Abstract
NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1) regulates systemic acquired resistance (SAR) in Arabidopsis thaliana, and current models propose that after treatment with salicylic acid (SA), Cys-82 and Cys-216 of NPR1 are reduced, leading to nuclear import. The interaction of nucleus-localized NPR1 with TGA transcription factors results in the activation of defense genes, including the SAR marker PATHOGENESIS-RELATED-1 (PR-1), and the deployment of SAR. Little is known about how TGA factors or NPR1 regulate transcription or whether a TGA-NPR1 complex forms on DNA. We show that TGA2 and NPR1 are recruited to PR-1 independently of each other and of SA treatment. Consistent with the result that a triple knockout in TGA2/5/6 derepresses PR-1, in vivo plant transcription assays revealed that TGA2 is not an autonomous transcription activator but is a transcriptional repressor in both untreated and SA-treated cells. However, after stimulation with SA, TGA2 is incorporated into a transactivating complex with NPR1, forming an enhanceosome that requires the core of the NPR1 BTB/POZ domain (residues 80 to 91) and the oxidation of NPR1 Cys-521 and Cys-529. These Cys residues are found in a new type of transactivation domain that we term Cys-oxidized. These data further our understanding of the mechanism by which TGA2 and NPR1 activate Arabidopsis PR-1.
INTRODUCTION
Plants, unlike animals, do not possess specialized cells for protection against invading pathogens. Instead, every plant cell must be capable of perceiving pathogens and mounting effective defense responses if the organism is to successfully protect itself from infection. Upon detection of an invading microbe, plant defense responses arise from the activation of signal transduction pathways that lead to global transcriptional reprogramming (Dangl and Jones, 2001; Durrant and Dong, 2004). Among the induced genes are pathogenesis-related (PR) genes, which are activated both at the site of infection and in uninfected parts of the plant in response to the pathogen-induced accumulation of salicylic acid (SA) (Ryals et al., 1996). Local and distal SA accumulations are mandatory for the deployment of a systemic long-lasting and broad-spectrum plant disease resistance response called systemic acquired resistance (SAR) (Ryals et al., 1996; Durrant and Dong, 2004; Pieterse and Van Loon, 2004). Exogenous application of SA, or SA analogs, including 2,6-dichloroisonicotinic acid (INA) and benzothiadiazole, termed chemical SAR, also triggers PR gene induction and SAR deployment (Ward et al., 1991).
The NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1) protein is the key regulator of SAR (Cao et al., 1994; Delaney et al., 1995). In resting cells of wild-type Arabidopsis thaliana, NPR1 is found in both the cytoplasm and the nucleus (Després et al., 2000). However, in an npr1-1 mutant line of Arabidopsis overexpressing an NPR1:green fluorescent protein (GFP) fusion protein, the NPR1 fusion is sequestered in the cytoplasm and localizes to the nucleus only after INA treatment (Kinkema et al., 2000). The cytoplasmic NPR1:GFP fusion protein is contained within an oligomer complex held together by disulfide bridges (Mou et al., 2003). Upon INA treatment, NPR1 Cys-82 and Cys-216 are presumably reduced and NPR1:GFP is released from this complex, resulting in the accumulation of protein monomers inside the nucleus (Mou et al., 2003).
Activation of PR genes during SAR, which requires the nuclear localization of NPR1 (Kinkema et al., 2000), is also dependent on a functionally redundant clade of three basic Leu zipper TGA transcription factors, TGA2, TGA5, and TGA6, that interact with NPR1 (Zhang et al., 1999; Després et al., 2000). A triple knockout of these TGA genes abolished PR-1 induction by INA, indicating that the gene products could act as transcriptional activators (Zhang et al., 2003). This conclusion is supported by a report in which a chimeric TGA2-GAL4:DB protein was used to study gene regulation and proposed to act as a transcriptional activator (Fan and Dong, 2002). However, in a finding that appears to be contradictory to the previous one, Zhang et al. (2003) showed that whether unstimulated or INA-treated, the triple knockout plants displayed higher levels of PR-1 (compared with levels found in the wild type without INA), which could indicate that the proteins of the TGA2-containing clade act as repressors of PR-1, presumably by binding to its promoter (Zhang et al., 2003). Furthermore, chromatin immunoprecipitation (ChIP) experiments have demonstrated that TGA2 physically interacts with the PR-1 promoter in an SA- and NPR1-dependent manner (Johnson et al., 2003), which would also contradict the hypothesis that TGA2 binds to the PR-1 promoter in the absence of SA (Zhang et al., 2003). Thus, it is not clear whether TGA2 is a transcriptional activator or a repressor. PR-1 is also positively regulated in an SA-dependent, but NPR1-independent, manner by the transcription factor WHY1 (Desveaux et al., 2004). Furthermore, PR-1 is negatively regulated by SUPPRESSOR OF NPR1-INDUCIBLE1 (SNI1) (Li et al., 1999), and ChIP experiments have shown an increase in histone H3 acetylation and methylation at the PR-1 promoter in sni1 mutant plants (Mosher et al., 2006). These data implicate chromatin structure in the regulation of PR-1 expression.
NPR1 and TGA factors (TGA1 and TGA2) show a direct physical interaction within the nucleus and in vitro (Subramaniam et al., 2001; Fan and Dong, 2002; Després et al., 2003). This interaction stimulates the DNA binding activity of TGA factors to their cognate cis-acting element in vitro (Després et al., 2000, 2003) and in vivo (Fan and Dong, 2002). However, despite the fact that NPR1 and TGA2 can form a ternary complex on the DNA (DNA-TGA2-NPR1 complex) in yeast (Weigel et al., 2005), it is unclear whether, when inside a plant nucleus, they will only interact in the nucleoplasm or whether they will form such a ternary complex. To date, there is no experimental evidence indicating that NPR1 is actually recruited to the PR-1 gene in vivo. Therefore, aside from its DNA binding enhancement activity on TGA factors, the biochemical role of NPR1 in NPR1-TGA complexes, if any, remains speculative.
NPR1 contains two protein–protein interaction motifs: ankyrin repeats (Cao et al., 1997; Ryals et al., 1997; Mosavi et al., 2004) and a BTB/POZ (for Broad complex, Tramtrack, and Bric-a-brac/Pox virus and Zinc finger) domain (Bardwell and Treisman, 1994; Aravind and Koonin, 1999). The ankyrin repeats mediate interactions with TGA factors, and their mutation abolishes NPR1-TGA complex formation, PR gene expression, and SAR (Cao et al., 1997; Ryals et al., 1997; Zhang et al., 1999; Després et al., 2000, 2003). The functional requirements of the NPR1 BTB/POZ in disease resistance are not yet understood.
Here, we demonstrate that TGA2 is not a transcriptional activator in resting or SA-treated cells, as it is unable to activate transcription when expressed on its own. We show that TGA2 and NPR1 can, independently of one another, physically interact with the PR-1 promoter in both resting and SA-treated cells. We also show that NPR1 contains an autonomous transactivation domain in its C terminus and acts as a coactivator in SA-treated cells, where it associates with TGA2 to create a transcriptional activating complex. NPR1 and TGA2 are sufficient to activate gene expression after stimulation of the cells with SA; thus, the DNA-TGA2-NPR1 ternary complex constitutes an SA-dependent enhanceosome. We demonstrate that the coactivator function of NPR1 requires the presence of the BTB/POZ core and the oxidation of Cys-521 and Cys-529, located in the transactivation domain of NPR1. Finally, using an in vivo labeling technique capable of distinguishing between the reduced and oxidized states of Cys residues, we determined that Cys-521 and Cys-529 are oxidized in both resting and SA-treated cells. The data presented here not only provide a mechanistic understanding of transcriptional regulation mediated by the TGA2-NPR1 complex but also help to elucidate the biochemical function of TGA2, a repressor of NPR1-mediated derepression, and NPR1, a coactivator, and to unravel the existence of a new type of eukaryotic transactivation domain that we term the Cys-oxidized transactivation domain.
RESULTS
Recruitment of TGA2 to the PR-1 Promoter Is Both SA- and NPR1-Independent
SA induction of the PR-1 gene is positively controlled by a clade of three TGA factors (TGA2, TGA5, and TGA6) with redundant functions. In triple TGA knockout plants, the levels of PR-1 transcripts were up to 50-fold higher compared with those in nonstimulated wild-type plants (Zhang et al., 2003). This was interpreted as a loss of TGA factor binding to a negative element in the PR-1 promoter; however, whether this effect was attributable to direct binding of the TGA factors to DNA was not addressed. If the interpretation of Zhang et al. (2003) is correct, their results would contradict those of Johnson et al. (2003), who demonstrated using ChIP that recruitment of TGA2 to the PR-1 promoter is both SA- and NPR1-dependent. These ChIPs were performed on endogenous TGA2 using an anti-TGA2 antibody raised against the N terminus. However, because ChIPs can generate false negatives when epitopes are inaccessible, we sought to determine whether the apparent lack of interaction between TGA2 and the PR-1 promoter in resting cells observed by Johnson et al. (2003) is attributable to the absence of antibody recognition, to masking of the epitope, or to the absence of TGA2.
