to act in conjunction with gibberellin (GA) to repress embryonic traits in primary roots during a very early stage of germination (6, 7). However, relatively little is known about the mechanisms involved in the termination of the seed maturation program and transition to active seedling growth.
In addition to the interactions of stage-specific developmental regulators and hormonal signals, recent studies suggest that metabolites such as sugars act as important signals regulating seed development and germination (8, 9). Although sugar-dependent regulation of expression of a variety of genes during seed germination can be mediated directly or indirectly by ABA (9), the role of sugar in the regulation of expression of seed maturation genes remains unclear.
Arabidopsis contains three proteins, HIGH-LEVEL EXPRESSION OF SUGAR-INDUCIBLE GENE 2 (HSI2), HSI2-Like 1 (HSL1), and HSI2-Like 2 (HSL2), that have B3 domains similar to those of ABI3, LEC2, and FUS3 (10). These proteins also include C-terminal ERF-associated amphiphilic repression (EAR) motifs, which mediate transcriptional repression (11). HSI2 functions as a transcriptional repressor of a sugar-inducible reporter gene (10). In the current work, we provide evidence that HSI2 and HSL1 are redundant inhibitors of sugar-inducible ectopic expression of seed maturation genes during seedling growth and that the two proteins play an essential role in regulating the transition from seed maturation to active seedling growth.
Results
Disruption of Both HSI2 and HSL1 Is Seedling-Lethal.
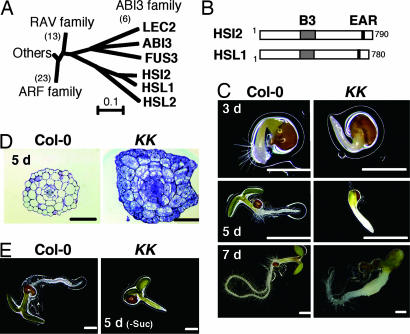
The three Arabidopsis proteins, HSI2, HSL1, and HSL2, contain B3 domains that are similar to those in ABI3, LEC2, and FUS3 (ref. 10 and Fig. 1A). In addition, the C-terminal regions of these three Arabidopsis proteins contain EAR motifs that mediate active transcriptional repression (refs. 10 and 11 and Fig. 1B). In addition to hsi2-2, a null mutant of HSI2 with a T-DNA insertion (10), we identified a homozygous hsl1-1 mutant in which a T-DNA was inserted within the 12th exon of HSL1 [supporting information (SI) Fig. 8]. Like hsi2-2, the hsl1-1 mutant grew normally and did not show visible phenotypes. When homozygous hsi2-2 and hsl1-1 plants were crossed, 8 of 144 F2 seeds developed abnormal seedlings and stopped growing ≈9 days after imbibition. We therefore selected hsi2-2−/−/hsl1-1+/− and hsi2-2+/−/hsl1-−/− plants by PCR-based genotyping. Of the seeds from self-pollinated hsi2-2−/−/hsl1-1+/− and hsi2-2+/−/hsl1-−/− plants, 62 of 252 (χ2 for a ratio of 3:1 = 0.085; P > 0.05) and 35 of 134 (χ2 for a ratio of 3:1 = 0.09; P > 0.05), respectively, developed defective seedlings. Defective seedlings cosegregated with the hsi2-2−/−/hsl1-1−/− genotype, whereas seedlings that contained at least one wild-type copy of HSI2 or HSL1 grew like the wild-type Col-0 seedlings (SI Fig. 8). These results suggest that HSI2 and HSL1 share a function that is essential for active seedling growth.
Fig. 1.
HSI2 subfamily of B3 domain proteins and phenotypic features of KK seedlings. (A) Amino acid sequences of B3 domain of Arabidopsis B3 domain proteins were aligned by using ClustalW, and a phylogenetic neighbor-joining tree was generated as described previously (10). (B) Schematic structures of HSI2 and HSL1. The locations of the B3 domain and EAR motif are shown. (C) Col-0 and KK seedlings germinated on medium containing 1% sucrose for 3, 5, or 7 days. (D) Cross-sections of hypocotyls of 5-day-old Col-0 and KK seedlings germinated on medium containing 1% sucrose and were stained with toluidine blue. (E) Col-0 and KK seedlings germinated on sucrose-free medium for 5 days. [Scale bars, 0.5 mm (C–E).]
