Abstract
Tocopherols (vitamin E) are lipophilic antioxidants that are synthesized by all plants and are particularly abundant in seeds. Two tocopherol-deficient mutant loci in Arabidopsis thaliana were used to examine the functions of tocopherols in seedlings: vitamin e1 (vte1), which accumulates the pathway intermediate 2,3-dimethyl-5-phytyl-1,4-benzoquinone (DMPBQ); and vte2, which lacks all tocopherols and pathway intermediates. Only vte2 displayed severe seedling growth defects, which corresponded with massively increased levels of the major classes of nonenzymatic lipid peroxidation products: hydroxy fatty acids, malondialdehyde, and phytoprostanes. In the absence of pathogens, the phytoalexin camalexin accumulated in vte2 seedlings to levels 100-fold higher than in wild-type or vte1 seedlings. Similarly, gene expression profiling in wild-type, vte1, and vte2 seedlings indicated that increased levels of nonenzymatic lipid peroxidation in vte2 corresponded to increased expression of many defense-related genes, which were not induced in vte1. Both biochemical and transcriptional analyses of vte2 seedlings indicate that nonenzymatic lipid peroxidation plays a significant role in modulating plant defense responses. Together, these results establish that tocopherols in wild-type plants or DMPBQ in vte1 plants limit nonenzymatic lipid peroxidation during germination and early seedling development, thereby preventing the inappropriate activation of transcriptional and biochemical defense responses.
INTRODUCTION
Tocopherols, which collectively constitute the essential dietary micronutrient vitamin E, are lipophilic antioxidants synthesized exclusively by plants, algae, and some cyanobacteria. There are four naturally occurring forms of tocopherols, α, β, γ, and δ, which differ in the presence and position of methyl groups on the chromanol ring. Because tocopherols are essential for human nutrition, tocopherol functions and chemistry have been studied most extensively in animal systems and in vitro. Although the biological activities of tocopherols may differ between plants and animals, the chemical properties of tocopherols are presumed to be the same in both kingdoms. Tocopherols scavenge a variety of reactive oxygen species (ROS) and free radicals and can quench singlet oxygen (Fukuzawa et al., 1982; Brigelius-Flohe and Traber, 1999; Wang and Quinn, 2000). The chromanol ring of tocopherol allows the donation of a hydrogen atom while maintaining a resonance-stabilized configuration (Liebler, 1993; KamalEldin and Appelqvist, 1996); other aromatic antioxidants must donate two hydrogen atoms to attain a stable state (Liebler and Burr, 2000). Donation of a hydrogen atom generates a tocopherol radical, which can be recycled back to the original tocopherol by direct interaction with ascorbate or another reductant (Liebler, 1993).
Although water-soluble antioxidants such as ascorbate and glutathione also scavenge ROS, in vitro and animal studies have established that tocopherols have the unique ability to protect polyunsaturated fatty acid (PUFA) acyl chains from oxidative damage because they are localized within membranes (reviewed in Schneider, 2005). ROS-generated radicals readily attack the double bonds of PUFAs, remove an electron, and create a lipid radical, which reacts with oxygen to form a lipid peroxyl radical (Figure 1A). In the densely packed environment of lipid bilayers or other lipid aggregates, lipid peroxyl radicals will in turn attack neighboring PUFAs, propagating a chain reaction of lipid peroxidation through the membrane (Figure 1A). Tocopherols scavenge lipid peroxyl radicals by donating a hydrogen atom to these radicals, converting them to lipid peroxides and thereby blocking the propagation of lipid peroxidation in membranes (Figure 1A). The lipid peroxides produced are either enzymatically or spontaneously converted to relatively inert hydroxy fatty acids or other products in vivo.
Figure 1.
Overview of Nonenzymatic Lipid Peroxidation and Tocopherol Scavenging.
(A) In step 1, free radicals attack the double bonds of PUFA acyl chains, creating a lipid radical that readily reacts with an oxygen molecule, forming a lipid peroxyl radical. In step 2, in the absence of tocopherols or other lipid-soluble antioxidants, such as DMPBQ in vte1, the lipid peroxyl radical can attack neighboring PUFAs, propagating a chain reaction. In step 3, tocopherols can scavenge lipid peroxyl radicals by donation of a hydrogen atom, thereby reducing the radical and blocking further propagation. In step 4, a linolenic acid (18:3) peroxyl radical can also react internally, forming a cyclic peroxyl radical, which spontaneously reacts with a second oxygen molecule and is subsequently reduced to phytoprostane G1 (PPG1). In step 5, PPG1 either spontaneously decays, forming MDA and other alkanes and alkenes, or forms other phytoprostanes. In step 6, lipid peroxides are converted to their corresponding hydroxy fatty acid either enzymatically or spontaneously.
(B) PPG1 spontaneously rearranges, forming PPD1, PPE1, and PPF1. PPD1 and PPE1 in turn rearrange to form dPPJ1 and PPA1, respectively. PPA1 can further rearrange to form PPB1. There are multiple stereoisomers and enantiomers for each phytoprostane family (e.g., PPE1, PPF1) that are not shown. Note the structural similarity between dPPJ1 and the enzymatically derived oxylipins jasmonic acid (JA) and its precursor 12-oxophytodienoic acid (OPDA).
Although a good deal is known about the chemistry and functions of tocopherols in animal systems and in vitro, much less is known about their functions in photosynthetic organisms (Fryer, 1992; Munne-Bosch and Alegre, 2002). Some important insights have been made in recent years from the analysis of tocopherol biosynthetic mutants in the cyanobacterium Synechocystis sp PCC6803 (Maeda et al., 2005; Sakuragi et al., 2006) and from the treatment of Chlamydomonas reinhardtii with an herbicide that disrupts both tocopherol and plastoquinone synthesis (Trebst et al., 2002, 2004). However, neither organism is multicellular, and understanding tocopherol functions in plants has only been possible with the isolation of mutants disrupting tocopherol synthesis in Arabidopsis thaliana, including vitamin e1 (vte1; tocopherol cyclase) and vte2 (homogentisate phytyl transferase) (Porfirova et al., 2002; Sattler et al., 2003, 2004; DellaPenna and Pogson, 2006). vte1 and vte2 mutants both lack tocopherols, but vte1 mutants accumulate the tocopherol pathway intermediate 2,3-dimethyl-5-phytyl-1,4-benzoquinone (DMPBQ), which lacks the characteristic chromanol ring of tocopherols but is still redox-active (Sattler et al., 2003, 2004). DMPBQ levels in vte1 mutant tissues are comparable to tocopherol levels in wild-type tissues, and DMPBQ is below the limit of detection in all wild-type and vte2 tissues (Sattler et al., 2003, 2004).
The vte mutants have allowed several groups to initiate studies of tocopherol functions in plants. Because tocopherols are only synthesized by photosynthetic organisms, are localized in chloroplasts, and accumulate during both abiotic and biotic stresses, it has been assumed that tocopherols play a central role in protecting chloroplast membranes from ROS generated by excessive light energy (Fryer, 1992; Munne-Bosch and Alegre, 2002). Surprisingly, mature vte1 and vte2 plants were indistinguishable from wild-type plants both under normal growth conditions (Porfirova et al., 2002; Sattler et al., 2003, 2004; Maeda et al., 2006) and during high light stress (Porfirova et al., 2002; Havaux et al., 2005; Maeda et al., 2006). It was only during extreme abiotic stress, when low temperature was combined with high light, that measurable physiological differences were observed between vte1 and wild-type plants (Havaux et al., 2005). Tocopherols have since been shown to play essential roles in low-temperature adaptation (Maeda et al., 2006), a response that is independent of light level. Thus, many of the functions deduced for tocopherols by comparing the responses of vte1 and wild-type plants with combined high-light and low-temperature treatment (Havaux et al., 2005) were likely attributable to the loss of low-temperature tocopherol functions (Maeda et al., 2006). These combined data suggest that the role of tocopherols in the response to high light is more limited than has been assumed in mature leaves.
In addition to temperature adaptation, tocopherols have also been show to play critical roles in seed and seedling physiology; both vte1 and vte2 have significantly reduced seed longevity compared with wild-type plants (Sattler et al., 2004). During germination, vte2 seedlings also displayed a range of defects, including impaired root growth, cotyledon expansion defects, and slowed storage lipid catabolism, all of which were correlated with dramatic increases in nonenzymatic lipid peroxides and the corresponding hydroxy fatty acids (Sattler et al., 2004). The vte2 mutant clearly demonstrates that an essential function of tocopherols is to control lipid peroxidation during germination and early seedling development. The observation that germinating vte1 seedlings were nearly identical to wild-type seedlings suggested that DMPBQ, which accumulates in vte1, can functionally replace tocopherol in this regard (Sattler et al., 2004).
In living organisms, nonenzymatic lipid peroxidation is a natural, continually occurring process, but under oxidative stress conditions, which enhance ROS and radical production, the levels of nonenzymatic lipid peroxidation can increase dramatically. In plants, nonenzymatic lipid peroxidation is known to increase during many abiotic stresses (Baryla et al., 2000) and in response to both bacterial and fungal pathogen attack (Muckenschnabel et al., 2001; Thoma et al., 2003). Linoleic (18:2) and linolenic (18:3) acids are highly abundant in plant membrane lipids and are the main reactants in nonenzymatic lipid peroxidation (Berger et al., 2001; Mueller, 2004; Weber et al., 2004). Peroxyl radicals, which are unstable and highly reactive, are formed on PUFA acyl chains and undergo a series of reactions resulting in at least three distinct classes of products (Figure 1) (Porter et al., 1995). The lipid peroxyl radical can be reduced to the corresponding hydroxy fatty acid, fragmented into a variety of reactive aldehydes including malondialdehyde (MDA), nonenal, hexanal, and hexenal, or cyclized to form a large class of compounds known as isoprostanes (Figure 1; phytoprostanes in plants) (Mueller, 1998). The initial phytoprostane formed in the membrane is PPG1, but it is highly unstable and either spontaneously rearranges to form other phytoprostanes or fragments, releasing MDA (Figure 1A). 18:2 peroxyl radicals primarily form hydroxy fatty acids and fatty acid fragments, whereas 18:3 peroxyl radicals form all three classes of compounds (Mueller, 2004; Weber et al., 2004).
Nonenzymatic lipid peroxidation products can also be separated into two groups based on chemical functionality: electrophiles that contain highly reactive α/β unsaturated carbonyl groups (Vollenweider et al., 2000; Stintzi et al., 2001; Almeras et al., 2003; Weber et al., 2004), and less reactive classes lacking α/β unsaturated carbonyl groups. Electrophiles, which include MDA and some of the phytoprostanes, can have deleterious effects on plant cells by covalently modifying thiol groups of peptides as well as reacting with other cellular components. Moreover, most nonenzymatic lipid peroxidation products initially accumulate within membranes and can disrupt the selective permeability of the lipid bilayer (Mueller, 2004).