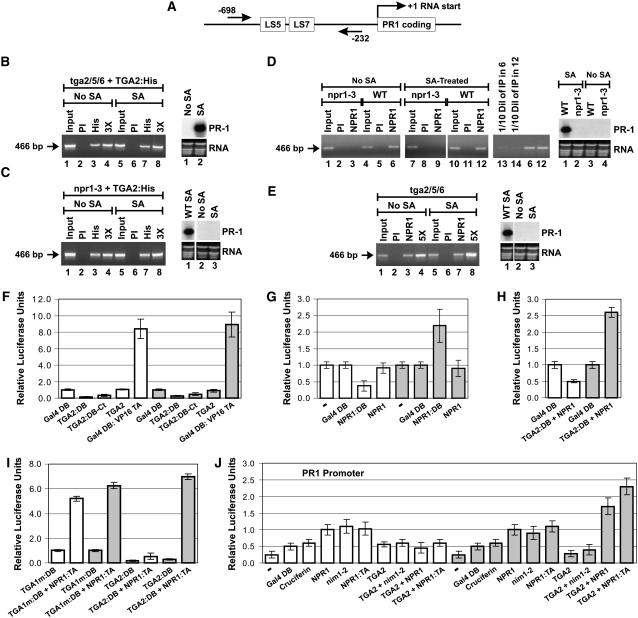
As a means of generating an alternative epitope, the TGA2 coding region was ligated to a 6-His tag (His), and the resulting fusion (TGA2:His), under the control of the cauliflower mosaic virus (CaMV) 35S promoter, was introduced into tga2/5/6 knockout plants. Figure 1A is a diagram of the PR-1 gene that shows the positions of the PCR primers used for all of the ChIP experiments. Figure 1B shows that a PCR product is present in the lanes corresponding to immunoprecipitations performed with the anti-His antibody (lanes 3 and 7), indicating that TGA2:His interacted with PR-1 in both untreated and SA-treated cells. Immunoprecipitation with preimmune serum (PI) did not lead to a detectable band (lanes 2 and 6). ChIP performed with the anti-His antibody on the untransformed tga2/5/6 mutant plant also did not lead to a detectable band (data not shown). The right section of Figure 1B shows the results of RNA gel blot analysis of PR-1, indicating that TGA2:His restored PR-1 inducibility in the tga2/5/6 mutant in an SA-dependent manner and that expression of PR-1 is not constitutive in these plants.
Figure 1.
NPR1 Is a Coactivator Required for Transcriptional Activation by a TGA2-NPR1 Complex in SA-Treated Cells Only.
(A) Graphic representation of the PR-1 gene. The straight arrows and the numbers indicate the positions of the PCR primers used for ChIP experiments. LS5 and LS7 are two DNA regions containing the TGA factor cognate binding sequence TGACG (Lebel et al., 1998).
(B) ChIPs of TGA2:His expressed in tga2/5/6 knockout plants. At right is an RNA gel blot illustrating that the TGA2:His protein complemented the tga2/5/6 mutation and restored PR-1 inducibility.
(C) ChIPs of TGA2:His expressed in npr1-3 mutant (npr1-3) Arabidopsis plants. At right is an RNA gel blot illustrating that the TGA2:His protein expressed in the npr1-3 mutation did not bring about expression of PR-1 regardless of whether tissues were treated or not with SA. An RNA gel blot from wild-type plants treated with SA is shown for comparison.
For (B) and (C), ChIPs were conducted with anti-His antibodies conjugated to agarose beads.
(D) ChIPs of NPR1 from wild-type (NPR1) and npr1-3 mutant (npr1-3) plants. At right is an RNA gel blot illustrating that the PR-1 gene is not expressed in the npr1-3 mutant and that expression in the wild-type plant is dependent upon SA treatment.
(E) ChIPs of NPR1 from tga2/5/6 knockout Arabidopsis plants. At right is an RNA gel blot illustrating that the PR-1 gene is not expressed in the tga2/5/6 mutant regardless of whether tissues have been treated or not with SA. An RNA gel blot from wild-type plants treated with SA is shown for comparison.
For (D) and (E), ChIPs were conducted with anti-NPR1 antibodies for which the specificity has been demonstrated previously (Després et al., 2000).
In (B) to (E), tissues were untreated (No SA) or treated for 6 h with 1 mM SA. PI indicates that ChIP was performed with preimmune serum. PCR was conducted with PR-1 promoter–specific primers. The arrow in each panel indicates the location of the PCR product. The NPR1-3 protein is a deletion version of NPR1 (Cao et al., 1997) that has lost the antigenic region used to raise the anti-NPR1 antibodies used in this study. The inputs represent 2% of the immunoprecipitated material (50-fold dilution). 3X and 5X indicate that the PCR was performed with three and five times the amount of immunoprecipitated material, respectively, to demonstrate that the PCR was in the linear range. In lanes 13 and 14 in (D), one-tenth of the amount of immunoprecipitated material used in lanes 6 and 12, respectively, was used to perform the PCR to demonstrate that the PCR was in the linear range. RNA stained with ethidium bromide is shown for loading comparison.
(F) Histograms illustrating the fact that TGA2 tethered to DNA through Gal4 DB fused to the N terminus (TGA2:DB) or C terminus (TGA2:DB-Ct) does not activate transcription, whereas a chimeric transcription activator composed of the Gal4 DB fused to the transactivation domain of viral protein 16 (Gal4 DB:VP16 TA) does. Gal4 DB represents the baseline level of transcription.
(G) Histograms illustrating the transcription activation of NPR1 tethered to DNA through Gal4 DB (NPR1:DB). NPR1 indicates the absence of fusion. – indicates that only the reporter and internal standard vectors were bombarded into the tissues; no effector was introduced.
(H) Histograms illustrating the effect of NPR1 on the transcriptional activity of TGA2:DB. NPR1 indicates that the protein is expressed without a fusion.
(I) Histograms illustrating the fact that TGA2 tethered to DNA through Gal4 DB (TGA2:DB) interacts very poorly with NPR1:TA in the absence of SA treatment.
In (F) to (I), Arabidopsis leaves were left untreated (white bars) or were treated for 24 h with 1 mM SA (gray bars). The constructs were transfected along with the 5X UASGAL4:firefly luciferase reporter and the CaMV 35S:renilla luciferase internal standard vectors. Data are reported as relative luciferase units. The fold activation represents the relative luciferase units obtained with the given protein or protein pair divided by the relative luciferase units obtained with the unfused Gal4 DB construct alone (baseline transcription). Values are from 25 samples and represent averages ± sd. Every bar represents five bombardments repeated five times (n = 25).
(J) Histograms illustrating the effect of NPR1 and nim1-2 on the transcriptional activity of the TGA2-NPR1 complex. All proteins were native (without fusion), with the exception of NPR1:TA (NPR1 fused to the VP16 transactivation domain), which was used to assess the level of interaction between NPR1 and TGA2 in the context of the PR-1 promoter. The reporter system was the Arabidopsis PR-1 promoter fused to firefly luciferase. The CaMV 35S promoter:renilla luciferase fusion was used as an internal standard. − indicates that no effector was bombarded along with the reporter and internal standard vectors. Cruciferin is an Arabidopsis storage protein used here to illustrate the background level of this system when expressing an unrelated protein. Gal4 DB served the same purpose. Arabidopsis leaves were left untreated (white bars) or were treated for 24 h with 1 mM SA (gray bars). Data are reported as relative luciferase units. Values are from 25 samples and represent averages ± sd. Every bar represents five bombardments repeated five times (n = 25).
These results indicate that the lack of interaction previously reported by Johnson et al. (2003) in the absence of SA might have been attributable to the masking, under certain conditions, of the N-terminal TGA2 epitope chosen by those authors. Therefore, it became relevant to test whether the same phenomenon was responsible for the lack of interaction reported between TGA2 and PR-1 in the npr1 background (Johnson et al., 2003). To do so, TGA2:His, under the control of the CaMV 35S promoter, was introduced in the npr1-3 mutant background and ChIPs were performed using the anti-His antibody. The results in Figure 1C show the presence of a PCR product in the lanes corresponding to immunoprecipitations performed with the anti-His antibody (lanes 3 and 7). This finding indicates that, in the absence of NPR1, TGA2:His interacted with PR-1 in both untreated and SA-treated cells. Immunoprecipitation with PI did not lead to a detectable band (lanes 2 and 6). ChIP performed with the anti-His antibody on the untransformed npr1-3 mutant plant did not lead to a detectable band (data not shown). As indicated by RNA gel blot analysis (right section of Figure 1C), expression of TGA2:His in the npr1-3 mutant background did not lead to the expression of PR-1, regardless of whether the cells were subjected to SA treatment.
Recruitment of NPR1 to the PR-1 Promoter Is Both SA- and TGA2/5/6-Independent
Because no experimental evidence exists to indicate that NPR1 can be recruited to the PR-1 promoter, it is unclear whether NPR1 is capable of forming a complex with TGA2 on DNA to modulate transcription. To address this question, we performed ChIP experiments with wild-type and npr1-3 mutant Arabidopsis plants before and after SA treatment (Figure 1D). The npr1-3 mutant was chosen as a negative control because this allele carries a premature stop codon (Cao et al., 1997), which removes the amino acid region used to raise the anti-NPR1 antibody (Després et al., 2000). The specificity of the anti-NPR1 antibody has been demonstrated previously (Després et al., 2000) and can also be seen in Figure 2C, where a band corresponding to NPR1 was detected in the wild-type plant (lane 1) but not in the npr1-3 mutant plant (lane 2). With the exception of input lanes (lanes 1 and 7), ChIP performed on the npr1-3 lines did not yield a band, regardless of whether cells were treated with SA or the immunoprecipitation antibodies were from PI or raised against NPR1. Conversely, ChIP performed on wild-type plants indicated that NPR1 interacted with PR-1 in both untreated cells and cells treated with SA (lanes 6 and 12). Immunoprecipitation with PI did not lead to a detectable band (lanes 5 and 11). RNA gel blot analysis (right section of Figure 1D) confirmed that PR-1 was not expressed in the npr1-3 mutant, nor was it induced in the wild-type plant before SA treatment.
Figure 2.
The BTB/POZ Domain of NPR1 Is Required for PR-1 Induction.