To examine the growth of seedlings of double knockouts (hsi2-2−/−/hsl1-1−/−), which we designated KK mutants, offspring seeds of hsi2-2−/−/hsl1-1+/− or hsi2-2+/−/hsl1-1−/− plants were germinated on agar medium containing 1% sucrose. The KK seedlings could be distinguished from seedlings with other genotypes 3–5 days after imbibition because they showed delayed root hair development and cotyledon opening compared with Col-0 seedlings (Fig. 1C). Hypocotyls of KK seedlings were swollen 5 days after imbibition, and the swollen hypocotyls developed a light yellow, rugged surface 7 days after imbibition. Root growth was severely inhibited in KK seedlings. Swollen and rugged KK hypocotyls further developed into yellow callus-like structures after 9 days, at which point most of the KK seedlings stopped growing. These phenotypes were observed repeatedly with seeds from both parents. In cross-sections of swollen hypocotyls from 5-day-old KK seedlings (Fig. 1D), the cortex cells showed marked expansion and proliferation, and the epidermal layer also showed a marked increase in cell number. Furthermore, like actively proliferating cells in meristems, cells of KK mutants showed intense staining with toluidine blue. Cross-sections of KK roots also showed similar structural anomalies (SI Fig. 9A). Approximately 8% of KK seedlings showed tricotyledons (see Fig. 2B), suggesting that the KK mutant also has some defects during embryogenesis.
Fig. 2.
Accumulation of seed storage compounds in KK seedlings. (A) Proteins extracted from 4-, 7-, and 9-day-old Col-0 and KK seedlings grown on medium containing 0% or 1% sucrose were analyzed by immunoblotting with anti-12S globulin and anti-oleosin S4 antibodies. Each lane contained 1 μg of protein. Proteins extracted from Col-0 seeds served as a control, and the size of protein molecular mass standards is indicated. (B) Five- and 12-day-old Col-0 and KK seedlings on medium containing 1% sucrose were stained with fat red 7B. (Scale bar, 0.5 mm.) (C) Total lipids were extracted from 4- and 9-day-old Col-0 and KK seedlings grown on medium containing 0% or 1% sucrose, and those from four seedlings were separated by TLC and stained with sulfuric acid. Lipids extracted from four Col-0 seeds served as a control, and 10 μg of glyceryl trilinoleate was used as a standard (Std).
In contrast to seeds germinated on sucrose-containing medium, most of the KK seedlings germinated on sucrose-free medium opened their green cotyledons 5 days after imbibition, similar to Col-0 seedlings (Fig. 1E); however, growth of the roots was inhibited, and most of the KK seedlings stopped growing. The KK seedlings germinated on medium containing glucose or mannitol were similar to those germinated in the presence or absence of sucrose, respectively, suggesting that sugar metabolism is required for the appearance of the swollen hypocotyl phenotype. The KK seedlings germinated on wet rockwool were also similar to those germinated on sugar-free medium (SI Fig. 9B). GA plays an important role in seed germination, and the penetrance of the “pickle root” phenotype of the pkl mutant is enhanced by uniconazole P, an inhibitor of GA synthesis (6). Neither GA nor uniconazole P, however, affected the phenotype of KK seedlings.
KK Seedlings Express Seed Maturation Genes and Accumulate Seed Storage Compounds.
We isolated RNAs from Col-0 and KK seedlings 4 days after imbibition on medium containing 1% sucrose, and then we compared their gene expression patterns by using an oligonucleotide microarray. From two pairs of independently isolated RNA samples, we identified 679 genes that showed >2-fold higher mRNA levels in KK compared with Col-0 plants in both sets of data. Of these genes, 35 showed >30-fold up-regulation (SI Table 1). These genes were enriched in genes involved in seed maturation, including five genes involved in the production of the seed-type oleosins of the oil body (12), three genes encoding 12S globulin seed storage proteins (13), four genes encoding 2S albumin seed storage proteins (14), and genes encoding transcription factors involved in seed development, such as LEC1-LIKE (LIL; 15) and DPBF2 (16). Examination of the AtGeneExpress microarray database (http://arabidopsis.org/info/expression/ATGenExpress.jsp) suggested that 32 of these 35 genes are specifically expressed during seed maturation in Col-0 plants. In addition, in KK seedlings, there was a 4- to 8-fold up-regulation of ABI3, FUS3, ENHANCED EM LEVEL (EEL; ref. 16) and WRINKLED1 (WRI1; refs. 17 and 18), which participate in the synthesis of storage proteins and lipids.