The ability of enzyme-derived lipid peroxidation products, the oxylipins (JA and OPDA) in plants (Figure 1B) and the eicosanoids (prostaglandins, leukotrienes, and thromboxanes) in animals, to act as signals and to regulate gene expression is well established (reviewed in Funk, 2001; Creelman and Mulpuri, 2002; Howe and Schilmiller, 2002). There is a growing body of evidence that certain nonenzymatic lipid peroxidation products can similarly act as signals and regulate gene expression in both the plant and animal kingdoms. In animals, nonenzymatic lipid peroxidation products, including isoprostanes, have been shown to act at low levels as signaling molecules that modulate the function of the immune and vascular systems (Bochkov et al., 2002; Hazen and Chisolm, 2002; Traidl-Hoffmann et al., 2005), whereas high levels are clearly detrimental (Leitinger, 2003). In plants, externally applied MDA and specific phytoprostanes (PPB1, PPA1, dPPJ1, and PPE1) have been shown to induce the expression of several stress-related genes (Vollenweider et al., 2000; Almeras et al., 2003; Thoma et al., 2003; Weber et al., 2004; Iqbal et al., 2005; Loeffler et al., 2005). To better understand the roles of tocopherols in plants, we examined nonenzymatic lipid peroxidation products in vte1 and vte2 and the effects of these mutations on global gene expression. These two mutants allow us to delineate the direct functions of tocopherols from those as a lipophilic antioxidant and to address how the absence of the plant's main lipophilic antioxidant affects gene expression and stress responses in Arabidopsis seedlings.
RESULTS
MDA Analysis
In a prior study, lipid-soluble peroxides and hydroxy octadecadienoic fatty acids (9- and 13-HODEs derived from 18:2) were found to be increased up to 5- and 100-fold, respectively, in germinating vte2 seedlings compared with wild-type or vte1-1 seedlings (Sattler et al., 2004). The stereoisomeric and enantiomeric compositions of the HODEs detected in vte2 seedlings were indicative of high levels of nonenzymatic lipid peroxidation (Sattler et al., 2004). However, hydroxy octadecatrienoic acids (HOTEs; 18:3-derived hydroxy fatty acids) were ∼20-fold less abundant than HODEs in vte2 seedlings (Sattler et al., 2004), even though 18:2 and 18:3 fatty acid levels in Arabidopsis seed are nearly equal in abundance and 18:3 autooxidizes far more readily than 18:2 in vitro and in vivo. Thus, the question remained, is 18:3 not being oxidized in vte2 seedlings, or are products other than HOTEs being formed?
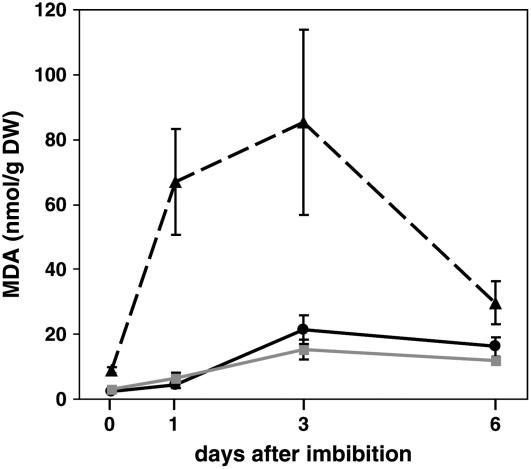
To address this issue, we measured a broader spectrum of nonenzymatic lipid peroxidation products in germinating seedlings. PUFA peroxyl radicals formed in the membrane during lipid peroxidation can subsequently decompose, resulting in an instantaneous release of MDA and other volatile alkane and alkene compounds. In Arabidopsis leaves, ∼75% of the MDA is formed from 18:3 (Weber et al., 2004). Total MDA levels in 0-, 1-, 3-, and 6-d-old seedlings were measured using gas chromatography–mass spectrometry (GC-MS) and found to be significantly increased in vte2 seedlings at all time points relative to wild-type or vte1 seedlings (Figure 2). In vte2 seedlings, MDA levels increased dramatically at 1 d to levels 14-fold higher than in either wild-type or vte1 seedlings. MDA levels peaked at 3 d in vte2 at levels fourfold higher than in either wild-type or vte1 seedlings. These data underscore the importance of tocopherols (or DMPBQ in vte1) in preventing the formation MDA during germination, an oxidatively intense period of the plant life cycle.
Figure 2.
MDA Levels in Germinating Seedlings.
Total (free and bound) MDA levels were measured by GC-MS (see Methods) in 0-, 1-, 3-, and 6-d-old seedlings of wild-type Columbia (solid black line), vte1-1 (solid gray line), and vte2-1 (dashed black line). Error bars represent sd (n = 6). DW, dry weight.
Phytoprostane Analysis
Another major group of nonenzymatic lipid peroxidation products, the phytoprostanes, are formed from 18:3 when linolenic peroxyl radicals cyclize to form PPG1. Spontaneous rearrangements result in the numerous species of phytoprostanes found in plant tissues (Figure 1B) (Mueller, 2004). We analyzed the formation of total F1-phytoprostanes (free and esterified PPF1) in 0-, 1-, 3-, and 6-d-old seedlings, because PPF1 are a class of nonreactive, chemically stable phytoprostane end products that are excellent markers for nonenzymatic lipid peroxidation (Imbusch and Mueller, 2000a). Similar to the results for MDA (Figure 2), lipid peroxides, and HODEs (Sattler et al., 2004), total PPF1 levels in wild-type and vte1 seedlings (Figure 3) were low and comparable to levels observed in mature leaves (Imbusch and Mueller, 2000a). By contrast, even in dry vte2 seeds collected immediately from plants, PPF1 levels were already increased (4.78 nmol/g seed) compared with those in the wild type, which were below the limit of detection (0.03 nmol/g seed). PPF1 levels in vte2 seeds increased almost ninefold after 5 d of imbibition at 6°C (42.5 nmol/g dry weight at 0 d of growth). At 1, 3, and 6 d of growth, PPF1 levels were 100- to 200-fold higher in vte2 relative to wild-type or vte1 seedlings, reaching a maximum level at 3 d before beginning to decline. At later time points, 14 and 35 d, the PPF1 levels in mature vte2 plants were similar to those in wild-type plants (Figure 3B), suggesting that PPF1 is turned over or that its production is limited by tocopherol-independent mechanisms in mature leaves. These data are consistent with a previous report showing that lipid peroxide levels in vte2 approached wild-type and vte1 levels by 18 d of growth (Sattler et al., 2004).
Figure 3.
Phytoprostane Levels in Germinating Seedlings.
(A) Total (free and esterified) phytoprostane F1 (PPF1) levels were measured by GC-MS (see Methods) in 0-, 1-, 3-, and 6-d-old seedlings of wild-type Columbia (solid black line), vte1-1 (solid gray line), and vte2-1 (dashed black line). The vte1-1 and Columbia levels are indistinguishable.
(B) Total (free and esterified) PPF1 levels were also measured in leaves of 14- and 35-d-old Columbia wild-type (black bars) and vte2-1 (white bars) plants.
Note that the y axis in (A) and (B) differs by 25-fold. Error bars represent sd (n = 3). DW, dry weight.
The extremely high phytoprostane levels in 3- and 6-d-old vte2 seedlings explain the relative differences in abundance between HODEs (derived from 18:2) and HOTEs (derived from 18:3), because 18:3 and HOTEs are prone to undergo double oxygenation as well as cyclization reactions after free radical attack. Therefore, phytoprostanes, MDA, and other oxidized lipids are formed instead of HOTEs in the absence of tocopherols (or DMPBQ in vte1) in vte2 seeds and seedlings.
JA, OPDA, and Oxylipin Analysis
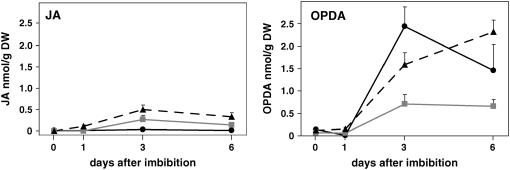
Enzymatically driven oxidation of 18:3 can produce a number of oxylipins, including the phytohormone JA and its precursor OPDA. To determine the levels of JA or OPDA in vte seedlings, quantitative analysis of jasmonates was performed by GC-MS on 0-, 1-, 3-, and 6-d-old seedlings. Free OPDA levels were similar in vte2 and wild-type seedlings at all time points but were twofold to fourfold lower in vte1 than in the wild type at 3 and 6 d (Figure 4). In the wild type, JA levels were near or below the detection limit (2 to 4 pmol/g dry weight) for all time points (Figure 4), and they were increased in both vte1 and vte2 relative to the wild type at 3 and 6 d. Although these results suggest that tocopherol deficiency modestly induces JA accumulation, the maximal JA level in vte2 of 0.49 nmol/g dry weight (corresponding to ∼38 pmol/g fresh weight) is still severalfold lower than the 171 pmol/g fresh weight in unwounded Arabidopsis leaf tissue and 200 times lower than the 8570 pmol/g fresh weight induced by wounding Arabidopsis leaf tissue (Weber et al., 1997).
Figure 4.
JA and OPDA Levels in Germinating Seedlings.
JA and OPDA levels were measured by GC-MS (see Methods) in 0-, 1-, 3-, and 6-d-old seedlings of wild-type Columbia (solid black lines), vte1-1 (solid gray lines), and vte2-1 (dashed black lines). Error bars represent sd (n = 3). DW, dry weight.
An oxylipin profiling method (Weber et al., 1997) was also applied to vte2-1 seedlings to assess the level of other known enzymatically generated oxylipins and to survey for the presence of potentially novel oxylipins in addition to JA and OPDA. This method is able to detect >20 different oxylipins as well as other stress-related compounds (Mueller et al., 2006). Only four compounds, none of which is an oxylipin, were highly increased in vte2 relative to the wild type and vte1. These compounds were two free fatty acids, oleic (18:1) and eicosenoic (20:1) acids, the phytoalexin camalexin, and its putative biosynthetic precursor indole 3-carboxylic acid. The levels of both 18:1 and 20:1 were ∼20-fold higher in 3- and 6-d-old vte2 seedlings relative to wild-type or vte1 seedlings.
Camalexin in vte2
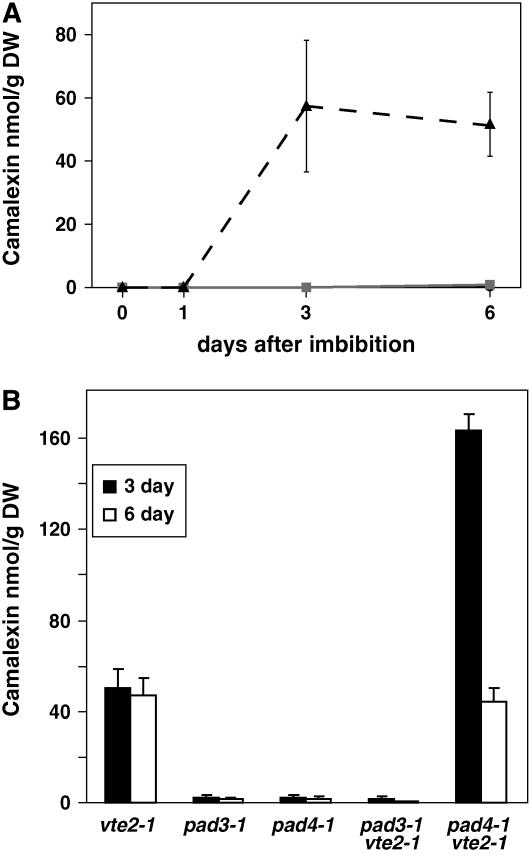
Camalexin in seedlings was quantified using an HPLC-based method. Camalexin was below the detection limit (0.5 nmol/g dry weight) in wild-type and vte1 seedlings at all time points and at 0 and 1 d in vte2. Camalexin levels in vte2 increased to ∼50 nmol/g dry weight at 3 and 6 d, a >100-fold increase relative to the wild type or vte1 (Figure 5A). Generally, camalexin is produced in response to plant pathogen attack, but its synthesis can also be induced by treating plants with redox-active heavy metals such as silver, copper, or iron (Mert-Turk et al., 2003; Glawischnig et al., 2004; Loeffler et al., 2005; Schuhegger et al., 2006). However, all seedlings were grown on sterile Murashige and Skoog medium, and pathogens or exogenous heavy metals were not present.