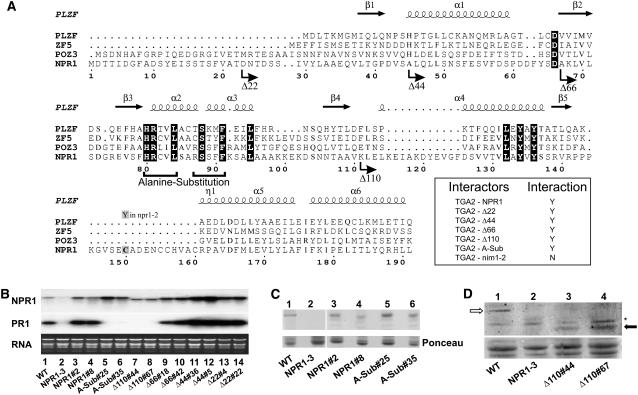
(A) Multiple alignment of selected BTB/POZ domains. Residues blocked in black are conserved among all sequences. Numbers refer to the amino acid position in NPR1. Straight arrows and coils indicate the positions of β-strands and helices in the PLZF crystal structure, respectively. α and η indicate α- and 310-helices, respectively, and β refers to β-strands. The bent arrows indicate the positions where the NPR1 deletion proteins begin. The horizontal brackets below the amino acid sequences of α2 and α3 indicate the residues that have been mutated to Ala in the Ala substitution mutant. Cys-150 bears a C-to-Y mutation in the npr1-2 mutant, which abolishes interaction with TGA2, PR gene activation, and deployment of SAR (Cao et al., 1997; Zhang et al., 1999; Després et al., 2000). The inset represents directed yeast two-hybrid assays using the filter test and the outcome of the experiments. nim1-2 is a mutant version of NPR1 that bears a His-to-Tyr replacement in one of the ankyrin repeats (Ryals et al., 1997), which abolishes interaction with TGA factors (Després et al., 2000, 2003). Y (yes) indicates an interaction, whereas N (no) indicates an absence of interaction (white color after 24 h of incubation with X-Gal).
(B) RNA gel blot analysis using NPR1 or PR-1 probes. RNA stained with ethidium bromide is shown for loading comparison. Lane 1 contains RNA from wild-type Arabidopsis, and lane 2 contains RNA from the npr1-3 mutant. The remaining lanes contain RNA from npr1-3 lines expressing the following constructs: wild-type NPR1 (lanes 3 and 4), the Ala substitution mutant (lanes 5 and 6), and the deletion mutants Δ110 (lanes 7 and 8), Δ66 (lanes 9 and 10), Δ44 (lanes 11 and 12), and Δ22 (lanes 13 and 14). Results from two independent transgenic lines are shown per construct. Specific line numbers follow the construct name.
(C) and (D) Top panels, immunoblot analysis of proteins from wild-type Arabidopsis, the npr1-3 mutant (NPR1-3), and the npr1-3 background lines expressing NPR1, the Ala substitution mutant, and Δ110 as described in (B). An anti-NPR1 antibody (Després et al., 2000) was used. Bottom panels, Ponceau staining of the membranes shown in the top panels. In (D), the open arrow indicates the position of the full-length NPR1 protein (66 kD), whereas the closed arrow indicates the position of the truncated protein Δ110 (54.4 kD). The asterisk indicates a protein interacting nonspecifically with the antibody.
Intuitively, knowing that NPR1 and TGA2 can interact with each other and because NPR1 does not contain a known DNA binding domain, one could expect the recruitment of NPR1 to the PR-1 promoter to be dependent on TGA2. To test this hypothesis, we performed ChIP experiments on the tga2/5/6 mutant plant using the anti-NPR1 antibody. The presence of a PCR product in the lanes corresponding to immunoprecipitations performed with the anti-NPR1 antibody (Figure 1E, lanes 3 and 7) indicates that NPR1 continues to interact with PR-1 in the absence of TGA2/5/6 in both untreated and SA-treated cells. Immunoprecipitation with PI did not lead to a detectable band (lanes 2 and 6). Note that formaldehyde, the cross-linker used in the ChIP experiments, can cross-link protein to DNA but also protein to protein (Buck and Lieb, 2004). Hence, recruitment of NPR1 to the PR-1 promoter does not indicate that NPR1 binds directly to DNA. The RNA gel blot shown in the right section of Figure 1E confirmed that PR-1 was not expressed in the tga2/5/6 mutant regardless of whether the cells were subjected to SA treatment.
NPR1 Is a Coactivator Required for Transcriptional Activation by a TGA2-NPR1 Complex in SA-Treated Cells Only
PR-1 is positively regulated by NPR1 (Cao et al., 1997; Ryals et al., 1997) and by TGA2/5/6 (Zhang et al., 2003). This prompted us to test whether TGA2 can act as a transcriptional activator. To do so, TGA2:DB (N-terminal fusion) and TGA2:DB-Ct (C-terminal fusion) were assayed using an in vivo plant transcription assay (Figure 1F). The baseline level of transcription was determined by transfecting leaves with Gal4 DB (not fused to any other protein or protein domain) along with a reporter construct consisting of a firefly luciferase gene under the control of five copies of the Gal4 upstream activating sequences (UAS) fused to a minimal promoter. Transfection with TGA2:DB or TGA2:DB-Ct did not result in reporter gene activation beyond the baseline level, regardless of whether cells were treated with SA or not. The same result was obtained with TGA2 that was not fused to Gal4 DB or any other foreign protein domain (TGA2). Transfection with Gal4 DB fused to a strong transactivation domain (Gal4 DB:VP16 TA) led to SA-independent expression of the reporter gene well above the baseline (Figure 1F, white versus gray bars). These results demonstrate that the reporter gene can indeed be activated under our experimental conditions and indicate that TGA2 is not a transcriptional activator, regardless of whether the cells are stimulated with SA. Indeed, we have found that TGA2 is capable of repressing an activated reporter gene before and after SA treatment (see Supplemental Figure 1 online), indicating that TGA2 is not a transcriptional activator but a repressor.
Knowing that NPR1 can be recruited to a promoter in vivo (Figures 1D and 1E), we tested whether NPR1 can activate transcription when tethered to DNA. To accomplish this, NPR1 was fused to Gal4 DB (NPR1:DB) and assayed using the in vivo plant transcription assay (Figure 1G). In untreated cells (white bars), NPR1:DB did not lead to gene activation beyond the baseline level. However, after SA treatment (gray bars), NPR1:DB activated transcription 2.2-fold above the baseline level. Expression of NPR1 without fusion to Gal4 DB (NPR1) did not lead to gene activation that was significantly different from the baseline level (P < 0.05), indicating that transactivation by NPR1:DB observed with SA was dependent on the recruitment of NPR1 to the promoter. These results indicate that NPR1 could potentially act as a transcriptional coactivator if recruited to a promoter via a DNA binding protein, such as TGA2.
We next addressed whether NPR1 could modulate the transcriptional properties of TGA2. When TGA2:DB was coexpressed with NPR1 (not fused to any foreign transcription activation or DNA binding domains), expression of the reporter gene in untreated cells did not increase beyond the baseline (Figure 1H, white bars). However, transcription increased 2.6-fold above the baseline level after SA treatment (Figure 1H, gray bars). Because neither TGA2:DB nor NPR1 activates transcription of the reporter gene on its own (Figures 1F and 1G) and NPR1 stimulates transcription when tethered to DNA (Figure 1G, NPR1:DB), the results in Figure 1H suggest that the transcriptional activation observed when NPR1 (unfused) is coexpressed with TGA2:DB is likely attributable to NPR1 being tethered, or recruited, to the DNA through TGA2:DB. Physical interaction between TGA2 and NPR1 at the reporter gene promoter and in the presence of SA was demonstrated using plant two-hybrid assays (Figure 1I, TGA2:DB + NPR1:TA). Together, these observations are consistent with the formation of a ternary complex between DNA, TGA2:DB, and NPR1, with NPR1 acting as a coactivator of TGA2 on the Gal4-based promoter.
Using plant two-hybrid assays (Figure 1I), we showed that, in the absence of SA (white bars), NPR1 fused to VP16 TA (NPR1:TA) also interacted with TGA2:DB (significant difference of P < 0.05 between TGA2:DB and TGA2:DB + NPR1:TA), but very poorly. A similar conclusion was reached based on data from a protein fragment complementation assay (Subramaniam et al., 2001). Thus, in addition to the fact that NPR1:DB (tethered to DNA) does not transactivate in the absence of SA, the very weak interaction between NPR1:TA and TGA2:DB in unstimulated cells may also account for the lack of transcriptional stimulation by NPR1. We also confirmed that, in the absence of SA, NPR1:TA is competent to interact with other proteins, as demonstrated by its interaction with a mutant version of TGA1 (Figure 1I, TGA1m:DB), which was shown previously to interact with NPR1 in the presence and absence of SA treatment (Després et al., 2003).
Next, we tested the transactivation properties of NPR1 and TGA2 in the context of the PR-1 promoter. DNA coding for native (unfused) proteins was delivered by biolistics as in Figures 1F to 1I, except that the reporter consisted of the luciferase gene under the control of the PR-1 promoter (Figure 1J). Relatively low levels of luciferase activity were detected after transfection of this reporter gene without effector plasmids (−). Transfection of two unrelated effectors, Gal4 DB, which does not bind PR-1 (there is no Gal4 binding site in the PR-1 promoter), and the seed storage protein cruciferin, increased reporter gene expression, which most likely represents the unspecific effect of expressing a protein in this system. Thus, expression levels observed with these proteins represent the baseline of this system. Whether cells were treated (gray bars) or not (white bars) with SA, NPR1 led to activation of the PR-1 promoter beyond the baseline level. TGA2, on the other hand, had no effect on the baseline activity of the promoter. However, in untreated cells (white bars), when NPR1 was coexpressed with TGA2, transcription values were brought back to the baseline level, indicating that TGA2 repressed the NPR1-dependent activation of PR-1. As observed with NPR1, protein nim1-2, a variant of NPR1 with a mutation in an ankyrin repeat that does not interact with TGA2 (Ryals et al., 1997; Després et al., 2000), also activated the PR-1 promoter in the absence of TGA2 in untreated and SA-treated cells. This finding suggests that the ankyrin repeats are unlikely to be involved in the recruitment of NPR1 to the PR-1 promoter. Furthermore, because nim1-2 does not interact with TGA2, this result is also consistent with a TGA2-independent recruitment of NPR1 to the PR-1 promoter, as was observed with ChIPs (Figure 1E). Coexpression of nim1-2 with TGA2 also restored transcription values to the baseline level. Coexpression of TGA2 and NPR1 in SA-treated tissues (gray bars) led to the activation of PR-1 beyond the baseline and significantly beyond what was observed with NPR1 alone (P < 0.05), confirming that NPR1 acts as a TGA2 coactivator on the PR-1 promoter. Also, coexpression of TGA2 with NPR1:TA established that the two proteins interact on the PR-1 promoter only in the presence of SA (gray bars), because values observed with TGA2 + NPR1:TA were significantly higher than those obtained with TGA2 + NPR1 (P < 0.05) or NPR1:TA alone (P < 0.05). Our results indicate that, in untreated cells, TGA2 represses the NPR1-dependent activation of PR-1 without the two proteins interacting with each other. However, after SA treatment, the two proteins interact to form a ternary complex with PR-1 DNA, in which NPR1 acts as a TGA2 coactivator.