In Col-0 seedlings germinated on sugar-free medium, the 12S globulin and oleosin S4 stored in mature dried seeds disappeared within 4 days after imbibition (Fig. 2A). Although germination on 1% sucrose medium delayed the degradation of oleosin S4, both 12S globulin and oleosin S4 disappeared within 7 days after imbibition. The KK seedlings grown on medium without sucrose also showed degradation of both 12S globulin and oleosin S4, although delayed degradation of oleosin S4 compared with Col-0 seedlings was observed. On the other hand, KK seedlings grown on 1% sucrose medium contained both 12S globulin and oleosin S4 even 7 and 9 days after imbibition.
The KK seedlings grown on medium containing sucrose were intensely stained with fat red 7B, which stains neutral lipids, especially in their hypocotyls, whereas Col-0 seedlings were not stained (Fig. 2B). To examine this result further, we extracted lipids from seedlings and separated them by TLC (Fig. 2C). Col-0 seedlings germinated on sugar-free medium lost most of the triacylglycerol (TAG) stored in seeds 4 days after imbibition, and degradation of seed TAG was delayed by sucrose in the medium (19). Although KK seedlings germinated on sugar-free medium still contained TAG 4 days after imbibition, most of the TAG was lost 9 days after imbibition. KK seedlings germinated on 1% sucrose medium contained large amounts of TAG between 4 and 9 days after imbibition. These profiles of TAG accumulation in Col-0 and KK seedlings were similar to those of oleosin S4 (Fig. 2A).
Expression of Seed Maturation Genes in KK Seedlings Occurs 4–5 Days After Imbibition.
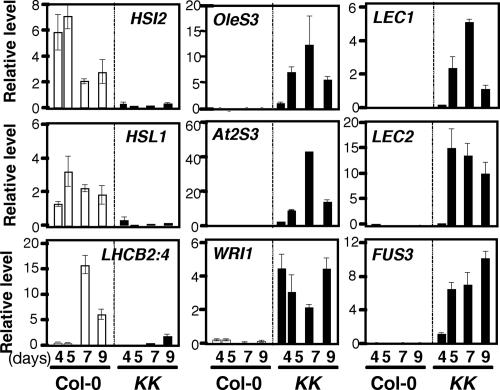
We next examined the changes in levels of various mRNAs in Col-0 and KK seedlings after day 4 postimbibition by real-time RT-PCR, with ACTIN2 mRNA as an internal control. Col-0 seedlings germinated on medium containing 1% sucrose expressed mRNAs for HSI2 and HSL1, and the level of mRNA for LHCB2:4, which is involved in photosynthesis, showed a steep increase between days 5 and 7, which is when cotyledon greening occurs (Fig. 3). In KK seedlings grown on medium containing 1% sucrose, expression of LHCB2:4 was severely diminished compared with Col-0 seedlings, and strong expression of seed maturation genes was observed (Fig. 3). Although OleS3 and At2S3 mRNAs were already >30-fold higher in 4-day-old KK seedlings compared with Col-0 seedlings (SI Table 1), the level of these mRNAs continued to increase until day 7. We also observed dramatic increases in the levels of mRNAs for LEC1, LEC2, and FUS3 from day 4 to 5. The timing of expression of these master regulators of seed development was similar to the appearance of swollen hypocotyls (Fig. 1C). On the other hand, the level of WRI1 mRNA was already high in KK seedlings at least at day 4 after imbibition. Unlike the other seed maturation genes examined here, WRI1 was expressed in vegetative tissues of Col-0 seedlings, albeit at levels lower than in siliques (17, 18).
Fig. 3.