Figure 5.
Camalexin Levels in Germinating Seedlings.
(A) Camalexin levels were measured by HPLC (see Methods) in 0-, 1-, 3-, and 6-d-old seedlings of wild-type Columbia (solid black line), vte1-1 (solid gray line), and vte2-1 (dashed black line).
(B) Camalexin was measured in 3-d-old (white bars) and 6-d-old (black bars) seedlings of vte2-1, pad3-1, pad4-1, and the pad3 vte2 and pad4 vte2 double mutants.
Error bars represent sd (n = 4). DW, dry weight.
To further examine the basis of camalexin production in vte2 seedlings, double mutants between vte2 and two camalexin-deficient mutants, phytoalexin-deficient3 (pad3) and pad4, were isolated. PAD3 is a cytochrome P450 protein in the camalexin biosynthetic pathway (Zhou et al., 1999; Schuhegger et al., 2006), whereas PAD4 has similarity to triacylglycerol lipases and is involved in defense signaling leading to camalexin biosynthesis (Jirage et al., 1999). As expected, 3- and 6-d-old pad3, pad4, and pad3 vte2 seedlings lacked camalexin (Figure 5B). However, pad4 vte2 seedlings contained camalexin at levels threefold higher than vte2 seedlings at 3 d and at levels comparable to vte2 at 6 d (Figure 5B), indicating that the induction of camalexin synthesis is PAD4-independent in vte2 seedlings. The presence of camalexin in vte2 seedlings in the absence of pathogen attack suggests that the high level of nonenzymatic lipid peroxidation in vte2 seedlings triggers a PAD4-independent signaling pathway leading to camalexin synthesis. This result suggests that tocopherols play a role in regulating biotic stress responses, such as camalexin synthesis, by modulating the level of nonenzymatic lipid peroxidation.
Expression Profiling in the vte Mutants
To gain further insight into how seedlings respond to the absence of tocopherols and to increased levels of lipid peroxidation products, we examined global gene expression profiles in 1- and 3-d-old vte1, vte2, and wild-type seedlings using an Affymetrix ATH1 GeneChip. The chip contained 22,810 probe sets, representing nearly the entire Arabidopsis genome, and 15,293 probe sets were called present for at least one of the conditions. As a whole, the expression of relatively few genes was either significantly increased or decreased (≥2.5-fold) relative to the wild type in either vte1 or vte2 at 1 or 3 d after imbibition. Our criteria for the expression of a gene to be considered increased (or decreased) relative to the wild type required that the probe set was called present for all three chips of the mutant (or called present in the wild type for decreased) and the signal log ratio was ≥1.3 (or ≤−1.3 for decreased) for all three comparisons of the mutant versus wild-type chips. In vte2 seedlings, the transcript levels for 21 and 160 probe sets were increased relative to the wild type at 1 and 3 d, respectively (see Supplemental Table 1 online). In vte1 seedlings, the transcript levels for 24 and 12 probe sets were increased relative to the wild type at 1 and 3 d, respectively (see Supplemental Table 1 online). In vte2, the transcript levels for 5 and 3 probe sets were decreased relative to the wild type at 1 and 3 d, respectively (see Supplemental Table 2 online). In vte1, the transcript levels for 0 and 6 probe sets were decreased relative to the wild type at 1 and 3 d, respectively (see Supplemental Table 2 online).
Interestingly, there was only a single probe set that was upregulated at 1 d in both vte1 and vte2 relative to the wild type, At5g39410, an expressed gene. There was only a single probe set that was downregulated in both vte1 and vte2 at 3 d, At3g62950, a glutaredoxin-like protein. At 3 d after imbibition, 6 of the 12 probe sets that were upregulated in vte1 were also upregulated in vte2. Genes for all six of these probe sets encode heat shock proteins (HSPs) and correspond to four classes of cytosolic HSPs (17, 70, 81, and 101). This set of six HSPs appears to represent the only genes whose regulation is similarly affected in vte1 and vte2 (i.e., tocopherol deficiency–specific) during early seedling growth and development (see Supplemental Table 1 online).
Curiously, at 1 d, the vte1 global expression pattern was consistent with a transient delay in germination and subsequent development, as had been reported previously in germinating vte1 seedlings (Sattler et al., 2004). Eight of the 24 probe sets upregulated at 1 d in vte1 are annotated as late embryogenesis abundant proteins. This expression profile may indicate a transient delay in vte1 development at 1 d but does not suggest an apparent cause for this delay. Evidence of a developmental delay is no longer apparent by 3 d in vte1.
The most dramatic changes observed in vte2 seedlings were at 3 d after the imbibition treatment. The majority of the 160 upregulated probe sets at day 3 were annotated as responsive to either abiotic or biotic stress. These transcripts encode gene products that are localized in nearly all subcellular compartments, including the cytosol, chloroplast, mitochondria, cell wall, and secretory pathway. Probe sets for 17 transcription factors were upregulated in the 3-d-old vte2 seedlings, including many that are stress-associated, such as NAMs, ZAT12, WRKYs, and DREB2-like (Dong et al., 2003; Rizhsky et al., 2004; Davletova et al., 2005; Vogel et al., 2005). Probe sets for nine HSPs were upregulated (including the six heat shock probe sets also upregulated in vte1), along with four probe sets related to proteolysis, which suggests increased protein damage in vte2. Probe sets for eight glutathione S-transferase (GST) genes, five cytochrome P450 genes, and eight oxidoreductase genes were also upregulated.
Probe sets for three Trp biosynthetic genes were upregulated in vte2 as well as the probe set for PAD3 (CYP71B15), which catalyzes the final step in camalexin synthesis (Zhou et al., 1999; Schuhegger et al., 2006) (Figure 6; see Supplemental Table 1 online). The probe sets for two additional cytochrome P450s, CYP79B2 and CYP79B3, which convert Trp to indole-3-acetaldoxime, an intermediate in the synthesis of camalexin, auxin, and indole glucosinolates (Glawischnig et al., 2004), were called increased for all three comparisons with the wild type, and the P values were highly significant (P = 2.5 × 10−6 and 1.2 × 10−8, respectively), but their signal:log ratios for at least one comparison were slightly below our cutoff of 1.3. The Trp metabolic pathway is upregulated specifically in vte2, evidently in response to the absence of tocopherols or DMPBQ during germination.
Figure 6.
The Trp Metabolic Pathway.
The pathway shows steps between the Trp precursor chorismate and the Trp-derived phytoalexin camalexin. The names in gray denote genes encoding pathway enzymes whose expression was called increased in 3-d-old vte2 seedlings relative to wild-type seedlings (expression data; see Supplemental Table 1 online). The number of enzymatic steps between indole-3-acetaldoxime and camalexin are not known. Enzymes are phosphoribosylanthranilate transferase (PAT1, also known as TRP1; At5g17990), indole-3-glycerol-phosphate synthase (IGPS; At2g04400), Trp synthase, α subunit (TSA1, also known as TRP3; At3g54640), CYP79B2 (At4g39950), CYP79B3 (At2g22330), and PAD3 (CYP71B15; At3g26830). CPADRP, 1-(o-carboxyphenylamino)-1′-deoxyribulose-5′-phosphate; IAOx, indole-3-acetaldoxime; I3GP, indole-3-glycerol-phosphate; NPRA, N-(5-phosphoribosyl)-anthranilate.
The 20 most highly upregulated transcripts in 3-d vte2 seedlings relative to the wild type are indicative of responses to both abiotic and biotic stresses (Table 1). These transcripts include the pathogen-related genes AIG1 and Defensin1.4 (Reuber and Ausubel, 1996) and a DREB2-like transcription factor. Members of the DREB class of transcription factors have been shown to activate transcriptional responses to abiotic stresses, including drought, salinity, and temperature (Liu et al., 1998; Shinozaki and Yamaguchi-Shinozaki, 2000). Two pEARLI genes, which encode proteins with a putative lipid-transfer motif, were highly upregulated in vte2 (Wilkosz and Schlappi, 2000). The pEARLI genes were originally isolated for their cold-responsiveness, but they have since been shown to be induced by a number of stresses (Bubier and Schlappi, 2004; Vogel et al., 2005). SAG13, a senescence-associated transcript (Lohman et al., 1994; Miller et al., 1999), was also highly upregulated in vte2, as was an iron deficiency–inducible metal transporter, IRT1 (Varotto et al., 2002). Transcripts of stress-protective enzymes, GST5, a peroxidase, and Glu decarboxylase were also induced in vte2 (Watahiki et al., 1995; Turano and Fang, 1998; Bouche et al., 2004).
Table 1.
The 20 Most Highly Upregulated Genes in vte2 Relative to the Wild Type
| Arabidopsis Genome Initiative Number | Description | Localization | Function | Fold Change | P Value |
|---|---|---|---|---|---|
| At1g65500 | Unknown protein | Unknown | Unknown | 150 | 6.98 × 10−04 |
| At4g12490 | pEARLI 1-like protein | Chloroplast | Lipid binding | 102 | 2.87 × 10−04 |
| At2g18660 | Expansin (EXPR3) | Extracellular | Cell wall | 86 | 1.70 × 10−03 |
| At2g38340 | DREB-like AP2 | Nucleus | Transcription factor | 86 | 5.23 × 10−03 |
| At1g33960 | AIG1 | Chloroplast | Response to pathogen | 65 | 3.46 × 10−03 |
| At1g19610 | Defensin (PDF1.4) | Endomembrane | Defense response | 47 | 2.00 × 10−04 |
| At1g56060 | Expressed protein | Unknown | Unknown | 46 | 3.58 × 10−04 |
| At1g26380 | FAD binding protein | Endomembrane | Oxidoreductase activity | 44 | 1.66 × 10−03 |
| At1g26410 | FAD binding protein | Endomembrane | Oxidoreductase activity | 43 | 2.53 × 10−03 |
| At5g05340 | Peroxidase | Endomembrane | Response to stress | 42 | 3.87 × 10−03 |
| At1g26390 | FAD binding protein | Endomembrane | Oxidoreductase activity | 41 | 3.70 × 10−03 |
| At2g30750 | Cytochrome P450 71A12 | Endomembrane | P450 | 35 | 2.35 × 10−03 |
| At1g02940 | Glutathione S-transferase, GST5 | Mitochondrion | Toxin catabolism | 31 | 6.98 × 10−04 |
| At2g29350 | SAG13 | Membrane | Senescence | 29 | 5.24 × 10−03 |
| At2g02010 | Putative Glu decarboxylase | Unknown | Glu metabolism | 28 | 5.50 × 10−03 |
| At4g19690 | Iron-responsive transporter (IRT1) | Membrane | Transport | 27 | 5.07 × 10−03 |
| At4g12480 | pEARLI 1 | Endomembrane | Lipid binding | 27 | 3.91 × 10−04 |
| At1g13340 | Expressed protein | Mitochondrion | Unknown | 27 | 2.68 × 10−03 |
| At1g26420 | FAD binding protein | Endomembrane | Oxidoreductase activity | 25 | 1.17 × 10−03 |
| At4g14365 | C3HC4-type RING finger | Unknown | Protein ubiquitination | 22 | 6.91 × 10−04 |
The 20 most highly upregulated genes in 3-d-old vte2 seedlings relative to wild-type seedlings are listed with the fold increase relative to the wild-type control and the P value calculated by the analysis of variance (see Methods). Localization and function were obtained from the Gene Ontology section of The Arabidopsis Information Resource. FAD, flavin adenine dinucleotide.