The BTB/POZ Domain of NPR1 Is Required for PR-1 Activation by SA
To determine the functional importance of the NPR1 BTB/POZ domain, we generated a series of rational mutants based on information available from other model systems. Of the four known structural classes of BTB domains (BTB Zinc Finger, Skp1, ElonginC, and T1), NPR1 is more similar to those associated with zinc fingers, the so-called long form (Stogios et al., 2005). We thus performed a small-scale multiple alignment (Figure 2A) of long-form BTB/POZ domains, including the one from human promyelocytic leukemia zinc finger (PLZF), the archetypical BTB/POZ domain (see Aravind and Koonin, 1999, for a more exhaustive alignment of 79 BTB/POZ domains, including that of NPR1). Also shown is a representation of the secondary structure of the PLZF BTB/POZ derived from its crystal structure (Ahmad et al., 1998).
Because the N-terminal region of the NPR1 BTB/POZ is longer than that of PLZF, we used the protein secondary structure prediction PSIPRED (Jones, 1999) and identified a potential β-strand formed by residues 19 to 22 (FVAT). Deletion of this putative structure generated the Δ22 mutant (Figure 2A). The next deletion, corresponding to the Δ44 mutant, removed β1, which has been shown to partially destabilize the PLZF dimer (Ahmad et al., 1998). Deletion mutant Δ66 removed all of the structural determinants (β1, α1, and D65) mandatory for BTB/POZ homodimerization (Ahmad et al., 1998). The α2 and α3 helices are buried within the BTB/POZ and constitute the monomer core of the domain (Ahmad et al., 1998). Ala substitution of the core in PLZF results in disruption of the BTB/POZ fold (Melnick et al., 2000). The core region is well conserved in NPR1, and of note, the sequence RSSFF (residues 87 to 91 of NPR1) is identical to the corresponding region in POZ3, and the sequence HRCVL (residues 80 to 84) is identical to the corresponding region in ZF5. Thus, to permit functional testing of the NPR1 BTB/POZ core without deleting other elements, the conserved residues in α2 and α3 were substituted to Ala (Figure 2A, alanine substitution brackets). Finally, because β2, β3, and β4 form a tertiary structure, an N-terminal deletion aimed at removing the core of the BTB/POZ was created after β4 but before the next structural element (Δ110 deletion mutant). The five NPR1 variants mutated in the BTB/POZ (Δ22, Δ44, Δ66, Δ110, and the Ala substitution) all interacted with TGA2 in yeast two-hybrid assays (Figure 2A, inset). Quantitative yeast two-hybrid tests confirmed that the five NPR1 mutants interacted with TGA2. However, the data also indicated that these mutants did not interact with TGA2 as well as did the full-length wild-type NPR1 (see Supplemental Figure 2 online).
To assess the biological significance of the NPR1 BTB/POZ in controlling PR-1 expression, we created and tested five cDNA constructs encoding the proteins depicted in Figure 2A. These were introduced, under the control of the CaMV 35S promoter, into the npr1-3 genetic background (Figure 2B). As a control, npr1-3 plants were transformed with the full-length wild-type NPR1 coding region fused to the CaMV 35S promoter (NPR1, lanes 3 and 4). Wild-type Arabidopsis accumulated PR-1 transcripts when treated with 0.5 mM SA for 16 h (lane 1), whereas npr1-3 plants (NPR1-3) did not (lane 2). PR-1 gene expression was restored in 21 of the 25 independent transgenic npr1-3 lines expressing NPR1 (lanes 3 and 4; data not shown), in all 23 lines expressing Δ22 (lanes 13 and 14; data not shown), in 35 of 38 lines expressing Δ44 (lanes 11 and 12; data not shown), in 18 of 24 lines expressing Δ66 (lanes 9 and 10; data not shown), but in none of the 31 and 40 independent lines expressing the Ala-substituted BTB/POZ (lanes 5 and 6; data not shown) and Δ110 (lanes 7 and 8; data not shown), respectively. Figures 2C and 2D indicate that the Δ110 and Ala substitution proteins were expressed in these lines. PR-1 transcripts were not detected in any of the lines tested in the absence of SA (data not shown).
Altogether, the results shown in Figure 2 indicate that although Ala substitution and Δ110 can interact with TGA2 (Figure 2A, inset; see Supplemental Figure 2 online), their expression cannot complement the npr1-3 mutation, demonstrating that the interaction of NPR1 with TGA2 is in itself not sufficient for biological activity and that the core of the NPR1 BTB/POZ, in the context of the full-length NPR1, is required for PR-1 induction.
The NPR1 BTB/POZ Core Is Required for the TGA2 Coactivator Function of NPR1 in SA-Treated Cells
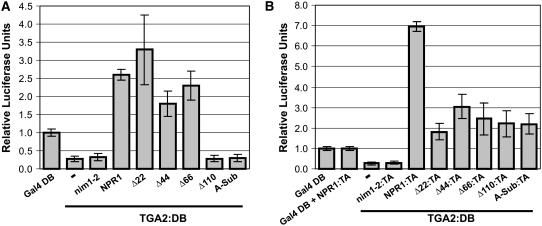
To establish a link between the complementation of PR-1 expression and the transactivation of the TGA2-NPR1 complex, we determined whether the deletions and the Ala substitution of the NPR1 BTB/POZ affected the capacity of this protein to act as a TGA2 coactivator. Deletions of the first 22, 44, or 66 amino acids of NPR1 did not substantially affect the capacity of NPR1 to convert TGA2:DB into an activator after treatment with SA (Figure 3A). However, deleting the first 110 amino acids or substituting the BTB/POZ core with Ala residues abolished transactivation of the coexpressed TGA2:DB (Figure 3A). The nim1-2 protein, which does not interact with TGA2, served as a negative control. In the absence of SA treatment, none of the mutants significantly altered the transactivation of TGA2:DB compared with results obtained with full-length NPR1 (Figure 1F); accordingly, data are not shown. We also tested the Ala substitution and Δ110 proteins for their capacity to interact with TGA2 in the plant two-hybrid system (Figure 3B), which evaluates interaction in the context of the promoter. The data indicate that Ala substitution and Δ110 fused to VP16 TA interacted with TGA2:DB with no significant differences in the level of interaction (P < 0.05) compared with Δ22, Δ44, and Δ66. However, the interaction of these five mutant proteins with TGA2 was significantly lower than that observed with wild-type NPR1 (P < 0.05). These results are consistent with those obtained with quantitative yeast two-hybrid assays (see Supplemental Figure 2 online). Together, the findings shown in Figure 3 indicate that amino acids located between residues 66 and 110 of NPR1, more precisely residues 80 to 84 and/or 87 to 91, which constitute the core of the BTB/POZ, are required for the TGA2 coactivator function of NPR1.
Figure 3.
The Core of the NPR1 BTB/POZ Is Required for the TGA2-Dependent Coactivator Function of NPR1 in SA-Treated Cells.
(A) Histograms illustrating the effect of NPR1 and the mutants described for Figure 2 on the transcriptional activity of the TGA2-NPR1 complex tethered to DNA through Gal4 DB fused to TGA2. Results obtained with TGA2:DB alone (−) are also shown.
(B) Histograms illustrating the interaction of NPR1 and the mutants described in (A) fused to the VP16 transactivation domain with TGA2 fused to the Gal4 DB. Results obtained with Gal4 DB alone (Gal4 DB), Gal4 DB coexpressed with NPR1:TA (Gal4 DB + NPR1:TA), and TGA2:DB alone (−) are also shown.
For (A) and (B), conditions were identical to those described for Figure 1. Data are reported as relative luciferase units. Values are from 25 samples and represent averages ± sd. Every bar represents five bombardments repeated five times (n = 25).
NPR1 Harbors a Cryptic Transactivation Domain in Its Last 80 Amino Acids
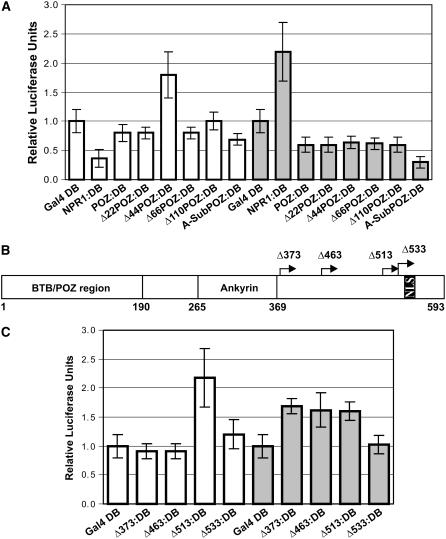
Given that the core of the NPR1 BTB/POZ is required for transactivation of the TGA2-NPR1 complex, we sought to determine whether this domain harbors autonomous transcriptional regulatory regions. To identify these potential regulatory regions, the NPR1 BTB/POZ (amino acids 1 to 190) was fused to Gal4 DB (POZ:DB) and assayed using the in vivo plant transcription assay (Figure 4A). In the absence (white bars) or presence (gray bars) of SA treatment, POZ:DB and variants, in which the first 22, 66, and 110 amino acids were deleted (Δ22POZ:DB, Δ66POZ:DB, and Δ110POZ:DB) or in which the core of the BTB/POZ was replaced with Ala residues (A-SubPOZ:DB), did not stimulate transcription beyond the baseline level (Gal4:DB). One of the most salient features of this experiment was the uncovering of a cryptic transactivation domain, revealed when the BTB/POZ was shortened by 44 amino acids at the N terminus (Δ44POZ:DB), suggesting that a repressing element is located between amino acids 22 and 44. However, in SA-treated cells, Δ44POZ:DB did not transactivate (Figure 4A, Δ44POZ:DB, gray versus white bars), indicating that the cryptic transactivation domain does not function when cells are induced with SA. Together, the results in Figure 4A indicate that the BTB/POZ domain cannot account for the transactivation properties of the full-length NPR1 tethered to DNA through the Gal4 DB.