Expression of HSI2, HSL1, and seed maturation genes in Col-0 and KK seedlings. Total RNAs were isolated from 4-, 5-, 7-, and 9-day-old Col-0 and KK seedlings grown on medium containing 1% sucrose. The levels of various mRNAs were determined by quantitative real-time RT-PCR using ACTIN2 mRNA as an internal reference. Results represent the average from two independent isolations of RNA ± SD.
Sugar-Dependent Expression of Seed Maturation Genes in KK Seedlings.
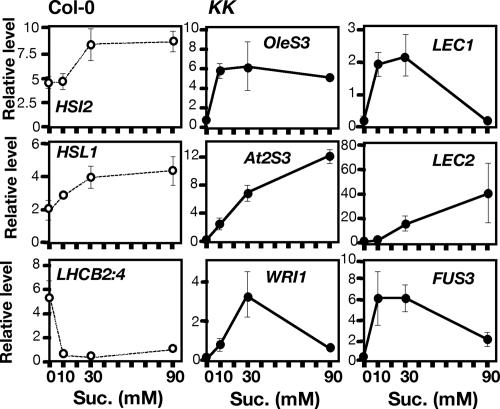
To examine the effects of sugar on the expression of seed maturation genes in KK seedlings, we changed the concentration of sucrose in the medium and isolated RNA from 5-day-old Col-0 and KK seedlings. In Col-0 seedlings, the presence of 30 mM (1.0% wt/vol) to 90 mM (3.1% wt/vol) sucrose enhanced the expression of both HSI2 and HSL1 nearly 2-fold compared with the absence of sucrose, whereas 10 mM sucrose was sufficient to repress the expression of LHCB2:4 (Fig. 4). Expression of six seed maturation genes examined in KK seedlings showed a strong response to sucrose, although with different dose dependences (Fig. 4). Specifically, the expression of OleS3, LEC1, and FUS3 was fully induced by 10 mM sucrose, whereas At2S3 and LEC2 showed a concentration-dependent increase in the level of expression up to 90 mM sucrose. Expression of WRI1 was maximal at 30 mM sucrose under these conditions. These results suggest that sugar-dependent accumulation of seed reserves in KK seedlings requires sugar-dependent expression of various seed maturation genes, which could be mediated directly or indirectly through multiple sugar-signaling pathways (9).
Fig. 4.
Effects of the concentration of sucrose in the medium on the expression of various genes in seedlings. Total RNA was isolated from 5-day-old Col-0 and KK seedlings grown on medium containing 0, 10, 30, and 90 mM sucrose. The levels of various mRNAs were determined by quantitative real-time RT-PCR using ACTIN2 mRNA as an internal reference. Results represent the average of data from two independent isolations of RNA ± SD.
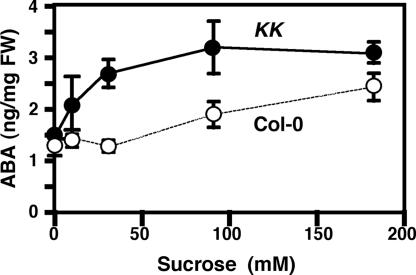
ABA plays an important role in the expression of seed maturation program and the maintenance of seed dormancy (4). Furthermore, the sugar-induced change in gene expression pattern in Arabidopsis seedlings is at least partly mediated by sugar-induced accumulation of ABA (9, 20). We examined the endogenous level of ABA in 7-day-old Col-0 and KK seedlings germinated on medium containing various concentrations of sucrose. In the wild-type Col-0 seedlings, the sugar-induced accumulation of ABA occurs with high concentrations of sugar (20, 21), and 10 or 30 mM sucrose in the medium did not affect the level of ABA (Fig. 5). The level of ABA in KK seedlings germinated in the absence of sucrose was similar to that in Col-0 seedlings. However, KK seedlings germinated on medium containing 10–30 mM sucrose contained higher levels of ABA compared with those germinated on medium without sugar. It seems that ABA plays a role at least partly in the sugar-dependent expression of various seed maturation genes in KK seedlings.
Fig. 5.
Endogenous level of ABA in Col-0 and KK seedlings. The ABA content in 7-day-old Col-0 and KK seedlings germinated on medium containing various concentrations of sucrose was determined. Results represent the average from three independent experiments ± SD.
Hypocotyls of KK Seedlings Exhibit Embryonic Properties.