To confirm the Affymetrix GeneChip results indicating that defense/pathogen-responsive genes were induced in vte2 seedlings, RNA gel blot analysis was performed on total RNA extracted from 3-d-old seedlings with two defense- or pathogen-related genes, OPR1 (At1g76680) and PAD3 (Figure 7A). These results show that both transcripts were specifically induced in vte2 relative to the wild type or vte1. Expression of the jasmonate biosynthetic gene AOS was not detectable in the wild type, vte1, or vte2 (Figure 7B) in the absence of exogenously administered methyl jasmonate (MeJA). However, the jasmonate biosynthetic gene OPR3 was detectable in all three genotypes and marginally induced in vte2 seedlings relative to wild-type or vte1 seedlings in the absence of exogenous MeJA (Figure 7B), which is consistent with slightly increased levels of JA in vte2 (Figure 4B). The expression of the JA-dependent gene VSP1 was also not detectable in wild-type, vte1, and vte2 seedlings in the absence of exogenous MeJA treatment. The JA-inducible gene CORI1 was also present in all three genotypes in the absence of exogenous MeJA (Figure 7B). Upon exogenous MeJA treatment, the expression of all four genes (AOS, CORI, OPR3, and VSP1) was strongly induced (Figure 7B). Together, these RNA gel blot results suggest that the levels of jasmonates in both vte1 and vte2 seedlings are not sufficient to induce JA-responsive gene expression in the seedlings, but that 3-d-old wild-type, vte1, and vte2 seedlings are capable of mounting a transcriptional response when JA levels are adequately increased.
Figure 7.
Expression of Defense-Related Transcripts in vte2 Seedlings.
(A) Ten micrograms of total RNA was extracted from 3-d-old seedlings (Columbia [Col], vte1-1, and vte2-1) and analyzed by RNA gel blot hybridization. PAD3 (At3g26830), OPR1 (At1g76680), and eIF4A (At3g13920) were detected using radiolabeled probes.
(B) Three-day-old seedlings (Columbia, vte1-1, and vte2-1) were treated with an aqueous solution with or without 10 μM MeJA (see Methods) and incubated for 3 h. Ten micrograms of total RNA was extracted from these seedlings and analyzed by RNA gel blot hybridization. The jasmonate-responsive genes AOS (At5g42650), OPR3 (At2g06050), CORI1 (At1g19670), and VSP1 (At5g24780) were detected using radiolabeled probes. eIF4A was used to determine equal loading.
DISCUSSION
Tocopherols and Germination
The phenotype conferred by vte2 clearly demonstrates that tocopherols are irreplaceable antioxidants during germination and early seedling growth that cannot be compensated for by other mechanisms. Indeed, in this regard, it was quite surprising that the expression of most genes involved in the biosynthesis and recycling of ascorbate and glutathione was similar in the wild type and vte2, expect for the induction in vte2 of 1 of 3 extracellular ascorbate oxidase genes (At4g39830) and 8 of the 52 GST genes in the genome. Likewise, genes encoding ascorbate peroxidases (8 genes) and the ROS-scavenging enzymes catalase (3 genes) and superoxide dismutase (11 genes) are also not induced in vte2 seedlings relative to wild-type seedlings. These results suggest either that the expression of genes encoding these ROS-scavenging enzymes is already at nearly maximal levels during germination and early seedling development or that young seedlings are unable to increase the expression of these genes in response to the compromised tocopherol antioxidant system of vte2. Alternatively, the seedling response to tocopherol deficiency may simply not include the induction of water-soluble antioxidant or ROS-scavenging systems, because the oxidative damage in vte2 is most likely confined to the lipid-soluble cellular compartments.
In comparing the global transcriptional response of vte1 and vte2 seedlings at 1 and 3 d after imbibition, the increased or decreased expression of only eight genes could be attributed specifically to the absence of tocopherol in both genotypes. Six genes are HSPs, some of which have been shown to act as chaperones, and are upregulated in response to a variety of abiotic stress conditions. These results suggest that as a whole, the Arabidopsis transcriptome has relatively few genes that respond directly to the presence of tocopherol at this stage of development. These observations are in contrast with studies in animal systems, in which direct, antioxidant-independent roles for tocopherols in modulating signaling pathways by inhibiting phospholipase A2 and protein kinase C activity and activating gene transcription have been reported (Tran et al., 1996; Yamauchi et al., 2001; Chandra et al., 2002; Clement et al., 2002). It is still possible that tocopherols have analogous antioxidant-independent functions at other stages of plant development, but this is clearly not a major function for tocopherols in seedlings. However, the induction of six HSP genes in both vte1 and vte2 indicates that there are biological consequences of tocopherol deficiency and that there may be a group of proteins that are specifically protected by tocopherols in germinating seedlings. Increased nonenzymatic lipid peroxidation was not required to trigger this HSP response, because vte1 and the wild type had nearly identical levels of lipid peroxides and lipid peroxidation products: HODEs (Sattler et al., 2004), MDA (Figure 2), and PPF1 (Figure 3). Based on these data, we can also conclude that the DMPBQ accumulated in vte1 can fully compensate for the absence of tocopherols in limiting the formation of nonenzymatic lipid peroxidation during germination and early seedling development. Whether DMPBQ plays a similar compensatory role in mature vte1 leaf tissue remains to be determined.
Tocopherols, Nonenzymatic Lipid Peroxidation, and the Control of Gene Expression
Although we find little evidence supporting a direct role for tocopherols in the regulation of gene expression, our analysis clearly indicates that, similar to in vitro and animal studies, tocopherols play a critical role in controlling nonenzymatic lipid peroxidation. The products generated by this pathway most likely play a key role in the global response observed in vte2 seedlings and likely in selected biological responses of wild-type plants. We observed high levels of lipid peroxidation products in vte2 seedlings, hydroxy fatty acids from 18:2 (Sattler et al., 2004) and 18:3 (MDA and PPF1), the predominant PUFAs in Arabidopsis seedlings (Figures 2 and 3). Hydroxy fatty acids and most phytoprostanes are relatively stable molecules, initially remaining esterified in the membrane, and are indicative of the oxidative history of a tissue. However, during membrane turnover and repair, phytoprostanes are released from membranes and rapidly metabolized (Imbusch and Mueller, 2000a; Mueller, 2004). By contrast, MDA is produced and released instantaneously by fragmentation of oxidized membrane lipids and is a member of a group of short-lived, lipid-derived reactive electrophile species (RES) produced in response to stress. For these reasons, MDA and PPF1 do not reach similar levels or follow the same temporal patterns of accumulation in vte2 seedlings.
In vte2 seedlings, MDA levels peaked at 3 d and declined rapidly to near wild-type and vte1 levels by 6 d. This rapid decline is consistent with prior studies, suggesting that plants have mechanisms to control the levels of MDA (Weber et al., 2004) and indicating that these mechanisms are tocopherol-independent in seedlings. RES, such as MDA, have strong biological activities in plants and stimulate the expression of genes involved in cell protection/detoxification (e.g., OPR1 [Almeras et al., 2003] and GST1 [Vollenweider et al., 2000; Loeffler et al., 2005]) as well as genes implicated in resistance to stresses such as heat shock and dehydration (Weber et al., 2004). GST1 has been shown to be stress- and MDA-inducible and is strongly upregulated in vte2. Likewise, OPR1, which is absent from the Affymetrix ATH1 GeneChip, is strongly upregulated in vte2 based on RNA gel blot analysis (Figure 7) and in response to a number of abiotic and biotic stress conditions (Biesgen and Weiler, 1999; Almeras et al., 2003; Weber et al., 2004; Mezzari et al., 2005).
In addition to MDA, the phytoprostanes, a second major class of nonenzymatic lipid peroxidation products derived from 18:3, were increased in vte2 seedlings to some of the highest levels ever observed in plants (Imbusch and Mueller, 2000a). The phytoprostane class measured, PPF1, has not been shown to have biological activity when administered exogenously, possibly because this class is polar and not readily taken up by cells, as is the case for the analogous F-type of mammalian prostaglandins in cells lacking prostaglandin uptake transporters (Schuster, 2002). However, the PPF1 class is an excellent marker for nonenzymatic lipid peroxidation and indicative that other classes of more labile and reactive phytoprostanes (PPA1, PPB1, PPD1, PPE1, PPG1, and dPPJ1) are likely to have been generated also (Imbusch and Mueller, 2000a). Exogenously supplied PPA1 and PPB1 have also been shown to trigger biological responses in plants, including activating mitogen-activated protein kinase kinase, inducing phytoalexin synthesis, and activating gene transcription (Thoma et al., 2003; Loeffler et al., 2005), including 17 GST genes. Previous work has suggested that some of the GSTs and P450s induced by phytoprostanes may function in detoxifying potentially damaging products of nonenzymatic lipid peroxidation and electrophilic xenobiotics (Baerson et al., 2005; Loeffler et al., 2005; Mezzari et al., 2005). In vte2 seedlings, 4 of these 17 GST genes were also upregulated relative to the wild type.
Within the phytoprostane family of compounds as well as in other lipid peroxidation products, there are several species that contain α/β unsaturated carbonyl groups and, like MDA, are electrophilic and reactive. There is substantial evidence that such electrophiles, which include PPA1, dPPJ1, β-hydroxyacrolein (a tautomeric form of MDA), and the JA biosynthetic intermediate OPDA, have potent effects on gene expression, possibly attributable to their electrophilic properties (Vollenweider et al., 2000; Stintzi et al., 2001; Almeras et al., 2003; Mueller, 2004; Weber et al., 2004). PPA1, dPPJ1, and OPDA are all cyclopentenones containing an unsaturated α/β carbonyl group within a five-carbon ring. Expression profiling of Arabidopsis suspension cells treated with either PPA1 or PPE1 affected the expression of ∼800 genes (S. Berger and M.J. Mueller, unpublished data). The expression of 59 of the 160 genes upregulated in vte2 seedlings at 3 d is also induced by either PPA1 or PPE1 treatment of Arabidopsis suspension cells. Together, these data indicate that nonenzymatic lipid peroxidation products generated in vivo play an important role in the transcriptional response observed in vte2 seedlings and provide further evidence that electrophilic lipid peroxidation products play a significant role in modulating gene expression and cellular responses in plants.