Figure 4.
NPR1 Harbors an Autonomous Transactivation Domain in the Last 80 Residues.
(A) Histograms illustrating the transcriptional activity of the NPR1 BTB/POZ domain, the deletion mutants of the BTB/POZ, and the Ala substitution mutant tethered to DNA through Gal4 DB. The deletion and the Ala substitution mutants were created starting with the NPR1 BTB/POZ domain. The BTB/POZ region represents the first 190 amino acids of NPR1.
(B) Scheme of NPR1 and the deletions analyzed in (C). Numbers preceded by Δ indicate the starting amino acid for that particular deletion mutant. NLS indicates the nuclear localization signal. Ankyrin represents the region containing the ankyrin repeats as defined by Pfam and SMART. Diagram is drawn to scale.
(C) Histograms illustrating the transcriptional activity of the NPR1 deletion mutants described for (B) tethered to DNA through Gal4 DB.
For (A) and (C), conditions were identical to those described for Figure 1. Data are reported as relative luciferase units. Values are from 25 samples and represent averages ± sd. Every bar represents five bombardments repeated five times (n = 25).
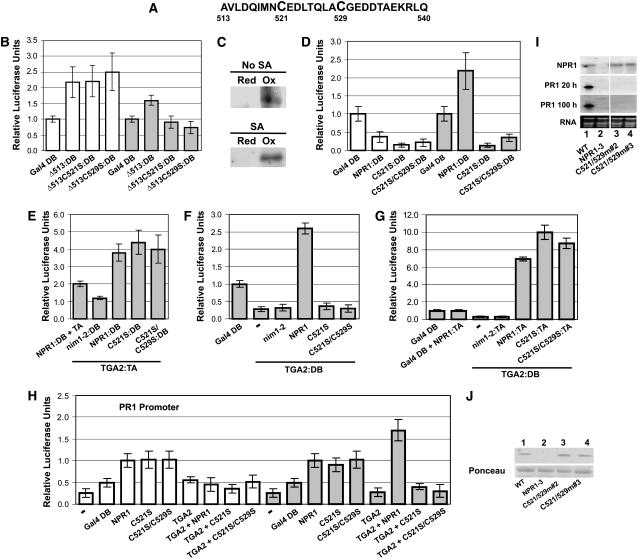
Having determined that the BTB/POZ domain does not harbor an autonomous transactivation domain active in SA-stimulated cells (Figure 4A), we set out to identify such domains in the C-terminal portion of NPR1. We created additional N-terminal deletions of NPR1 (Figure 4B): one at amino acid 373, which occurs right after the ankyrin repeats, as predicted by Pfam (Finn et al., 2006) and SMART (Letunic et al., 2006); one after residue 463, which is the end point of sequence similarity with Drosophila ankyrin 2 (GenBank accession number AAN12046.1); one at position 513, which corresponds to the beginning of the last stretch of negatively charged and hydrophobic residues, a signature of transactivation domains (Cress and Triezenberg, 1991); and finally, one right before the nuclear localization signal (Kinkema et al., 2000), at amino acid 533. These constructs were fused to Gal4 DB and assayed using the in vivo plant transcription assay (Figure 4C). In unstimulated cells (Figure 4C, white bars), deletion of the first 373 or 463 amino acids of NPR1 (Δ373:DB and Δ463:DB) did not show gene activation beyond the baseline level. However, further deletion to residue 513 (Δ513:DB) resulted in gene activation 2.2-fold above the baseline level, indicating that a repressing region had been deleted, thus exposing a cryptic transactivation domain. Extending the deletion to position 533 (Δ533:DB) reduced gene activity to the baseline level, emphasizing the importance of residues 513 to 533 for transactivation.
In SA-stimulated cells (Figure 4C, gray bars), deletion of the first 373, 463, or 513 amino acids of NPR1 (Δ373:DB, Δ463:DB, or Δ513:DB) resulted in gene activation 1.6-fold above the baseline level. Extending the deletion to position 533 (Δ533:DB) reduced gene activity to the baseline level, again indicating the importance of residues 513 to 533 for transactivation. The results in Figure 4C demonstrate that, in addition to amino acids 22 to 44 in the BTB/POZ, NPR1 possesses a second repression region, located between position 463 and 513, and active in unstimulated cells only, as these regions do not bring about repression in the SA-treated cells. Furthermore, a transactivation domain, active in uninduced as well as in SA-stimulated cells, requires residues located between positions 513 and 533.
Oxidation of NPR1 Cys Residues 521 and 529 Is Required for the Activity of the Transactivation Domain in SA-Treated Cells Only
Inspection of the region containing the C-terminal transactivation domain of NPR1 reveals that it contains two Cys residues (Figure 5A) at positions 521 (Cys-521) and 529 (Cys-529). Because Cys residues can be subjected to redox modifications that affect protein function, we first set out to determine whether Cys-521 and Cys-529 were required for the transactivation of the last 80 amino acids of NPR1 tethered to DNA (Δ513:DB). Cys-521 and Cys-529 (the only two NPR1 Cys residues found in Δ513:DB) were individually mutated to Ser, an amino acid similar to Cys in size and structure but lacking the ability for redox modifications. Hence, Ser can mimic the reduced form of Cys and preserve the capability for hydrogen bonding.
Figure 5.
Oxidation of Cys-521 and Cys-529 Correlates with Transcriptional Activation of the PR-1 Gene by the TGA2-NPR1 Complex.
(A) Sequence of amino acids located between positions 513 and 540.
(B) Histograms illustrating the transcriptional activity of the Δ513 deletion mutant of NPR1 and the effect of mutating Cys-521 or Cys-529 within the context of the Δ513 protein. Proteins were tethered to DNA through Gal4 DB.
(C) Blot analysis of NPR1Δ513 immunoprecipitate used to assess the in vivo redox status of residues Cys-521 and Cys-529 present in cells of Arabidopsis leaves treated for 24 h with SA (SA) or left untreated (No SA). Red indicates immunoprecipitates from proteins labeled for reduced Cys residues, and Ox indicates immunoprecipitates from proteins labeled for oxidized Cys (see Methods).
(D) Histograms illustrating the transcriptional activity of the full-length NPR1 and the effect of mutating Cys-521 or simultaneously Cys-521 and Cys-529 within the context of the full-length NPR1. Proteins were tethered to DNA through Gal4 DB.
(E) Histograms illustrating the interaction of NPR1 with the Cys-521 or the Cys-521 and Cys-529 mutants described for (D) with TGA2 fused to the VP16 transactivation domain. nim1-2, which does not interact with TGA2, was also expressed with TGA2:DB as a negative control. NPR1, nim1-2, and the mutants described for (D) were all fused to the Gal4 DB. NPR1:DB was also expressed along with the VP16 transactivation domain (NPR1:DB + TA) as another negative control.
(F) Histograms illustrating the effect of NPR1, Cys-521, or the Cys-521 and Cys-529 mutants described for (D) on the transcriptional activity of TGA2:DB. All proteins, except TGA2:DB, were expressed without a fusion.
(G) Histograms illustrating the interaction of NPR1, Cys-521, or the Cys-521 and Cys-529 mutants described for (D) all fused to the VP16 transactivation domain with TGA2 fused to the Gal4 DB.
For (B), (D), (E), (F), and (G), conditions were identical to those described for Figure 1. Data are reported as relative luciferase units. Values are from 25 samples and represent averages ± sd. Every bar represents five bombardments repeated five times (n = 25).
(H) Histograms illustrating the effect of NPR1, nim1-2, and the Cys-521 or the Cys-521 and Cys-529 mutants on the transcriptional activity of the TGA2-NPR1 complex. All proteins were native (without fusion). The reporter system was the Arabidopsis PR-1 promoter fused to luciferase. The CaMV 35S promoter:renilla luciferase fusion was used as an internal standard. − indicates that no effector was bombarded along with the reporter and internal standard vectors. Arabidopsis leaves were left untreated (white bars) or were treated for 24 h with 1 mM SA (gray bars). Data are reported as relative luciferase units. Values are from 25 samples and represent averages ± sd. Every bar represents five bombardments repeated five times (n = 25).
(I) RNA gel blot analysis using NPR1 or PR-1 probes. RNA stained with ethidium bromide is shown for loading comparison. Lane 1 contains RNA from wild-type Arabidopsis, and lane 2 contains RNA from the npr1-3 mutant. Lanes 3 and 4 contain RNA from two independent npr1-3 transgenic lines expressing NPR1 bearing Cys-to-Ser mutations at positions 521 and 529. Specific line numbers follow the construct name. PR1 20 h and PR1 100 h represent 20 h and 100 h of autoradiography, respectively. All lanes are from the same gel and blot.
(J) Top panel, immunoblot analysis of proteins from wild-type Arabidopsis, the npr1-3 mutant (NPR1-3), and the npr1-3 background lines expressing the mutant described for (I). An anti-NPR1 antibody (Després et al., 2000) was used. Bottom panel, Ponceau staining of the membranes shown in the top panel.
The constructs bearing a mutated Cys were fused to Gal4 DB and assayed using the in vivo plant transcription assay (Figure 5B). In resting cells (Figure 5B, white bars), mutation of the Cys residue at position 521 (Δ513C521S:DB) or 529 (Δ513C529S:DB) had no effect on gene activation, with levels similar to those in Δ513:DB (no difference at P = 0.05), indicating that redox modulation of Cys-521 and Cys-529 does not play a role in transactivation under noninduced conditions. However, in SA-treated tissues (Figure 5B, gray bars), Δ513C521S:DB and Δ513C529S:DB did not lead to gene activation beyond the baseline level, and values were significantly different from those in Δ513:DB (P < 0.05). These results indicate that, in the context of the last 80 amino acids of NPR1, Cys-521 and Cys-529 are required for transactivation only after SA treatment. To establish the redox status of Cys-521 and Cys-529, we used a labeling technique that distinguishes between protein sulfhydryls (reduced Cys residues) and disulfides (oxidized Cys residues) (see Després et al., 2003, for a flow chart and description of the method). The results (Figure 5C) indicate that Cys residues in the last 80 amino acids of NPR1 are predominantly oxidized, regardless of whether the cells have been treated with SA.