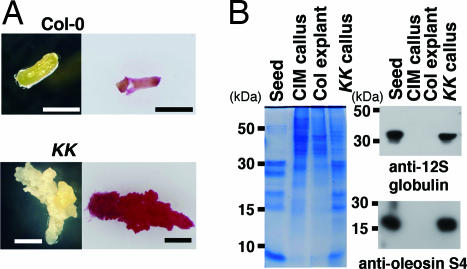
When hypocotyls of 7-day-old KK seedlings were excised and placed on hormone-free medium containing 1% sucrose, all of the 48 excised hypocotyls continued active callus-like growth. These callus-like tissues were intensely stained with fat red 7B (Fig. 6A). None of the excised Col-0 hypocotyls showed callus-like growth. Calluses developed from excised KK hypocotyls (KK callus) showed a profile of protein expression similar to mature seeds, and immunoblot analysis confirmed the presence of a large amount of 12S globulin and oleosin S4 (Fig. 6B). These proteins were not detected in cultured Col-0 hypocotyls (Col explant) or callus derived from Col-0 explants after they were grown on callus-inducing medium (CIM). These results suggest that KK seedlings have embryonic activities.
Fig. 6.
Calluses developed from hypocotyls of KK seedlings. (A) Hypocotyls of 7-day-old Col-0 and KK seedling were excised and cultured on medium containing 1% sucrose for 14 days. (Right) The photographs were taken after staining with fat red 7B. (Scale bar, 1 mm.) (B) Proteins were extracted from Col-0 and KK hypocotyls that had been excised and cultured for 14 days (Col explant and KK callus, respectively), callus from Col-0 explants on CIM (CIM callus), and Col-0 seeds (Seed). Equal amounts of proteins (1 mg) were separated by SDS/PAGE and stained with Coomassie brilliant blue (Left) or analyzed by immunoblotting with anti-12S globulin and anti-oleosin S4 antibodies (Right). The size of the protein molecular mass standards is indicated to the left of the gels.
Discussion
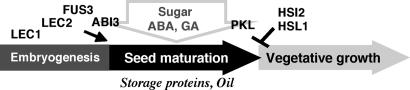
The present work revealed a novel function of HSI2 and HSL1 in preventing the expression of embryonic traits after seed germination (Fig. 7). The pkl mutant develops pickle roots that express embryonic traits at the tips of primary roots, and the appearance of this phenotype is markedly enhanced by inhibitor of GA synthesis (6). PKL is a CHD3 chromatin-remodeling factor (22), and it has been suggested that expression of embryonic genes is turned off during a very early stage of germination (≈48 h after imbibition) in a PKL- and GA-dependent manner, ensuring activation of seedling development (6, 7). In contrast to the pkl mutant, appearance of embryonic traits in KK seedlings occurs without inhibition of GA synthesis, and the presence of GA does not suppress this phenotype. Furthermore, expression of most of embryonic and seed maturation genes in KK seedlings occurred after day 4–5 of seed germination, and degradation of seed reserves took place in KK seedlings grown on sugar-free medium. Thus, whereas PKL is required to turn off embryonic traits during the early stage of germination, HSI2 and HSL1 seem to be required to prevent the expression of embryonic traits after seedling establishment.
Fig. 7.
Schematic illustration of the role of HSI2, HSL1, and other genes in Arabidopsis seed development.
The appearance of a swollen hypocotyl phenotype and the accumulation of seed reserves in KK seedlings depended on the presence of metabolizable sugar in the medium. The fact that the expression of various seed maturation genes in KK seedlings changed in response to the concentration of sucrose in the medium suggests that sugar is required not only as a source of energy and carbon skeleton for reserve synthesis but also as a signal for activating the expression of seed maturation genes. The sugar-hypersensitive response of ABA suggests that ABA could be involved in these sugar-dependent processes in KK seedlings. HSI2 functions as an active transcriptional repressor of the sugar-inducible reporter gene (10), and expression of HSI2 and HSL1 in Col-0 seedlings was up-regulated by sucrose. We therefore suggest that HSI2 and HSL1 prevent ectopic expression of seed maturation genes in cells that encounter high-sugar status during vegetative growth. In particular, because ectopic expression of LEC1 and LEC2 can provoke the embryonic program (2, 5), derepression of these master regulatory genes during the vegetative phase of plants must be strictly avoided. Identification of direct targets of HSI2 and HSL1 is essential to understand the mechanisms involved in the repression of seed maturation genes during seedling growth.
