Abstract
XA21 is a receptor-like kinase protein in rice (Oryza sativa) that confers gene-for-gene resistance to specific races of the causal agent of bacterial blight disease, Xanthomonas oryzae pv oryzae. We identified XA21 binding protein 3 (XB3), an E3 ubiquitin ligase, as a substrate for the XA21 Ser and Thr kinase. The interaction between XB3 and the kinase domain of XA21 has been shown in yeast and in vitro, and the physical association between XB3 and XA21 in vivo has also been confirmed by coimmunoprecipitation assays. XB3 contains an ankyrin repeat domain and a RING finger motif that is sufficient for its interaction with the kinase domain of XA21 and for its E3 ubiquitin ligase activity, respectively. Transgenic plants with reduced expression of the Xb3 gene are compromised in resistance to the avirulent race of X. oryzae pv oryzae. Furthermore, reduced levels of Xb3 lead to decreased levels of the XA21 protein. These results indicate that Xb3 is necessary for full accumulation of the XA21 protein and for Xa21-mediated resistance.
INTRODUCTION
Plant immunity is often governed by dominant resistance (R) genes. Over the past decade, a large number of R genes have been characterized from diverse plant species (Dangl and Jones, 2001; Staskawicz et al., 2001). The majority of these encode proteins with a nucleotide binding site and leucine-rich repeats (NB-LRRs). The NB-LRR class of R proteins can be divided into two subgroups based on the N-terminal domains: coiled-coil NB-LRRs carrying a coil-coil structure and TIR-NB-LRRs containing the TIR domain that was originally identified in the intracellular regions of the Drosophila melanogaster transmembrane receptor Toll and the mammalian interleukin 1 receptor (IL-1R). In the evolutionarily conserved animal innate immunity pathways, the TIR domains of Toll and IL-1R form similar protein complexes, including the Ser and Thr kinase Pelle or IRAK (Hoffmann and Reichhart, 2002). While it is still unclear whether the TIR domain of the R proteins also recruits a Pelle-like kinase in the defense response, a number of receptor-like kinases (RLKs) containing a Pelle-related kinase domain have been implicated in plant disease resistance (Song et al., 1995; Brueggeman et al., 2002; Scheer and Ryan, 2002; Godiard et al., 2003; Sun et al., 2004; Zipfel et al., 2004; Diener and Ausubel, 2005; Llorente et al., 2005; Chen et al., 2006). These RLKs appear to be equivalent to the receptor/kinase complexes in animal innate immunity.
Ubiquitin-mediated protein modification regulates many cellular processes, including homeostasis, development, cell division, growth, and hormone and stress responses (Smalle and Vierstra, 2004). Ubiquitin is a small peptide that often directs its conjugated target proteins for degradation. Ubiquitin is activated by the ubiquitin-activating enzyme E1, transferred to the ubiquitin-conjugating enzyme E2, and finally linked to a target substrate by the ubiquitin ligase E3 (Smalle and Vierstra, 2004). Characterization of a number of E3 proteins in both animal and plant systems indicates that the zinc binding domain RING (for Really Interesting New Gene) finger (RF) is essential for many ubiquitin-mediated protein modification events (Joazeiro and Weissman, 2000; Osterlund et al., 2000). Ubiquitinated proteins can then be degraded by the 26S proteasome or can assume a role in other proteolysis-independent processes (Ben-Neriah, 2002; Smalle and Vierstra, 2004). It has been suggested that polyubiquitination of the RF-containing protein TRAF6 activates the downstream protein kinase TAK1 in the IL-1–mediated innate immunity pathway (Wang et al., 2001).
The ubiquitin-mediated protein modification system also plays a role in plant defense mechanisms. For example, SGT1 and RAR1 are required for the function of multiple R genes (Shirasu et al., 1999; Austin et al., 2002; Azevedo et al., 2002; Liu et al., 2002a; Tor et al., 2002; Tornero et al., 2002). SGT1 associates with RAR1 and SKP1, a component of the SCF-type of E3 complexes. Suppression of the tobacco (Nicotiana benthamiana) SKP1 gene leads to the loss of N-mediated resistance to Tobacco mosaic virus (Liu et al., 2002a). Furthermore, it has been shown that SGT1 interacts with the NB-LRR protein MLA1 in the yeast two-hybrid system (Bieri et al., 2004). Other E3s that have been implicated in plant defense responses include the F-box proteins COI1, SON1, and ACRE189 (Xie et al., 1998; Kim and Delaney, 2002; Rowland et al., 2005); the U-box proteins SPL11, ACRE276/PUB17, and CMPG1 (Zeng et al., 2004; Rowland et al., 2005; Gonzalez-Lamothe et al., 2006; Yang et al., 2006); and the RF proteins RIN2 and RIN3 (Kawasaki et al., 2005). RIN2 and RIN3 interact with the Arabidopsis thaliana R protein RPM1 in yeast. Despite the key role established for defense signaling, a physical interaction between an E3 and R proteins has not been shown in vivo.