We next tested the effect of their mutations in the context of the full-length NPR1 tethered to DNA by the Gal4 DB (Figure 5D). In unstimulated cells (white bars), NPR1:DB did not lead to transactivation beyond baseline levels regardless of whether the Cys residues were mutated. However, after SA treatment, in contrast with what was observed with NPR1:DB, mutations of these Cys residues abolished transactivation and values were significantly different from those of wild-type NPR1:DB (P < 0.05). Plant two-hybrid experiments confirmed that C521S:DB and C529S:DB were expressed and retained the capacity to interact with TGA2 to an extent comparable to wild-type NPR1:DB (Figure 5E). These results suggest that the TGA2 coactivator function of NPR1 may require Cys-521 and Cys-529.
Transcriptional Activation of the PR-1 Gene and TGA2 Coactivator Function of NPR1 Require Cys-521 and Cys-529 of NPR1
Finally, we sought to determine whether mutating Cys-521 and Cys-529 would affect the TGA2 coactivator function of NPR1. We first tested the role of these Cys residues in the context of the Gal4 promoter and observed that mutation of Cys-521 or the double mutation Cys-521/Cys-529 abolished the capacity of the TGA2-NPR1 complex to transactivate (Figure 5F). Plant two-hybrid experiments confirmed that constructs C521S and C521S/C529S retained the capacity to interact with TGA2 to an extent comparable to wild-type NPR1 (Figure 5G) in the configuration in which TGA2 is fused to the Gal4 DB. Next, we tested the role of these Cys residues in the context of the PR-1 promoter. DNA coding for native (unfused) proteins was delivered by biolistics along with the luciferase reporter gene under the control of the PR-1 promoter (Figure 5H). As observed with NPR1, proteins C521S and C521S/C529S activated the PR-1 promoter in the absence of TGA2 regardless of whether cells were treated with SA, indicating that these residues of NPR1 are not required for recruitment to the promoter. Under induced (gray bars) and noninduced (white bars) conditions, mutation of these Cys residues did not bring about transactivation of the complex, as values were not significantly different from those obtained with TGA2 alone (at P = 0.05). The results observed after SA treatment (gray bars) were significantly different (P < 0.05) from those obtained with the TGA2-NPR1 complex, which activated the PR-1 promoter, as values were significantly greater (P < 0.05) than those observed with TGA2 alone or NPR1 alone, with and without mutated Cys residues.
To further confirm the biological significance of Cys-521 and Cys-529 of NPR1 in controlling PR-1 expression, an NPR1 construct harboring the double mutation at Cys-521 and Cys-529 was introduced, under the control of the CaMV 35S promoter, into the npr1-3 genetic background (Figure 5I, C521/529m). Wild-type Arabidopsis expressed PR-1 transcript when treated with 0.5 mM SA for 16 h (lane 1), whereas npr1-3 plants did not (lane 2). PR-1 gene induction was not restored in any of the 19 independent transgenic npr1-3 lines expressing C521/529m (lanes 3 and 4; data not shown). Figure 5J indicates that protein C521/529m was expressed at levels similar to those observed with NPR1. None of the lines tested expressed PR-1 in the absence of SA (data not shown).
DISCUSSION
Our study has demonstrated that TGA2 is not a transcriptional activator whether cells are resting or SA-treated. Furthermore, our data argue that, upon SA treatment, PR-1 is upregulated by a transactivation complex composed of at least TGA2 and NPR1. First, ChIP in wild-type Arabidopsis confirmed that NPR1 is recruited to the PR-1 promoter in both nontreated and SA-treated cells. Second, despite the fact that TGA2 is not a transactivator, NPR1 associates with TGA2 in SA-stimulated cells to form a transcriptional activating complex, both on a heterologous (5X Gal4 UAS) and a native (PR-1) promoter. Third, genetic complementation analyses of rationally designed site-directed and deletion mutants of the NPR1 BTB/POZ established a role for the core of this domain in activating PR-1. This finding is important because it establishes a direct correlation between complementation of PR-1 expression and transactivation on the heterologous promoter of a complex containing TGA2 and these NPR1 BTB/POZ mutants. Fourth, a Cys-oxidized transactivation domain in the C terminus of NPR1 is also required for the activation of PR-1 by the TGA2-NPR1 complex. This emphasizes again the correlation between transactivation of the TGA2-NPR1 complex and the activation of PR-1. We thus conclude that, in SA-treated cells, NPR1 is a TGA2 coactivator essential for PR-1 induction.
TGA2 Is Required for Transcriptional Repression of PR-1 in Uninduced Cells
The observation that, under uninduced and INA-induced conditions, the triple knockout tga2/5/6 mutant displayed levels of PR-1 expression 50-fold higher than in the wild-type suggested that TGA2, and members of its clade, could act as transcriptional repressors (Zhang et al., 2003), which implied that they can bind the PR-1 promoter independently of treatment with SA. Our ChIP results using TGA2 fused to a 6-His tag (Figures 1B and 1C) indeed indicate that recruitment of TGA2 to the PR-1 promoter is both SA- and NPR1-independent and thus suggest that the derepression of PR-1 observed by Zhang et al. (2003) in the tga2/5/6 knockout plant is attributable to the lack of direct binding of these TGA factors to PR-1. However, this contradicts a report in which ChIP indicated that binding of TGA2 to PR-1 is both NPR1- and SA-dependent (Johnson et al., 2003). Because the N-terminal region of TGA2 used by Johnson et al. (2003) to raise the anti-TGA2 antibody contains 28% Ser and Thr, two phosphorylatable amino acids, it is plausible that phosphorylation of a number of these residues could contribute to a decrease in the antibody–antigen interaction. To reconcile these apparently incongruous results, we propose that the data of Johnson et al. (2003) together with our data suggest that the N-terminal region of TGA2 is either inaccessible to the antibody or that the epitope is posttranslationally modified when cells are unstimulated or in the absence of NPR1.
In an in vivo transcription system based on the PR-1 promoter (Figure 1J), we demonstrated that, in uninduced cells, TGA2 repressed transcription of the PR-1 promoter activated only by the expression of NPR1 (Figure 1J, white bars, NPR1 compared with TGA2 + NPR1). However, TGA2 was unable to repress the baseline level of PR-1 expression, as defined by expression of the unrelated proteins Gal4 DB and cruciferin (Figure 1J). Therefore, in the context of PR-1 and in uninduced cells, TGA2 may only serve to repress the activating effect resulting from the recruitment of NPR1 to the promoter. TGA2:DB can also repress transcription from a LexA:VP16-activated synthetic promoter (see Supplemental Figure 1 online). These data further emphasize the fact that TGA2 is not a transcriptional activator on its own. The repressing effect of TGA2 observed on the PR-1 promoter activated by NPR1 is likely independent of an interaction between TGA2 and NPR1, because there is no detectable interaction between these two proteins in the context of the PR-1 promoter (Figure 1J, white bars, TGA2 compared with TGA2 + NPR1:TA). This is further substantiated by the observation that the activating effect resulting from the recruitment of nim1-2 (an NPR1 mutant version that does not interact with TGA2) to PR-1 is also repressed by TGA2 (Figure 1J, white bars, nim1-2 compared with TGA2 + nim1-2).
As determined by RNA gel blot analysis, overexpression of TGA2:His did not lead to the constitutive expression of PR-1 in plants used for ChIP experiments (Figure 1). These results are consistent with those reported for native TGA2 (Kim and Delaney, 2002) and a TGA2:Gal4 DB chimeric protein (Fan and Dong, 2002). By contrast, a chemically inducible TGA2:GFP fusion was shown to lead to the accumulation of PR-1 transcripts in the absence of benzothiadiazole treatment (Kang and Klessig, 2005). These results are difficult to explain in light of the results presented here (see Supplemental Figure 1 online) demonstrating that TGA2 is a transcriptional repressor. It is possible that the chemically inducible nature of the transgene used by Kang and Klessig (2005) resulted in the accumulation of vastly higher amounts of proteins than was achieved in our study or in those of Fan and Dong (2002) and Kim and Delaney (2002), leading to unspecific effects. Another possibility, not tested by Kang and Klessig (2005), is that the particular TGA2:GFP fusion they created fortuitously generated a transactivation domain, leading to SA-independent activation of PR-1. Finally, those authors did not assess whether the TGA2:GFP fusion is recruited to PR-1 in planta. Therefore, the possibility that this fusion exerted its effect on PR-1 through an indirect mechanism cannot be ruled out.
NPR1 Is a Transcriptional Coactivator in SA-Stimulated Cells
When tethered to DNA through the Gal4 DB, NPR1 activates transcription only after cells have been stimulated with SA. However, the finding that expression of NPR1 without fusion to the Gal4 DB does not lead to transcriptional modulation indicates that recruitment to the promoter is required for transcriptional activation. In the absence of a fusion to the Gal4 DB, NPR1 can be recruited to the heterologous Gal4 promoter via TGA2:DB. This recruitment leads to transactivation of the TGA2-NPR1 complex in SA-treated cells (Figure 1H), thus defining NPR1 as a coactivator. Remarkably, the TGA2-NPR1 complex is sufficient to activate the heterologous (Figure 1H) and the PR-1 (Figure 1J) promoters in an SA-dependent manner. Therefore, the complex behaves as an SA-regulated enhanceosome exposing a unique activating interface (Thanos and Maniatis, 1995; Merika and Thanos, 2001). Transactivation of the TGA2-NPR1 enhanceosome requires the core of the NPR1 BTB/POZ domain, because deletion beyond it (Δ110) or its mutation (Ala substitution) abolishes the function of the enhanceosome both on a transiently delivered heterologous promoter (Figure 3A) and on the endogenous PR-1 gene (Figure 2B). Transactivation of the TGA2-NPR1 enhanceosome is also dependent on the oxidation of Cys-521 and Cys-529 of NPR1 (Figure 5).