Our finding of sugar-dependent expression of a variety of embryonic genes including LEC1, LEC2, FUS3, and WRI in KK seedlings is intriguing in light of the emerging role of sugar signaling in seed development. Spatial and temporal transitions in sugar status play important roles in legume seed development (8), and sugar-dependent regulation of expression of many genes in seedlings and other vegetative tissues has been well characterized (9, 23). However, the role of sugar signaling in the regulation of gene expression during seed maturation is not well established. Further analysis of the role of sugar on the expression of seed maturation genes in KK seedlings might shed new light on the impact of sugar signaling in seed development.
Because of the agronomic and industrial importance of seed oils, much effort has been devoted to the qualitative and quantitative improvement of oil production in seeds. In KK mutants, seed oil accumulates in hypocotyls and in callus tissues derived from hypocotyls, which could provide a new way for producing plant seed oils in nonseed tissues.
Materials and Methods
Plant Materials and Growth Conditions.
Arabidopsis thaliana (L.) Heynh. ecotype Columbia (Col-0) was used as the wild-type plant. The hsi2-2 mutant with a T-DNA insertion in HSI2 has been described previously as Δhsi2 (10). The hsl1-1 mutant with a T-DNA insertion in HSL1, formerly HSI2-L1 (10), was obtained from the sequence-tagged T-DNA insertion line SALK_059568 (Salk Institute Genomic Analysis Laboratory, La Jolla, CA). Unless otherwise indicated, seeds were sterilized in sterile water, kept at 4°C for 4 days in the dark, and sown on 0.3% (wt/vol) gellan gum plates containing Murashige and Skoog medium (Wako, Tokyo, Japan) (pH 5.7) supplemented with 100 mg/liter myo-inositol, 10 mg/liter thiamine HCl, 1 mg/liter nicotinic acid, 1 mg/liter pyridoxine HCl, and 1% sucrose. Plates were incubated in a growth chamber at 22°C under continuous fluorescent light at an intensity of 65 μmol m−2 s−1. CIM contained 2.3 μM 2,4-dichlorophenoxy acetic acid and 0.46 μM N6-furfuryladenine (24).
Microscopy.
Five-day-old seedlings were fixed in 1:1:18:20 (vol/vol) formaldehyde/acetic acid/ethanol/H2O for 16 h at 4°C, dehydrated in a graded series of ethanol (50–99.5%), and embedded in Technovit 7100 resin (Kulzer, Wehrheim, Germany) according to the manufacturer's instructions. The samples were cut into 5-μm sections with a tungsten knife on an RM 2125RT rotary microtome (Leica, Vienna, Austria) and dried on coverslips. The sections were stained with 0.05% toluidine blue (Merck, Darmstadt, Germany) for 1 min at room temperature and washed in distilled water for 5 min before observation with a BX60 microscope (Olympus, Tokyo, Japan).
Quantitative Real-Time RT-PCR.
Total RNA was isolated from plants by using an RNeasy plant mini kit (Qiagen, Valencia, CA), and levels of various mRNAs were determined by quantitative real-time RT-PCR using an iCycler iQ with iQ SYBR green supermix (Bio-Rad, Hercules, CA) as described previously (25). The primer sets for HSI2, HSL1, and ACTIN2 mRNAs were described by Tsukagoshi et al. (10); those for OleS3, At2S3, LEC1, and LEC2 mRNAs were described by Mendoza et al. (26); and that for WRI1 mRNA was described by Masaki et al. (18). The primer set for LHCB2:4 mRNA was 5′-GTAAAGGTCCGATCGAAAATCTGT-3′ and 5′-TTATCCGATCAAACTCTATTTTCCG-3′. For each pair of primers, a melting-curve analysis of the PCR product was performed to confirm that the PCR produced only one fragment. The threshold cycles at which the fluorescence of the PCR product–SYBR green complex first exceeded the background level were determined, and the relative template concentration to control was determined based on the standard curve for each transcript. The level of each mRNA was normalized by the level of ACTIN2 mRNA.