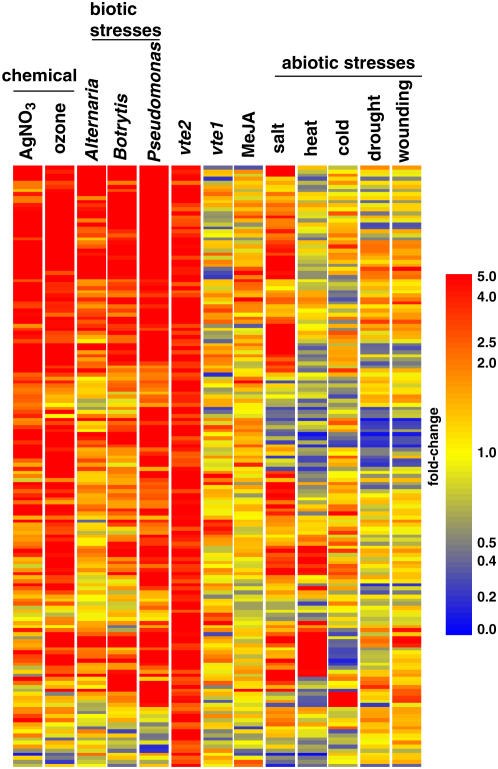
The 160 transcripts induced in 3-d-old vte2 seedlings relative to wild-type seedlings (see Supplemental Table 1 online) were examined for their responses to both abiotic and biotic conditions using GENEVESTIGATOR (Zimmermann et al., 2004, 2005) (Figure 8). Most of these genes were unchanged in expression in 3-d-old vte1 seedlings, which further highlights how dissimilar the tocopherol-deficient vte1 and vte2 transcription profiles are to each other (Figure 8). Generally, many of the vte2-1–induced genes were also induced in response to biotic stress (Pseudomonas, Alternaria, and Botrytis infection), but surprisingly few of these genes were induced in response to abiotic stress treatments, including drought, wounding, heat, or cold stress. In addition, many of the 160 vte2-1–induced genes were also induced by ozone, salt, and silver nitrate but not by MeJA treatment. Ozone is itself a ROS and readily penetrates membranes, causing nonenzymatic lipid peroxidation of PUFAs (Pauls and Thompson, 1980; Pryor and Church, 1991), whereas exposure of plants to heavy metals such as silver nitrate is also known to induce nonenzymatic lipid peroxidation (Baryla et al., 2000; Imbusch and Mueller, 2000b; Thoma et al., 2004).
Figure 8.
Expression of the Upregulated Genes in vte2 Seedlings under Different Conditions.
Genes called increased in 3-d-old vte2 seedlings relative to wild-type seedlings were examined using the Meta-Analyzer feature of GENEVESTIGATOR (Zimmermann et al., 2004) to assess their the response to various conditions or treatments relative to each corresponding negative control. Red indicates increased, blue indicates decreased, and yellow indicates no change (see Methods). The genes were clustered using hierarchical clustering based upon their response to the condition or treatment.
The meta-analysis in Figure 8 suggests that from a biological perspective, the transcriptome of vte2 seedlings responds as if under pathogen attack and that the nonenzymatic lipid peroxidation occurring in vte2 seedlings plays a role in triggering aspects of plant responses to pathogens. The fact that the majority of the genes induced in both vte2 and during pathogen attack are also not induced by MeJA treatment suggests that nonenzymatic lipid peroxidation is a component of previously described JA-independent responses of plants to pathogens (Kloek et al., 2001; Roetschi et al., 2001; Almeras et al., 2003; Bohman et al., 2004). Most surprisingly, this analysis also suggests that abiotic stresses such as drought, heat, wounding, or cold stress do not have a strongly regulated nonenzymatic lipid peroxidation component. Although tocopherols show increased accumulation under drought and heat (Munne-Bosch and Alegre, 2002), tocopherols may not play as crucial a role in protecting plants from these abiotic stresses as has long been assumed. A plausible alternative explanation is that increased tocopherol levels in response to these abiotic stresses may limit nonenzymatic lipid peroxidation and thereby prevent the inappropriate induction of biotic stress responses. Further analysis of the response of tocopherol-deficient plants to abiotic stresses is needed to test this intriguing hypothesis.
Induction of Trp Metabolism
Trp provides the indole ring for the biosynthesis of a variety of important plant compounds, including auxin, and the indole glucosinolates and phytoalexins (Glawischnig et al., 2004), which are important for defending plants against insect herbivory and pathogens, respectively. In plants, Trp biosynthetic genes are upregulated in response to a variety of biotic and xenobiotic stresses as well as to amino acid starvation (Zhao and Last, 1996; Zhao et al., 1998) and in response to electrophilic vinyl ketones (Almeras et al., 2003). The expression of PAD3 (CYP71B15), a P450 involved in camalexin biosynthesis, is upregulated in vte2 seedlings (Figures 6 and 7) and in response to PPB1 treatment (Loeffler et al., 2005). PAD3 catalyzes the final step in camalexin synthesis, acting downstream of CYP79B2 and CYP79B3 (Glawischnig et al., 2004; Schuhegger et al., 2006), whose expression levels were likewise increased in vte2 relative to the wild type, although slightly below our 1.3 signal:log ratio criterion. Because several Trp biosynthetic genes are upregulated in vte2 seedlings (Figure 6), our data suggest that nonenzymatic lipid peroxidation may be a key signal for the increase of Trp metabolism in response to biotic stress in the wild type. Interestingly, the spontaneous lesion mutant acd2 also accumulates camalexin in the absence of pathogens (Zhao and Last, 1996), although any possible connection between nonenzymatic lipid peroxidation and the acd2 mutant remains to be determined.
PAD4 has similarity to triacylglycerol lipases (Jirage et al., 1999) and has been shown to be a member of a defense signaling complex together with EDS1 and SAG101 (Feys et al., 2001, 2005). pad4 mutants are unable to fully induce several defense responses, including camalexin synthesis in response to specific pathogens (Glazebrook et al., 1997). Interestingly, the camalexin level in the pad4 vte2 double mutant was significantly higher than that in vte2 at 3 d and nearly equal to that in vte2 at 6 d, indicating that camalexin synthesis in the vte2 mutant is either independent of or repressed by PAD4. These data support the model that both PAD4-dependent and -independent pathways lead to camalexin synthesis (Glazebrook et al., 1997). Our data further define an important role for nonenzymatic lipid peroxidation in inducing the PAD4-independent pathway, but they do not exclude the possibility that nonenzymatic lipid peroxidation might also be involved in the PAD4-dependent pathway. The oxidative burst associated with incompatible host–pathogen interactions would create the biochemical environment necessary to form a variety of nonenzymatic lipid peroxidation products and trigger downstream defense responses such as camalexin synthesis. Our data suggest that tocopherols play a role in modulating such defense signals in seedlings under normal conditions or in locations removed from the site of pathogen attack. Whether such biotic interactions are also affected in mature vte2 leaves remains to be determined.
Nonenzymatic versus Enzymatic Lipid Peroxidation Pathways
Both plants and animals have evolved enzymatic pathways for the production of lipid peroxidation products (octadecanoids in plants and eicosanoids in animals), and the molecular, genetic, and biochemical roles of these compounds in mediating/modulating biological responses, including defense responses, have been well studied. In plants, the best-described oxylipin is JA, which affects a wide range of signal transduction pathways, including reproductive development, pathogen defense, and insect defense (Howe and Schilmiller, 2002). However, there has been comparably little analysis of the products of nonenzymatic lipid peroxidation as signaling compounds, probably because the production of these compounds occurs independently of enzymatic control and leads to a wider range of products. Undoubtedly, nonenzymatic lipid peroxidation occurs ubiquitously in organisms because it requires only PUFAs, oxygen, and free radicals. In plants, these nonenzymatic and enzymatic pathways appear to have diverged, and vte2 seedlings are strong genetic evidence that the two pathways have separate roles.
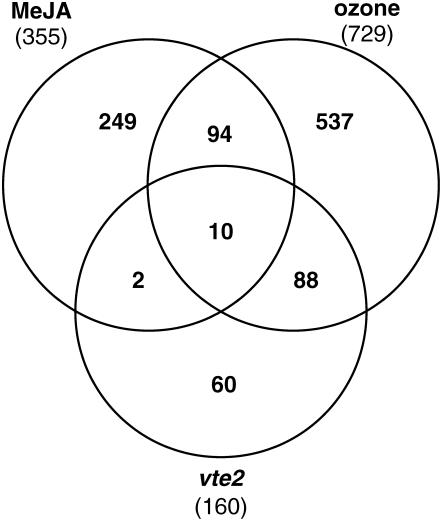
The potential of RES to act as regulators of gene expression in plants has, until now, lacked genetic validation (Farmer et al., 2003). The data presented here provide a strong genetic demonstration of RES activity in vivo and underscore the potential of nonenzymatic lipid peroxidation products to act as signals. In addition, these data provide clear genetic evidence separating enzymatically derived oxylipins and nonenzymatic lipid peroxidation products in signaling pathways. The levels of JA were increased slightly in both vte1 and vte2, and the levels of OPDA were similar in vte2 and wild-type seedlings, which suggests that JA and OPDA are not major factors contributing to the vte2 seedling phenotype. The observation that well-characterized JA-inducible genes (AOS, CORI1, and VSP1) were not altered in expression in vte2, vte1, and wild-type seedlings but were similarly induced upon exogenous MeJA treatment in 3-d-old seedlings of all genotypes supports this conclusion. These genes were clearly responsive to JA at this stage of development, but the endogenous JA levels in all three genotypes were insufficient for their induction (Figure 7B). vte2 was crossed to the JA signaling mutant coi1, and the progeny of vte2 coi1/+ (heterozygous for coi1) were not altered in their development compared with vte2 seedlings alone, indicating that JA signaling is not a key factor in the vte2 seedling phenotype (S.E. Sattler, unpublished data). Finally, analysis of public GeneChip data for MeJA and ozone-induced transcripts (≥2.5-fold relative to their respective controls; see Methods) showed substantial overlap (61%) between vte2 and ozone-induced genes, but only a small number (7.5%) of vte2-induced genes were in common with MeJA-induced genes (Figure 9). It should also be stressed that the MeJA-treated tissue (7-d-old seedlings) were developmentally more similar to 3-d-old vte2 seedlings than were the 2-week-old plants used for ozone treatment. Together, these results suggest that enzymatic oxylipins, including JA and ODPA, play at best a minor role in the vte2 seedling phenotype and that the major responses of vte2 seedlings are mediated by various products of nonenzymatic lipid peroxidation.
Figure 9.
Comparison of Upregulated Genes in MeJA, Ozone, and vte2 Data Sets.
Global expression profiles were obtained from public databases (NASCArray Affywatch) for MeJA-treated 7-d-old seedlings and 2-week-old ozone-treated plants and their corresponding negative controls. Genes showing increased expression relative to their respective controls were identified as described in Methods. The 355 genes called increased by MeJA treatment and the 729 genes called increased by ozone treatment were compared with the 160 genes called increased in 3-d-old vte2 seedlings relative to wild-type seedlings.