Although NPR1 behaves as a transcriptional activator in SA-treated cells when tethered to DNA on the Gal4-based promoter, this may not be the case when NPR1 is recruited to the PR-1 promoter. First, addition of a strong transactivation domain (VP16) to NPR1 (NPR1:TA) did not lead to further activation of the transiently delivered PR-1 promoter compared with unfused NPR1 transfected alone (Figure 1J). Second, mutation of Cys-521/Cys-529, which abolishes the transactivation properties of NPR1, activated PR-1 to the same extent as the wild-type NPR1 (Figure 5H). These results could suggest that in the architectural context of the PR-1 promoter, the PR-1–activating effect of NPR1 observed over the baseline level may be derepression as opposed to activation; that is to say, the effect may be attributable to chromatin structure modification instead of an active recruitment of the basal transcription machinery by NPR1. The discrepancy between the results observed on the Gal4-based and PR-1 promoters could arise from dissimilarity in the architecture of the two promoters. It could also arise from dissimilarity in the architecture of protein complexes caused by allosteric effects of DNA (Lefstin and Yamamoto, 1998), because in one case NPR1 interacts with DNA through a heterologous DB, and in the other case NPR1 is recruited to PR-1 through an unidentified DB or through an unknown DNA binding protein, itself recruited to PR-1. Therefore, although ChIPs demonstrated that NPR1 is recruited to PR-1 in both resting and SA-treated cells (Figures 1D and 1E), its role on the uninduced PR-1 is unclear. However, it seems reasonable to think that NPR1 interacts with PR-1 as a ready-to-go latent coactivator. This is consistent with the fact that overexpression of NPR1 does not lead to constitutive PR-1 expression; transcription still requires activation by SA (Cao et al., 1998). The nature of this switch remains elusive.
It has been proposed that the role of NPR1, in a wild-type plant, is to inactivate the repressing effect of SNI1 on PR genes (Li et al., 1999). As such, in the sni1 npr1-1 double mutant, PR gene expression is restored and is inducible. This also led to the proposal that induction of PR genes requires the activation of TGA factors in an SA-dependent but NPR1-independent manner. However, it is clear from the results presented here that TGA2, the prototype of the TGA2/5/6 clade, does not display any autonomous transactivation properties regardless of whether cells are treated with SA, but it requires an association with NPR1 to display such activities. Furthermore, the transactivating capacity of the TGA2-NPR1 complex is dependent upon the functionality of a transactivation domain found in the C terminus of NPR1. Thus, in the case of the PR-1 gene, it is unlikely that the role of NPR1 is simply to inactivate SNI1. Instead, we propose that the sni1 mutation, in the sni1 npr1 double mutant background, might activate pathways that regulate PR-1 in an NPR1- and TGA2/5/6-independent manner. Indeed, it has been shown that PR-1 is regulated in an SA-dependent but NPR1-independent manner by the transcription factor WHY1 (Desveaux et al., 2004). In resting cells, WHY1 is held inactive by an inhibitor, which prevents it from binding to DNA. Upon SA treatment, WHY1 is released from the effects of this inhibitor, which allows it to be recruited to its cognate DNA (Desveaux et al., 2004). It is thus possible that SNI1 plays a role in the WHY1-dependent pathway leading to PR-1 induction, as opposed to the TGA2-NPR1–dependent pathway. However, in the absence of ChIP data indicating that SNI1 and WHY1 are themselves recruited to the PR-1 promoter, it is unclear whether their effects on the PR-1 promoter are direct or indirect. Furthermore, in the absence of data indicating that SNI1 can interact physically with TGA2 or NPR1, it is very difficult to place, with any confidence, this protein in a model of PR-1 regulation.
The Cys-Oxidized Transactivation Domain: A New Type of Transactivation Domain
Cys residues in eukaryotic transcription factors have been demonstrated to be the target of redox regulation. In most cases, Cys residues affect DNA binding activity, which is abolished when these are oxidized (Abate et al., 1990; Toledano and Leonard, 1991; Lando et al., 2000). However, in a few instances, oxidation has been shown to control homodimerization (Benezra, 1994) and to inhibit nuclear export (Kuge et al., 2001). When one eliminates cases in which effects on transactivation are attributable to the modulation of DNA binding activity (as opposed to the modulation of transactivation per se), there is only one example in the literature in which a transactivation domain is controlled by Cys redox. However, in that instance, oxidation abolished transactivation (Morel and Barouki, 2000). It thus appears that NPR1 is a rare example of a transactivation domain positively regulated by oxidized Cys residues (Cys-521 and Cys-529). Remarkably, despite the fact that Cys-521 and Cys-529 are oxidized regardless of whether cells are exposed to SA (Figure 5C), these Cys residues only modulate transactivation in SA-stimulated cells (Figure 5B). This finding suggests that different factors mediating contact between the NPR1-transactivating domain and the basal transcription machinery operate in noninduced and SA-stimulated cells.
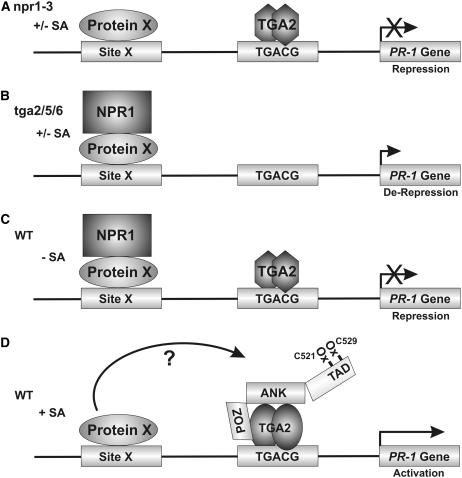
The results reported here constitute a significant advancement of our knowledge of plant disease resistance by elucidating the molecular function of TGA2 as a transcriptional repressor of NPR1-dependent derepression of PR-1 and of NPR1 as a coactivator of TGA2, and by establishing the existence of an SA-regulated enhanceosome composed of at least TGA2 and NPR1. Figure 6 presents a model that summarizes the results reported here on the regulation of PR-1.
Figure 6.
Working Model for the Regulation of PR-1 by the TGA2-NPR1 Enhanceosome.
(A) In an npr1 mutant plant such as npr1-3, there is no NPR1-dependent derepression of PR-1 and there is no incorporation of TGA2 into a TGA2-NPR1 enhanceosome. PR-1 is repressed. Because NPR1 is recruited to PR-1 independently from TGA2 and NPR1 does not contain a known DNA binding domain, we postulate that in a wild-type plant NPR1 is recruited through an unknown protein (Protein X) binding to an unknown DNA element (Site X). Although TGA3 has been shown to be recruited to PR-1, its interaction with the promoter is both NPR1- and SA-dependent (Johnson et al., 2003). Therefore, it is unlikely that NPR1 would be recruited by TGA3 or that TGA3 is the postulated Protein X. Another possible scenario to explain the recruitment of NPR1 to PR-1 is that NPR1 interacts directly with DNA using an unidentified DNA binding domain. Further experimentation is required to distinguish between these two possibilities.
(B) In the tga2/5/6 triple knockout (Zhang et al., 2003), NPR1 is recruited to the PR-1 promoter, which becomes derepressed. In these plants, the TGA2-NPR1 enhanceosome is not recruited to the PR-1 promoter because of the absence of TGA2, TGA5, and TGA6.
(C) In a wild-type plant unstimulated with SA, both NPR1 and TGA2 are recruited to the PR-1 promoter independent of each other. However, under resting conditions, NPR1 and TGA2 do not interact with each other. Again here, NPR1 is postulated to be recruited through an unknown protein (Protein X).
(D) In the presence of SA, NPR1 forms an enhanceosome with TGA2. Transactivation of the complex requires the oxidation of Cys-521 and Cys-529, which are found within the confines of a transactivation domain (TAD) in the C terminus of NPR1. The BTB/POZ domain of NPR1 is hypothesized to interact with TGA2. NPR1 is postulated to be transferred from the unknown Protein X to TGA2. However, it is possible that NPR1, Protein X, and TGA2 all interact at the same time. The question mark illustrates this fact. The exact nature of the enhanceosome remains undetermined, but it contains at the very least NPR1 and TGA2.
METHODS
Plant Transcription Assays and Two-Hybrid Assays
All procedures for the yeast two-hybrid system were described previously (Després et al., 2000). All procedures for the plant two-hybrid assays, the reporter gene vector, the internal standard vector, and the VP16:NPR1 construct were described previously (Després et al., 2003). TGA2, NPR1, the Ala substitution, and deletion mutants of NPR1 were created by PCR using appropriate primers and cloned in-frame with the GAL4 DB or VP16 TA contained in pBI524 to create N-terminal fusion proteins as described (Després et al., 2003). The unfused versions of TGA2 and NPR1 were cloned into pBI524 lacking the GAL4 DB or VP16 TA. To create the PR-1 promoter–luciferase reporter gene fusion, the −1293 promoter fragment (Lebel et al., 1998) was amplified by PCR and used to substitute the 5X UASGAL4 fragment in the luciferase–nopaline synthase polyadenylation signal reporter plasmid. Every bar in each graph represents five bombardments repeated five times (n = 25). Arabidopsis thaliana ecotype Columbia was used throughout this study.
ChIP of NPR1 and TGA2
ChIP was performed as we described previously (Chakravarthy et al., 2003). The specificity of the anti-NPR1 antibody has been demonstrated (Després et al., 2000). The anti-His antibody was from Santa Cruz Biotechnology (sc-803 AC). The PCR primer pair specific to the PR-1 promoter was as follows: 5′-ATGGGTGATCTATTGACTGTTT-3′ and 5′-GTAGCTTTGCCATTGTTGAT-3′. To confirm that the PCR product generated was indeed a fragment of the PR-1 promoter, it was gel-excised, cloned, and sequenced.
Plant Growth Conditions and Transformation
Conditions for the growth of Arabidopsis (Columbia) and npr1-3 plants (Cao et al., 1997) and methods for plant transformation, the selection of transgenic individuals, and RNA gel blot hybridization were described previously (Liu et al., 2005).