Microarray Analysis.
Total RNA was isolated from the 4-day-old seedlings of Col-0 and the KK mutant. Cy3- and Cy5-labeled cDNA probes were synthesized and hybridized to the Arabidopsis 3 oligonucleotide microarray (Agilent Technologies, Palo Alto, CA) according to the manufacturer's instructions. The microarray analysis was performed with two independently isolated RNA samples. Feature extraction and image analysis software (Agilent Technologies) was used to locate and delineate each spot in the array and to integrate each spot's intensity, filtering, and normalization.
SDS/PAGE and Immunoblot Analysis.
SDS/PAGE and immunoblot analysis were carried out by using antibodies specific for 12S globulin and oleosin S4 as described by Shimada et al. (27).
Staining with Fat Red 7B.
Tissues were stained with a 0.1% (wt/vol) solution of fat red 7B (Sigma, St. Louis, MO) as described by Brundrett et al. (28). Tissue samples after staining were examined under a SZX12 stereomicroscope (Olympus).
ABA and Lipid Analyses.
ABA in seedlings was determined as described previously (10). For the identification of TAG, four seedlings or seeds were homogenized in 1 ml of 2:1 (vol/vol) chloroform/methanol (2:1, vol/vol) and centrifuged at 20,000 × g for 5 min. The supernatants were lyophilized, and the remaining materials were redissolved in 20 μl of chloroform. Lipid samples were spotted on a Silica Gel 60 F254 HPTLC sheet (Merck) and developed with 4:1:0.05 (vol/vol) hexane/diethylether/acetic acid. Glyceryl trilinoleate (Sigma) was run as a TAG standard. Lipids were visualized by spraying the plate with sulfuric acid and heating at 120°C for 5 min.
Supplementary Material
Acknowledgments
We thank S. Ishiguro and T. Hattori of Nagoya University for helpful discussions, T. Shimada and I. Nishimura of Kyoto University for antibodies specific to 12S globulin and oleosin S4, and the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutant. This work was supported in part by the 21st Century Centers of Excellence program from the Ministry of Education, Science, Sports, and Culture of Japan (to K.N.). H.T. was supported by a Research Fellowship from the Japan Society for the Promotion of Science for Young Scientists.
Abbreviations
- ABA
abscisic acid
- ABI3
ABSCISIC ACID-INSENSITIVE 3
- CIM
callus-inducing medium
- EAR
ERF-associated amphiphilic repression
- FUS3
FUSCA 3
- GA
gibberellin
- HSI2
HIGH-LEVEL EXPRESSION OF SUGAR-INDUCIBLE GENE 2
- HSL1
HSI2-Like 1
- HSL2
HSI2-Like 2
- LEC
LEAFY COTYLEDON
- PKL
PICKLE
- TAG
triacylglycerol
- WRI1
WRINKLED1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: Microarray data reported in this paper have been deposited in the public microarray database (www.ebi.ac.uk/arrayexpress) with accession no. E-MEXP-542.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607940104/DC1.