METHODS
Plants and Growth Conditions
vte1-1 and vte2-1 were isolated and described previously (Sattler et al., 2003, 2004). pad3-1 and pad4-1 were obtained from the ABRC. Arabidopsis thaliana plants were grown at 22°C under a 16-h photoperiod (120 μmol photons·m−2·s−1 or μE) in a vermiculite and potting soil mixture and fertilized with 1× Hoagland solution. Alternatively, seedlings were grown on Petri plates containing 0.5× Murashige and Skoog basal medium (Sigma-Aldrich) and 1.0% phytoagar under a 12-h photoperiod (120 μmol photons·m−2·s−1 ) after 5 d of imbibition at 6°C to break dormancy, as described previously (Sattler et al., 2004). For the MeJA treatment, 3-d-old seedlings were sprayed with 10 μM MeJA (Sigma-Aldrich) in 0.05% Tween 20 and incubated for 3 h. For the negative control, 3-d-old seedlings were sprayed with 0.05% Tween 20 and incubated for 3 h.
Oxylipin Profiling
Seedlings were harvested from plates, weighed, and immediately frozen in liquid nitrogen. A total of 500 mg of frozen tissue was ground in a mortar to a fine powder and added to 3.5 mL of ice-cold methanol containing 100 ng of tetrahydro-12-OPDA and 100 ng of dihydrojasmonic acid as internal standards and 12.5 μL of trimethyl phosphite (Fluka). The solution was immediately homogenized with a Polytron (Kinematica) for 1 min on ice. Extraction of fatty acids was continued by rotating the samples in tubes for 2 h at 4°C. Ice-cold water (1.5 mL) was added. After centrifugation, 80 μL of 1 M NH4OH was added, and the supernatant was passed through an 8-mL C18-SPE column (JT Baker; Bakerbond 8ml 500 mg spe Glass Octadecyl), which had been prewashed with methanol followed by methanol:water (70:30, v/v). The column was washed with an additional 7 mL of methanol:water (75:25, v/v), both eluates were combined and acidified with 120 μL of 10% formic acid, and the volume was adjusted to 50 mL using water. The column was reconditioned with 6 mL of methanol containing 40 μL of 10% formic acid, followed by 5 mL of methanol, 6 mL of diethyl ether, 5 mL of methanol, and finally twice with 7 mL of water. Elute was loaded on the column and washed with ethanol:water (15:85, v/v). The oxylipins were eluted with 10 mL of diethyl ether, dried over anhydrous MgSO4, and evaporated under a stream of N2 at 35°C. Oxylipins were resusupended in 60 μL of methanol and methylated with 450 μL of diazomethane in ether, dried under N2, and redissolved in 120 μL of acetonitrile. Samples were derivatized with 60 μL of N,O-bis(trimethylsilyl)trifluoroacetamide (Pierce) for 30 min at 60°C, dried under N2, and redissolved in 500 μL of hexane. The samples were loaded onto an 8-mL Florisil-SPE column (JT Baker; Bakerbond 8ml 500 mg spe Glass Florisil), which had been prewashed first with diethyl ether (3 mL) and then with hexane (10 mL). The loaded column was washed with 4 mL of hexane, and the oxylipins were eluted with 7 mL of ethyl acetate:diethyl ether (1:1, v/v). After evaporation of the solvent under N2, the samples were taken up in 10 μL of hexane and stored at −80°C. Oxylipins were analyzed by GC-MS (Hewlett-Packard 5890 gas chromatograph linked to a Hewlett-Packard 5927 mass spectrometer, electron ionization mode, 70-eV electron potential, 11-p.s.i. column head pressure, HP-5MS column 30 m × 0.25 mm). The temperature gradient was 100°C for 1 min, 100 to 160°C at 20°C/min, 160 to 238°C at 3°C/min, and 238 to 300°C at 30°C/min, as described previously (Weber et al., 1997). Camalexin was identified in oxylipin profiles by its characteristic retention time (23.70 min) and mass spectrum: m/z (%) = 200 (M+, 100), 173 (4), 142 (30), 115 (17), 83 (25), 58 (50). Similarly, for indole 3-carboxylic acid, retention time = 14.41 min, m/z (%) = 175 (M+, 53), 144 (100), 116 (22), 89 (21), 63 (9).
Camalexin Analysis
For quantitative camalexin analysis, seedlings were harvested and immediately frozen in liquid nitrogen. A total of 500 mg of frozen seedlings was ground in a mortar to a fine powder, added to 3.5 mL of 80% methanol, and incubated at 80°C for 20 min. After centrifugation, the supernatant was removed, the volume was reduced to 1 mL by drying with N2 gas, and 0.5 mL of water was added. The solution was extracted twice with 1.5 mL of chloroform, and samples were vacuum-dried and resuspended in 100 μL of hexane:isopropanol (93:7, v/v). HPLC analysis was performed using an Agilent 1100 series HPLC apparatus on a ReliaSil Silica 5-μm, 250 × 4.6-mm normal phase column (Column Engineering) at 30°C with a flow rate of 1 mL/min and hexane:isopropanol (93:7, v/v). Camalexin was detected with an excitation wavelength of 318 nm and an emission wavelength of 385 nm using a fluorescence detector. A camalexin standard (a gift from Ray Hammerschmidt, Michigan State University) was used to identify the compound based on HPLC retention time and to construct standard curves for quantification.
MDA Analysis
Approximately 200 mg of seedlings was harvested and stored in liquid nitrogen until MDA quantification. Total MDA levels were determined by GC-MS according to a protocol modified from Weber et al. (2004). Tissues were ground in liquid nitrogen and resuspended in 0.4 mL of PBS containing 1 mM deferoxamine mesylate (Sigma-Aldrich), 0.08 mM butylated hydroxytoluene (Sigma-Aldrich), and 0.5 nmol of the internal standard [2H2]MDA. After rapid vortexing, 15 μL of 6.6 N sulfuric acid was immediately added and the sample was incubated for 10 min on ice. After hydrolysis of the bound MDA pool, proteins were precipitated with 112 μL of 0.3 M sodium tungstate (Fluka) and the samples were centrifuged. The supernatant was combined with 450 μL of buffer containing 0.58 M sodium phosphate, 0.21 M sodium citrate, and 75 μL of pentafluorophenylhydrazine (5 mg/mL; Fluka) in a new Eppendorf tube and incubated for 30 min at room temperature while shaking. The reaction was stopped by adding 15 μL of 9 N sulfuric acid, and the MDA–pentafluorophenylhydrazine products were extracted with 125 μL of isooctane (Fluka). Preparation of the internal standard [2H2]MDA from [2H2]1,1,3,3-tetraethoxypropane (CDN Isotopes) and GC-MS analyses were performed as described previously (Weber et al., 2004).
PPF1 and Jasmonate Analysis
Total levels of PPF1 (free and esterified) were determined after alkaline hydrolysis of plant extracts containing [18O3]PPF1 as an internal standard. Extraction, saponification, derivatization, and quantitation of PPF1 by GC-MS in negative ion chemical ionization mode were performed as described (Imbusch and Mueller, 2000a, 2000b). Levels of free OPDA and JA were determined as described (Parchmann et al., 1997).
RNA Extraction and Affymetrix GeneChip
Approximately 100 mg of seedlings was frozen in liquid nitrogen, and total RNA was isolated using the RNAqueous RNA extraction kit and the Plant RNA Isolation Aid (Ambion) according to the manufacturer's protocol. Biotinylated target RNA (copy RNA) was prepared from 15 μg of total RNA using the Affymetrix GeneChip One-Cycle Target Labeling kit (Affymetrix) according to the manufacturer's protocols. The three biological replicates for each time point and genotype were hybridized to the Affymetrix Arabidopsis ATH1 GeneChip (Affymetrix) and analyzed as described previously (Fowler and Thomashow, 2002).
Affymetrix GeneChip Data Analysis
The Affymetrix Microarray Suite 5.0 was used to analyze the microarray data. Output from all GeneChip hybridizations was scaled globally such that its average intensity was equal to an arbitrary target intensity of 500, allowing comparisons between GeneChips. Signal (gene expression) values and signal:log ratios were calculated from the GeneChip fluorescence intensity data. The software was also used to determine whether each gene was present, marginal, or absent (Detection) and whether the signal:log ratio represented a genuine change in mRNA accumulation (Change; no change, increased, or decreased). Signal:log ratios and change calls were determined for vte1 and vte2 compared with the corresponding wild-type sample of the same age. Microsoft Access was used as the database management software and to filter and query the data. Probe sets that met the following criteria were selected for further analysis. Those determined to be upregulated in the vte mutants were selected as having, at any time point, an absolute call of present in all three samples, difference calls of increase for all three Change comparisons, and signal:log ratios of at least 1.3 for all three Change comparisons. Those determined to be downregulated in the vte mutants were selected as having, at any time point, an absolute call of present in all three wild-type samples, difference calls of decrease for all three Change comparisons, and signal:log ratios of at least −1.3 for all three Change comparisons. Statistically significant changes in mRNA abundance were determined using the statistical package with GeneSpring 7.2 (Silicon Genetics). The GeneChip data were imported into GeneSpring and normalized to respective control samples as specified by the manufacturer for single-color array data. Statistical significance was determined by analysis of variance using a P value of 0.05 as the cutoff. The Benjamini and Hochberg false-discovery rate was used as the multiple comparison correction for type I family-wise error.
The expression data for the 160 induced genes in vte2 under different stress conditions and treatments (Figure 8) were obtained from the meta-analyzer feature of GENEVESTIGATOR (https://www.genevestigator.ethz.ch), which was imported into GeneSpring 7.2 to perform hierarchical clustering (standard correlation) including vte1 and vte2 3-d data sets. For Figure 9, the Affymetrix ATH1 data for MeJA-treated seedlings (NASCARRAYS-174) and ozone-treated plants (NASCARRAYS-26) were obtained from the public database (NASCArray Affywatch; http://affymetrix.Arabidopsis.info/AffyWatch.html). The GeneChips were analyzed as described above. The lists of induced genes for MeJA and ozone treatments were determined by the following criteria: the probe set were called present in all replicates of the treatment (ozone or MeJA), called increased for all replicates relative the untreated controls, and the signal-log ratio was ≥1.3 (2.5-fold) for all replicates relative to untreated controls. The statistical analysis was performed as described above.
RNA Gel Blot Analysis
RNA gel blots with 10 μg of total RNA per lane were prepared, transferred, and hybridized as described previously (Eccles et al., 1992). Probes were prepared from PCR products: for PAD3, forward (5′-CCGGTGAATCTTGAGAGAGCC-3′) and reverse (5′-GATCAGCTCGGTCATTCCCC-3′) (Zhou et al., 1999); for OPR1, forward (5′-TACAGCTCAAGGATATCAAGA-3′) and reverse (5′-GAAACTTATTACATCTTATATAA-3′) (He and Gan, 2001); for AOS, forward (5′-GGGAGCGATTGAGAAAATGG-3′) and reverse (5′-CGACGAGAAATTAACGGAGC-3′); for CORI1, forward (5′-GCCGCCGGGAGGGCAAGTGG-3′) and reverse (5′-CACATGCAACTTAGGACATG-3′); for OPR3, forward (5′-GGGCTCGAGATGACGGCGGCACAAGGGAA-3′) and reverse (5′-CCCACTAGTTCAGAGGCGGGAAAAAGGAG-3′); and for At VSP1, forward (5′-GTTGGTGTGACAAAATGG-3′) and reverse (5′-GCGGTGAAGATATATGC-3′) (Berger et al., 1995). Probes were randomly labeled with [α-32P]dCTP according to the manufacturer's protocol (Invitrogen), and detection of radioisotope-labeled probes was as described previously (Vogel et al., 2005). Blots were stripped and probed with eIF4A to assess equal loading of RNA samples (Taylor et al., 1993).