In Vivo Determination of the Cys Redox Status of NPR1 Δ513
Because of very low amounts of proteins in the biolistics assays, 80 bombardments were performed with the Δ513:TA constructs. After a 24-h incubation period with or without SA, proteins were extracted from Arabidopsis leaves, separated into two aliquots, and processed immediately and in parallel as described previously (Després et al., 2003). Immunoprecipitations were performed with an anti-VP16 antibody (sc-7545 AC; Santa Cruz Biotechnology). The VP16 TA does not contain any Cys residues.
Statistical Methods
All pooled data are expressed as averages, and error bars represent sd. When data from two independent populations are compared, statistical significance was assessed using a two-tailed Student's t test.
Accession Numbers
Sequence data from this article can be found in the GenBank data library under accession numbers GI:486933, GI:1773295, GI:2291257, and GI:1399185 and in the Protein Data Bank data library under accession code 1buo.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. TGA2 Represses a LexA:VP16-Activated Synthetic Promoter.
Supplemental Figure 2. The BTB/POZ Mutants of NPR1 Interact with TGA2 in Yeast Two-Hybrid Assays.
Supplementary Material
Acknowledgments
We thank Xin Li for providing seeds of the TGA2/5/6 knockout plants, Jun Murata for help with the plant biolistic assay system, Maureen Anderson, Catherine DeLong, Catherine Chubak, and Errol Su for technical assistance, and the DNA Technology Unit of the Plant Biotechnology Institute for oligonucleotide synthesis and DNA sequencing. This research was supported by National Research Council Plant Biotechnology Institute core funding (P.R.F.), the National Research Council Genomics and Health Initiative (P.R.F.), the National Science and Engineering Research Council (NSERC) discovery grant program (C.D.), the Canada Foundation for Innovation (C.D.), the Ontario Innovation Trust (C.D.), and the NSERC graduate scholarship program (A.R. and P.B.). C.D. is a Canada Research Chair in Molecular Plant Pathology. This is National Research Council Canada publication 46599.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Pierre R. Fobert (pierre.fobert@nrc-cnrc.gc.ca).
Online version contains Web-only data.
References
- Abate, C., Patel, L., Rauscher, F.J., III, and Curran, T. (1990). Redox regulation of fos and jun DNA-binding activity in vitro. Science 249 1157–1161. [DOI] [PubMed] [Google Scholar]
- Ahmad, K.F., Engel, C.K., and Prive, G.G. (1998). Crystal structure of the BTB domain from PLZF. Proc. Natl. Acad. Sci. USA 95 12123–12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind, L., and Koonin, E.V. (1999). Fold prediction and evolutionary analysis of the POZ domain: Structural and evolutionary relationship with the potassium channel tetramerization domain. J. Mol. Biol. 285 1353–1361. [DOI] [PubMed] [Google Scholar]
- Bardwell, V.J., and Treisman, R. (1994). The POZ domain: A conserved protein-protein interaction motif. Genes Dev. 8 1664–1677. [DOI] [PubMed] [Google Scholar]
- Benezra, R. (1994). An intermolecular disulfide bond stabilizes E2A homodimers and is required for DNA binding at physiological temperatures. Cell 79 1057–1067. [DOI] [PubMed] [Google Scholar]
- Buck, M.J., and Lieb, J.D. (2004). ChIP-chip: Considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics 83 349–360. [DOI] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, A.S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88 57–63. [DOI] [PubMed] [Google Scholar]
- Cao, H., Li, X., and Dong, X. (1998). Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl. Acad. Sci. USA 95 6531–6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy, S., Tuori, R.P., D'Ascenzo, M.D., Fobert, P.R., Despres, C., and Martin, G.B. (2003). The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell 15 3033–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cress, W.D., and Triezenberg, S.J. (1991). Critical structural elements of the VP16 transcriptional activation domain. Science 251 87–90. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411 826–833. [DOI] [PubMed] [Google Scholar]
- Delaney, T.P., Friedrich, L., and Ryals, J.A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 92 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després, C., Chubak, C., Rochon, A., Clark, R., Bethune, T., Desveaux, D., and Fobert, P.R. (2003). The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15 2181–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després, C., DeLong, C., Glaze, S., Liu, E., and Fobert, P.R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12 279–290. [PMC free article] [PubMed] [Google Scholar]
- Desveaux, D., Subramaniam, R., Després, C., Mess, J.N., Levesque, C., Fobert, P.R., Dangl, J.L., and Brisson, N. (2004). A “Whirly” transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Dev. Cell 6 229–240. [DOI] [PubMed] [Google Scholar]
- Durrant, W.E., and Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42 185–209. [DOI] [PubMed] [Google Scholar]
- Fan, W., and Dong, X. (2002). In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 14 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, R.D., et al. (2006). Pfam: Clans, web tools and services. Nucleic Acids Res. 34 D247–D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C., Boden, E., and Arias, J. (2003). Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell 15 1846–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.T. (1999). Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292 195–202. [DOI] [PubMed] [Google Scholar]
- Kang, H.G., and Klessig, D.F. (2005). Salicylic acid-inducible Arabidopsis CK2-like activity phosphorylates TGA2. Plant Mol. Biol. 57 541–557. [DOI] [PubMed] [Google Scholar]
- Kim, H.S., and Delaney, T.P. (2002). Over-expression of TGA5, which encodes a bZIP transcription factor that interacts with NIM1/NPR1, confers SAR-independent resistance in Arabidopsis thaliana to Peronospora parasitica. Plant J. 32 151–163. [DOI] [PubMed] [Google Scholar]
- Kinkema, M., Fan, W., and Dong, X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12 2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge, S., Arita, M., Murayama, A., Maeta, K., Izawa, S., Inoue, Y., and Nomoto, A. (2001). Regulation of the yeast Yap1p nuclear export signal is mediated by redox signal-induced reversible disulfide bond formation. Mol. Cell. Biol. 21 6139–6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando, D., Pongratz, I., Poellinger, L., and Whitelaw, M.L. (2000). A redox mechanism controls differential DNA binding activities of hypoxia-inducible factor (HIF) 1alpha and the HIF-like factor. J. Biol. Chem. 275 4618–4627. [DOI] [PubMed] [Google Scholar]
- Lebel, E., Heifetz, P., Thorne, L., Uknes, S., Ryals, J., and Ward, E. (1998). Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J. 16 223–233. [DOI] [PubMed] [Google Scholar]
- Lefstin, J.A., and Yamamoto, K.R. (1998). Allosteric effects of DNA on transcriptional regulators. Nature 392 885–888. [DOI] [PubMed] [Google Scholar]
- Letunic, I., Copley, R.R., Pils, B., Pinkert, S., Schultz, J., and Bork, P. (2006). SMART 5: Domains in the context of genomes and networks. Nucleic Acids Res. 34 D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Zhang, Y., Clarke, J.D., Li, Y., and Dong, X. (1999). Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98 329–339. [DOI] [PubMed] [Google Scholar]
- Liu, G., Holub, E.B., Alonso, J.M., Ecker, J.R., and Fobert, P.R. (2005). An Arabidopsis NPR1-like gene, NPR4, is required for disease resistance. Plant J. 41 304–318. [DOI] [PubMed] [Google Scholar]
- Melnick, A., Ahmad, K.F., Arai, S., Polinger, A., Ball, H., Borden, K.L., Carlile, G.W., Prive, G.G., and Licht, J.D. (2000). In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol. Cell. Biol. 20 6550–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merika, M., and Thanos, D. (2001). Enhanceosomes. Curr. Opin. Genet. Dev. 11 205–208. [DOI] [PubMed] [Google Scholar]
- Morel, Y., and Barouki, R. (2000). The repression of nuclear factor I/CCAAT transcription factor (NFI/CTF) transactivating domain by oxidative stress is mediated by a critical cysteine (Cys-427). Biochem. J. 348 235–240. [PMC free article] [PubMed] [Google Scholar]
- Mosavi, L.K., Cammett, T.J., Desrosiers, D.C., and Peng, Z.Y. (2004). The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 13 1435–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher, R.A., Durrant, W.E., Wang, D., Song, J., and Dong, X. (2006). A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell 18 1750–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou, Z., Fan, W., and Dong, X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113 935–944. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M., and Van Loon, L.C. (2004). NPR1: The spider in the web of induced resistance signaling pathways. Curr. Opin. Plant Biol. 7 456–464. [DOI] [PubMed] [Google Scholar]
- Ryals, J., Weymann, K., Lawton, K., Friedrich, L., Ellis, D., Steiner, H.Y., Johnson, J., Delaney, T.P., Jesse, T., Vos, P., and Uknes, S. (1997). The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor I kappa B. Plant Cell 9 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J.A., Neuenschwander, U.H., Willits, M.G., Molina, A., Steiner, H.Y., and Hunt, M.D. (1996). Systemic acquired resistance. Plant Cell 8 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stogios, P.J., Downs, G.S., Jauhal, J.J., Nandra, S.K., and Prive, G.G. (2005). Sequence and structural analysis of BTB domain proteins. Genome Biol. 6 R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam, R., Desveaux, D., Spickler, C., Michnick, S.W., and Brisson, N. (2001). Direct visualization of protein interactions in plant cells. Nat. Biotechnol. 19 769–772. [DOI] [PubMed] [Google Scholar]
- Thanos, D., and Maniatis, T. (1995). Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 83 1091–1100. [DOI] [PubMed] [Google Scholar]
- Toledano, M.B., and Leonard, W.J. (1991). Modulation of transcription factor NF-kappa B binding activity by oxidation-reduction in vitro. Proc. Natl. Acad. Sci. USA 88 4328–4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, E.R., Uknes, S.J., Williams, S.C., Dincher, S.S., Wiederhold, D.L., Alexander, D.C., Ahl-Goy, P., Metraux, J.P., and Ryals, J.A. (1991). Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, R.R., Pfitzner, U.M., and Gatz, C. (2005). Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell 17 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Fan, W., Kinkema, M., Li, X., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 96 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Tessaro, M.J., Lassner, M., and Li, X. (2003). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15 2647–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.