References
- 1.Harada JJ. In: Cellular and Molecular Biology of Plant Seed Development. Larkins BA, Vasil IK, editors. The Netherlands: Kluwer, Dordrecht; 1997. pp. 545–592. [Google Scholar]
- 2.Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. Cell. 1998;93:1195–1205. doi: 10.1016/s0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- 3.Harada JJ. Curr Opin Plant Biol. 1999;2:23–27. doi: 10.1016/s1369-5266(99)80005-3. [DOI] [PubMed] [Google Scholar]
- 4.Vicente-Carajosa J, Caronerd J. Int J Dev Biol. 2005;49:645–651. doi: 10.1387/ijdb.052046jc. [DOI] [PubMed] [Google Scholar]
- 5.Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ. Proc Natl Acad Sci USA. 2001;98:11806–11811. doi: 10.1073/pnas.201413498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogas J, Cheng JC, Sung ZR, Somerville C. Science. 1997;277:91–94. doi: 10.1126/science.277.5322.91. [DOI] [PubMed] [Google Scholar]
- 7.Li HC, Chuang K, Henderson JT, Rider SD, Bai Y, Zhang H, Fountain M, Gerber J, Ogas J. Plant J. 2005;44:1010–1022. doi: 10.1111/j.1365-313X.2005.02602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber H, Borisjuk L, Wobus U. Annu Rev Plant Biol. 2005;56:253–279. doi: 10.1146/annurev.arplant.56.032604.144201. [DOI] [PubMed] [Google Scholar]
- 9.Rolland F, Baena-Gonzalez E, Sheen J. Annu Rev Plant Biol. 2006;57:675–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- 10.Tsukagoshi H, Saijo T, Shibata D, Morikami A, Nakamura K. Plant Physiol. 2005;138:675–685. doi: 10.1104/pp.104.057752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohta M, Matui K, Hiratsu K, Shinshi H, Ohme-Takagi M. Plant Cell. 2001;13:1959–1968. doi: 10.1105/TPC.010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim HU, Hsieh K, Ratnayake C, Huang AH. J Biol Chem. 2002;277:22677–22684. doi: 10.1074/jbc.M109298200. [DOI] [PubMed] [Google Scholar]
- 13.Pang PP, Pruitt RE, Meyerowits EM. Plant Mol Biol. 1988;11:805–820. doi: 10.1007/BF00019521. [DOI] [PubMed] [Google Scholar]
- 14.Krebbers E, Herdies L, Clercq AD, Seurinck J, Leemans J, Damme J, Seura M, Gheysen G, Montagu M, Vandekerckhove J. Plant Physiol. 1988;87:859–866. doi: 10.1104/pp.87.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ. Plant Cell. 2003;15:5–18. doi: 10.1105/tpc.006973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bensmihen S, Rippa S, Lambert G, Jublot D, Pautot V, Granier F, Giraudat J, Parcy F. Plant Cell. 2002;14:1391–1403. doi: 10.1105/tpc.000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cernac A, Benning C. Plant J. 2004;40:575–585. doi: 10.1111/j.1365-313X.2004.02235.x. [DOI] [PubMed] [Google Scholar]
- 18.Masaki T, Mitsui N, Tsukagoshi H, Nishii T, Morikami A, Nakamura K. Plant Cell Physiol. 2005;46:547–556. doi: 10.1093/pcp/pci072. [DOI] [PubMed] [Google Scholar]
- 19.To JPC, Reiter WD, Gibson SI. BMC Plant Biol. 2002;2:4–14. doi: 10.1186/1471-2229-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, León P. Genes Dev. 2000;14:2085–2096. [PMC free article] [PubMed] [Google Scholar]
- 21.Price J, Li T-C, Kang SG, Na JK, Jang J-C. Plant Physiol. 2003;132:1424–1438. doi: 10.1104/pp.103.020347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogas J, Kaufmann S, Henderson J, Somerville C. Proc Natl Acad Sci USA. 1999;96:13839–13844. doi: 10.1073/pnas.96.24.13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koch KE. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:509–540. doi: 10.1146/annurev.arplant.47.1.509. [DOI] [PubMed] [Google Scholar]
- 24.Yasutani I, Ozawa S, Nishida T, Sugiyama M, Komamine A. Plant Physiol. 1994;105:815–822. doi: 10.1104/pp.105.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoine M, Ohto M, Onai S, Mita S, Nakamura K. Plant J. 2006;47:49–62. doi: 10.1111/j.1365-313X.2006.02771.x. [DOI] [PubMed] [Google Scholar]
- 26.Mendoza M, Dubreucq M, Miquel M, Caboche M, Lepiniec L. FEBS Lett. 2005;579:4666–4670. doi: 10.1016/j.febslet.2005.07.037. [DOI] [PubMed] [Google Scholar]
- 27.Shimada T, Yamada K, Kataoka M, Nakaune S, Koumoto Y, Kuroyanagi M, Tabata S, Kato T, Shinozaki K, Seki M, et al. J Biol Chem. 2003;278:32292–32299. doi: 10.1074/jbc.M305740200. [DOI] [PubMed] [Google Scholar]
- 28.Brundrett MC, Kendrick B, Peterson CA. Biotech Histochem. 1991;66:111–116. doi: 10.3109/10520299109110562. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.