Accession Numbers
The Affymetrix GeneChip data were deposited at Gene Expression Omnibus (accession number GSE4847; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4847) in compliance with MIAME standards.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Genes Increased in Expression in the vte Mutants Relative to the Wild Type.
Supplemental Table 2. Genes Decreased in Expression in the vte Mutants Relative to the Wild Type.
Supplementary Material
Acknowledgments
We thank Jeff Landgraff, Heather Van Buskirk, and Jonathan Vogel for their advice on transcript profiling and data analysis and Annette Thelen of the Research Support Technology Facility at Michigan State University for performing GeneChip hybridization and scanning. We also thank Ray Hammerschmidt for providing a camalexin standard. E.E.F. was supported by the Swiss National Science Foundation National Center of Competence in Research Plant Survival program and by National Science Foundation Grant 3100A0-101711. M.J.M. was supported by the Deutsch Forschungsgemeinschaft Sonderforschungsbereich 367 (Project A2). This work was supported by National Science Foundation Grant MCB-023529 to D.D.P.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Dean DellaPenna (dellapen@msu.edu).
Online version contains Web-only data.
References
- Almeras, E., Stolz, S., Vollenweider, S., Reymond, P., Mene-Saffrane, L., and Farmer, E.E. (2003). Reactive electrophile species activate defense gene expression in Arabidopsis. Plant J. 34 202–216. [DOI] [PubMed] [Google Scholar]
- Baerson, S.R., Sanchez-Moreiras, A., Pedrol-Bonjoch, N., Schulz, M., Kagan, I.A., Agarwal, A.K., Reigosa, M.J., and Duke, S.O. (2005). Detoxification and transcriptome response in Arabidopsis seedlings exposed to the allelochemical benzoxazolin-2(3H)-one. J. Biol. Chem. 280 21867–21881. [DOI] [PubMed] [Google Scholar]
- Baryla, A., Laborde, C., Montillet, J.L., Triantaphylides, C., and Chagvardieff, P. (2000). Evaluation of lipid peroxidation as a toxicity bioassay for plants exposed to copper. Environ. Pollut. 109 131–135. [DOI] [PubMed] [Google Scholar]
- Berger, S., Bell, E., Sadka, A., and Mullet, J.E. (1995). Arabidopsis thaliana Atvsp is homologous to soybean VspA and VspB, genes encoding vegetative storage protein acid phosphatases, and is regulated similarly by methyl jasmonate, wounding, sugars, light and phosphate. Plant Mol. Biol. 27 933–942. [DOI] [PubMed] [Google Scholar]
- Berger, S., Weichert, H., Porzel, A., Wasternack, C., Kuehn, H., and Feussner, I. (2001). Enzymatic and non-enzymatic lipid peroxidation in leaf development. Biochim. Biophys. Acta 1533 266–276. [DOI] [PubMed] [Google Scholar]
- Biesgen, C., and Weiler, E.W. (1999). Structure and regulation of OPR1 and OPR2, two closely related genes encoding 12-oxophytodienoic acid-10,11-reductases from Arabidopsis thaliana. Planta 208 155–165. [DOI] [PubMed] [Google Scholar]
- Bochkov, V.N., Kadl, A., Huber, J., Gruber, F., Binder, B.R., and Leitinger, N. (2002). Protective role of phospholipid oxidation products in endotoxin-induced tissue damage. Nature 419 77–81. [DOI] [PubMed] [Google Scholar]
- Bohman, S., Staal, J., Thomma, B., Wang, M.L., and Dixelius, C. (2004). Characterisation of an Arabidopsis-Leptosphaeria maculans pathosystem: Resistance partially requires camalexin biosynthesis and is independent of salicylic acid, ethylene and jasmonic acid signalling. Plant J. 37 9–20. [DOI] [PubMed] [Google Scholar]
- Bouche, N., Fait, A., Zik, M., and Fromm, H. (2004). The root-specific glutamate decarboxylase (GAD1) is essential for sustaining GABA levels in Arabidopsis. Plant Mol. Biol. 55 315–325. [DOI] [PubMed] [Google Scholar]
- Brigelius-Flohe, R., and Traber, M.G. (1999). Vitamin E: Function and metabolism. FASEB J. 13 1145–1155. [PubMed] [Google Scholar]
- Bubier, J., and Schlappi, M. (2004). Cold induction of EARLI1, a putative Arabidopsis lipid transfer protein, is light and calcium dependent. Plant Cell Environ. 27 929–936. [Google Scholar]
- Chandra, V., Jasti, J., Kaur, P., Betzel, C., Srinivasan, A., and Singh, T.P. (2002). First structural evidence of a specific inhibition of phospholipase A(2) by alpha-tocopherol (vitamin E) and its implications in inflammation: Crystal structure of the complex formed between phospholipase A(2) and alpha-tocopherol at 1.8 angstrom resolution. J. Mol. Biol. 320 215–222. [DOI] [PubMed] [Google Scholar]
- Clement, S.A., Tan, C.C., Guo, J.L., Kitta, K., and Suzuki, Y.J. (2002). Roles of protein kinase C and alpha-tocopherol in regulation of signal transduction for GATA-4 phosphorylation in HL-1 cardiac muscle cells. Free Radic. Biol. Med. 32 341–349. [DOI] [PubMed] [Google Scholar]
- Creelman, R.A., and Mulpuri, R. (August 12, 2002). The oxylipin pathway in Arabidopsis. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0012, http://www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed]
- Davletova, S., Schlauch, K., Coutu, J., and Mittler, R. (2005). The zinc-finger protein zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol. 139 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPenna, D., and Pogson, B.J. (2006). Vitamin synthesis in plants: Tocopherols and carotenoids. Annu. Rev. Plant Biol. 57 711–738. [DOI] [PubMed] [Google Scholar]
- Dong, J., Chen, C., and Chen, Z. (2003). Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 51 21–37. [DOI] [PubMed] [Google Scholar]
- Eccles, M.R., King, F.J., and Cole, M.D. (1992). A colony-stimulating factor 1 (CSF-1) receptor/platelet-derived growth factor-beta receptor gene fusion confers CSF-1 independence and tumorigenicity on a c-myc-immortalized monocyte cell line. Mol. Cell. Biol. 12 386–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer, E.E., Almeras, E., and Krishnamurthy, V. (2003). Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Curr. Opin. Plant Biol. 6 372–378. [DOI] [PubMed] [Google Scholar]
- Feys, B.J., Moisan, L.J., Newman, M.A., and Parker, J.E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J., Wiermer, M., Bhat, R.A., Moisan, L.J., Medina-Escobar, N., Neu, C., Cabral, A., and Parker, J.E. (2005). Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler, S., and Thomashow, M.F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer, M.J. (1992). The antioxidant effects of thylakoid vitamin-E (alpha-tocopherol). Plant Cell Environ. 15 381–392. [Google Scholar]
- Fukuzawa, K., Tokumura, A., Ouchi, S., and Tsukatani, H. (1982). Antioxidant activities of tocopherols on Fe2+-ascorbate-induced lipid peroxidation in lecithin liposomes. Lipids 17 511–513. [DOI] [PubMed] [Google Scholar]
- Funk, C.D. (2001). Prostaglandins and leukotrienes: Advances in eicosanoid biology. Science 294 1871–1875. [DOI] [PubMed] [Google Scholar]
- Glawischnig, E., Hansen, B.G., Olsen, C.E., and Halkier, B.A. (2004). Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc. Natl. Acad. Sci. USA 101 8245–8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J., Zook, M., Mert, F., Kagan, I., Rogers, E.E., Crute, I.R., Holub, E.B., Hammerschmidt, R., and Ausubel, F.M. (1997). Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics 146 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux, M., Eymery, F., Porfirova, S., Rey, P., and Dormann, P. (2005). Vitamin E protects against photoinhibition and photooxidative stress in Arabidopsis thaliana. Plant Cell 17 3451–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen, S.L., and Chisolm, G.M. (2002). Oxidized phosphatidylcholines: Pattern recognition ligands for multiple pathways of the innate immune response. Proc. Natl. Acad. Sci. USA 99 12515–12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y., and Gan, S. (2001). Identical promoter elements are involved in regulation of the OPR1 gene by senescence and jasmonic acid in Arabidopsis. Plant Mol. Biol. 47 595–605. [DOI] [PubMed] [Google Scholar]
- Howe, G.A., and Schilmiller, A.L. (2002). Oxylipin metabolism in response to stress. Curr. Opin. Plant Biol. 5 230–236. [DOI] [PubMed] [Google Scholar]
- Imbusch, R., and Mueller, M.J. (2000. a). Analysis of oxidative stress and wound-inducible dinor isoprostanes F-1 (phytoprostanes F-1) in plants. Plant Physiol. 124 1293–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbusch, R., and Mueller, M.J. (2000. b). Formation of isoprostane F-2-like compounds (phytoprostanes F- 1) from alpha-linolenic acid in plants. Free Radic. Biol. Med. 28 720–726. [DOI] [PubMed] [Google Scholar]
- Iqbal, M., Evans, P., Lledo, A., Verdaguer, X., Pericas, M.A., Riera, A., Loeffler, C., Sinha, A.K., and Mueller, M.J. (2005). Total synthesis and biological activity of 13,14-dehydro-12-oxo-phytodienoic acids (deoxy-J(1)-phytoprostanes). ChemBioChem 6 276–280. [DOI] [PubMed] [Google Scholar]
- Jirage, D., Tootle, T.L., Reuber, T.L., Frost, L.N., Feys, B.J., Parker, J.E., Ausubel, F.M., and Glazebrook, J. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KamalEldin, A., and Appelqvist, L.A. (1996). The chemistry and antioxidant properties of tocopherols and tocotrienols. Lipids 31 671–701. [DOI] [PubMed] [Google Scholar]
- Kloek, A.P., Verbsky, M.L., Sharma, S.B., Schoelz, J.E., Vogel, J., Klessig, D.F., and Kunkel, B.N. (2001). Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 26 509–522. [DOI] [PubMed] [Google Scholar]
- Leitinger, N. (2003). Oxidized phospholipids as modulators of inflammation in atherosclerosis. Curr. Opin. Lipidol. 14 421–430. [DOI] [PubMed] [Google Scholar]
- Liebler, D.C. (1993). The role of metabolism in the antioxidant function of vitamin-E. Crit. Rev. Toxicol. 23 147–169. [DOI] [PubMed] [Google Scholar]
- Liebler, D.C., and Burr, J.A. (2000). Antioxidant reactions of alpha-tocopherolhydroquinone. Lipids 35 1045–1047. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffler, C., Berger, S., Guy, A., Durand, T., Bringmann, G., Dreyer, M., von Rad, U., Durner, J., and Mueller, M.J. (2005). B-1-Phytoprostanes trigger plant defense and detoxification responses. Plant Physiol. 137 328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman, K.N., Gan, S.S., John, M.C., and Amasino, R.M. (1994). Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiol. Plant. 92 322–328. [Google Scholar]
- Maeda, H., Sakuragi, Y., Bryant, D.A., and DellaPenna, D. (2005). Tocopherols protect Synechocystis sp. strain PCC 6803 from lipid peroxidation. Plant Physiol. 138 1422–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, H., Song, W., Sage, T.L., and DellaPenna, D. (2006). Tocopherols play a crucial role in low-temperature adaptation and phloem loading in Arabidopsis. Plant Cell 18 2710–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mert-Turk, F., Bennett, M.H., Mansfield, J.W., and Holub, E.B. (2003). Camalexin accumulation in Arabidopsis thaliana following abiotic elicitation or inoculation with virulent or avirulent Hyaloperonospora parasitica. Physiol. Mol. Plant Pathol. 62 137–145. [Google Scholar]
- Mezzari, M.P., Walters, K., Jelinkova, M., Shih, M.C., Just, C.L., and Schnoor, J.L. (2005). Gene expression and microscopic analysis of Arabidopsis exposed to chloroacetanilide herbicides and explosive compounds. A phytoremediation approach. Plant Physiol. 138 858–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J.D., Arteca, R.N., and Pell, E.J. (1999). Senescence-associated gene expression during ozone-induced leaf senescence in Arabidopsis. Plant Physiol. 120 1015–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenschnabel, I., Goodman, B.A., Deighton, N., Lyon, G.D., and Williamson, B. (2001). Botrytis cinerea induces the formation of free radicals in fruits of Capsicum annuum at positions remote from the site of infection. Rapid communication. Protoplasma 218 112–116. [DOI] [PubMed] [Google Scholar]
- Mueller, M.J. (1998). Radically novel prostaglandins in animals and plants: The isoprostanes. Chem. Biol. 5 R323–R333. [DOI] [PubMed] [Google Scholar]
- Mueller, M.J. (2004). Archetype signals in plants: The phytoprostanes. Curr. Opin. Plant Biol. 7 441–448. [DOI] [PubMed] [Google Scholar]
- Mueller, M.J., Mene-Saffrane, L., Grun, C., Karg, K., and Farmer, E.E. (2006). Oxylipin analysis methods. Plant J. 45 472–489. [DOI] [PubMed] [Google Scholar]
- Munne-Bosch, S., and Alegre, L. (2002). The function of tocopherols and tocotrienols in plants. CRC Crit. Rev. Plant Sci. 21 31–57. [Google Scholar]
- Parchmann, S., Gundlach, H., and Mueller, M.J. (1997). Induction of 12-oxo-phytodienoic acid in wounded plants and elicited plant cell cultures. Plant Physiol. 115 1057–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls, K.P., and Thompson, J.E. (1980). In vitro simulation of senescence-related membrane damage by ozone-induced lipid peroxidation. Nature 283 504–506. [DOI] [PubMed] [Google Scholar]
- Porfirova, S., Bergmuller, E., Tropf, S., Lemke, R., and Doermann, P. (2002). Isolation of an Arabidopsis mutant lacking vitamin E and identification of a cyclase essential for all tocopherol biosynthesis. Proc. Natl. Acad. Sci. USA 99 12495–12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, N.A., Caldwell, S.E., and Mills, K.A. (1995). Mechanisms of free-radical oxidation of unsaturated lipids. Lipids 30 277–290. [DOI] [PubMed] [Google Scholar]
- Pryor, W.A., and Church, D.F. (1991). Aldehydes, hydrogen peroxide, and organic radicals as mediators of ozone toxicity. Free Radic. Biol. Med. 11 41–46. [DOI] [PubMed] [Google Scholar]
- Reuber, T.L., and Ausubel, F.M. (1996). Isolation of Arabidopsis genes that differentiate between resistance responses mediated by the RPS2 and RPM1 disease resistance genes. Plant Cell 8 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky, L., Davletova, S., Liang, H., and Mittler, R. (2004). The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J. Biol. Chem. 279 11736–11743. [DOI] [PubMed] [Google Scholar]
- Roetschi, A., Si-Ammour, A., Belbahri, L., Mauch, F., and Mauch-Mani, B. (2001). Characterization of an Arabidopsis-Phytophthora pathosystem: Resistance requires a functional PAD2 gene and is independent of salicylic acid, ethylene and jasmonic acid signalling. Plant J. 28 293–305. [DOI] [PubMed] [Google Scholar]
- Sakuragi, Y., Maeda, H., Dellapenna, D., and Bryant, D.A. (2006). α-Tocopherol plays a role in photosynthesis and macronutrient homeostasis of the cyanobacterium Synechocystis sp. PCC 6803 that is independent of its antioxidant function. Plant Physiol. 141 508–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler, S.E., Cahoon, E.B., Coughlan, S.J., and DellaPenna, D. (2003). Characterization of tocopherol cyclases from higher plants and cyanobacteria. Evolutionary implications for tocopherol synthesis and function. Plant Physiol. 132 2184–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler, S.E., Gilliland, L.U., Magallanes-Lundback, M., Pollard, M., and DellaPenna, D. (2004). Vitamin E is essential for seed longevity, and for preventing lipid peroxidation during germination. Plant Cell 16 1419–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C. (2005). Chemistry and biology of vitamin E. Mol. Nutr. Food Res. 49 7–30. [DOI] [PubMed] [Google Scholar]
- Schuhegger, R., Nafisi, M., Mansourova, M., Petersen, B.L., Olsen, C.E., Svatos, A., Halkier, B.A., and Glawischnig, E. (2006). CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiol. 141 1248–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster, V.L. (2002). Prostaglandin transport. Prostaglandins Other Lipid Mediat. 68–69 633–647. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3 217–223. [PubMed] [Google Scholar]
- Stintzi, A., Weber, H., Reymond, P., Browse, J., and Farmer, E.E. (2001). Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc. Natl. Acad. Sci. USA 98 12837–12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, C.B., Bariola, P.A., delCardayre, S.B., Raines, R.T., and Green, P.J. (1993). RNS2: A senescence-associated RNase of Arabidopsis that diverged from the S-RNases before speciation. Proc. Natl. Acad. Sci. USA 90 5118–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma, I., Krischke, M., Loeffler, C., and Mueller, M.J. (2004). The isoprostanoid pathway in plants. Chem. Physiol. Lipids 128 135–148. [DOI] [PubMed] [Google Scholar]
- Thoma, I., Loeffler, C., Sinha, A.K., Gupta, M., Krischke, M., Steffan, B., Roitsch, T., and Mueller, M.J. (2003). Cyclopentenone isoprostanes induced by reactive oxygen species trigger defense gene activation and phytoalexin accumulation in plants. Plant J. 34 363–375. [DOI] [PubMed] [Google Scholar]
- Traidl-Hoffmann, C., Mariani, V., Hochrein, H., Karg, K., Wagner, H., Ring, J., Mueller, M.J., Jakob, T., and Behrendt, H. (2005). Pollen-associated phytoprostanes inhibit dendritic cell interleukin-12 production and augment T helper type 2 cell polarization. J. Exp. Med. 201 627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, K., Wong, J.T., Lee, E., Chan, A.C., and Choy, P.C. (1996). Vitamin E potentiates arachidonate release and phospholipase A(2) activity in rat heart myoblastic cells. Biochem. J. 319 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebst, A., Depka, B., and Hollander-Czytko, H. (2002). A specific role for tocopherol and of chemical singlet oxygen quenchers in the maintenance of photosystem II structure and function in Chlamydomonas reinhardtii. FEBS Lett. 516 156–160. [DOI] [PubMed] [Google Scholar]
- Trebst, A., Depka, B., Jager, J., and Oettmeier, W. (2004). Reversal of the inhibition of photosynthesis by herbicides affecting hydroxyphenylpyruvate dioxygenase by plastoquinone and tocopheryl derivatives in Chlamydomonas reinhardtii. Pest Manag. Sci. 60 669–674. [DOI] [PubMed] [Google Scholar]
- Turano, F.J., and Fang, T.K. (1998). Characterization of two glutamate decarboxylase cDNA clones from Arabidopsis. Plant Physiol. 117 1411–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varotto, C., Maiwald, D., Pesaresi, P., Jahns, P., Salamini, F., and Leister, D. (2002). The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. Plant J. 31 589–599. [DOI] [PubMed] [Google Scholar]
- Vogel, J.T., Zarka, D.G., Van Buskirk, H.A., Fowler, S.G., and Thomashow, M.F. (2005). Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 41 195–211. [DOI] [PubMed] [Google Scholar]
- Vollenweider, S., Weber, H., Stolz, S., Chetelat, A., and Farmer, E.E. (2000). Fatty acid ketodienes and fatty acid ketotrienes: Michael addition acceptors that accumulate in wounded and diseased Arabidopsis leaves. Plant J. 24 467–476. [DOI] [PubMed] [Google Scholar]
- Wang, X., and Quinn, P.J. (2000). The location and function of vitamin E in membranes. Review. Mol. Membr. Biol. 17 143–156. [DOI] [PubMed] [Google Scholar]
- Watahiki, M.K., Mori, H., and Yamamoto, K.T. (1995). Inhibitory effects of auxins and related substances on the activity of an Arabidopsis glutathione-S-transferase isozyme expressed in Escherichia coli. Physiol. Plant. 94 566–574. [Google Scholar]
- Weber, H., Chetelat, A., Reymond, P., and Farmer, E.E. (2004). Selective and powerful stress gene expression in Arabidopsis in response to malondialdehyde. Plant J. 37 877–888. [DOI] [PubMed] [Google Scholar]
- Weber, H., Vick, B.A., and Farmer, E.E. (1997). Dinor-oxo-phytodienoic acid: A new hexadecanoid signal in the jasmonate family. Proc. Natl. Acad. Sci. USA 94 10473–10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkosz, R., and Schlappi, M. (2000). A gene expression screen identifies EARLI1 as a novel vernalization-responsive gene in Arabidopsis thaliana. Plant Mol. Biol. 44 777–787. [DOI] [PubMed] [Google Scholar]
- Yamauchi, J., Iwamoto, T., Kida, S., Masushige, S., Yamada, K., and Esashi, T. (2001). Tocopherol-associated protein is a ligand-dependent transcriptional activator. Biochem. Biophys. Res. Commun. 285 295–299. [DOI] [PubMed] [Google Scholar]
- Zhao, J.M., and Last, R.L. (1996). Coordinate regulation of the tryptophan biosynthetic pathway and indolic phytoalexin accumulation in Arabidopsis. Plant Cell 8 2235–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J.M., Williams, C.C., and Last, R.L. (1998). Induction of Arabidopsis tryptophan pathway enzymes and camalexin by amino acid starvation, oxidative stress, and an abiotic elicitor. Plant Cell 10 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, N., Tootle, T.L., and Glazebrook, J. (1999). Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11 2419–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hennig, L., and Gruissem, W. (2005). Gene-expression analysis and network discovery using Genevestigator. Trends Plant Sci. 10 407–409. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.