The rice (Oryza sativa) RLK XA21 confers resistance against Xanthomonas oryzae pv oryzae (Xoo) and is predicted to recognize a molecule secreted from Xoo (Song et al., 1995; Goes da Silva et al., 2004). The kinase domain of XA21 is capable of autophosphorylating multiple Ser and Thr residues (Liu et al., 2002b). Autophosphorylation of Ser-686, Thr-688, and Ser-689 in the juxtamembrane (JM) domain has recently been implicated in the stability of the XA21 protein (Xu et al., 2006). Interestingly, these three autophosphorylated residues are located in a putative proteolytic cleavage motif (P/GX 5-7 P/G) (named XA21CS1). Cleavage of XA21 at XACS1 results in a 100-kD product. We have demonstrated that c-Myc–tagged wild-type XA21 and its kinase mutants (XA21K736E and XA21S686A/T688A/S689A) can be degraded to a similar 100-kD product but to a much greater extent with the mutants. We have proposed that XA21 is cleaved within or near XA21CS1 by a developmentally controlled proteolytic activity and that autophosphorylation of XA21 stabilizes the R protein.
Here, we report that the kinase domain of XA21 interacts with the RF-containing protein XB3 (for XA21 binding protein 3) in vitro and in vivo. The RF domain of XB3 ubiquitinates MBP-XB3RF, indicating that XB3 is an E3. Moreover, XB3 is specifically transphosphorylated by the kinase domain of XA21. Reduction of Xb3 leads to a decrease in the level of the XA21 protein and of Xa21-mediated disease resistance at the adult stage. Finally, we show that Xa21-mediated resistance is affected by gene dosage of Xa21. These results indicate that XB3 physically interacts with XA21 and is positively involved in Xa21-mediated immunity.
RESULTS
Xb3 Encodes a Putative E3 Protein with Multiple Domains
A truncated kinase domain of XA21 (XA21K690) was used to screen a yeast two-hybrid rice cDNA library to identify XA21-interacting proteins. XA21K690 spans the entire intracellular region of XA21 (XA21K) except the first 13 amino acids of the JM domain that contains the autophosphorylated Ser-686, Thr-688, and Ser-689 (Figure 1A). Seven classes of XA21K690-interacting proteins were identified (data not shown). Among them, XB3 is encoded by the cDNA 98-2 that contains a complete open reading frame and 65 bp of 5′ untranslated region.
Figure 1.
XB3 Interacts with XA21K in Yeast.
(A) Schematic representation of XA21. Domains were as described by Song et al. (1995). The DraIII site used for cloning the c-Myc or Protein A (ProA) tag is indicated. The transmembrane domain is in black, while the JM domain is represented by the dotted box, and its sequence is shown above. The XA21CS1 site in the JM domain is underlined. The autophosphorylated residues within XA21CS1 are highlighted in bold. The position of the invariable Lys-736 (K736) residue is shown. Two versions of the kinase domain of XA21 used in library screening (XA21K690) and subsequent confirmation (XA21K) are indicated below. The first amino acid residue of XA21K690 is in italics.
(B) XB3 specifically interacts with XA21K in yeast. Indicated proteins were expressed as the DNA binding domain (BD) or activation domain (AD) fusions of the GAL4 transcription factor in yeast. Cells capable of growing on selective medium (SD/-Leu-Trp) were replicated on the SD/-Leu-Trp-His medium to examine the presence of interacting proteins.
(C) Predicted amino acid sequence of the Xb3 gene product. The deduced protein domains are indicated as follows: (I) putative N-myristoylation site (underlined); (II) ankyrin repeats (the highly conserved amino acids in the ankyrin domain are in red); (III) unknown; (IV) RF (the eight conserved Cys and His residues that are important for zinc chelating in the RF are in blue); and (V) C-terminal tail.
In addition to XA21K690, XB3 interacted with XA21K and the autophosphorylation mutant XA21KS686A/T688A/S689A (Figure 1B; Xu et al., 2006), indicating that these three autophosphorylated residues are not required for the binding of XB3 in yeast. By contrast, XB3 failed to interact with the catalytically inactive mutant XA21KK736E (Figure 1B; Liu et al., 2002b). Additionally, XA21K did not interact with the closest rice homolog of XB3, XBOS31 (69% identity and 76% similarity) (see Supplemental Figure 1 and Supplemental Table 1 online). Thus, the interaction between XA21K and XB3 is specific.
The open reading frame of Xb3 and its 5′ sequence were fused in frame to the activation domain of the GAL4 transcription factor in the cDNA 98-2. XB3 has 450 amino acids (Figure 1C). The N terminus carries a putative N-myristoylation site (Resh, 1999). Amino acids 11 to 305 carry eight imperfect copies of ankyrin repeats that have been implicated in protein–protein interactions (Sedgwick and Smerdon, 1999). Following the ankyrin domain is a region (amino acids 323 to 371) sharing the conserved Cys and His residues characteristic of RF motifs. Because a number of RF-containing proteins carry E3 activities (Lorick et al., 1999), we hypothesized that XB3 is an E3 enzyme. The C terminus of XB3 has the potential to form a coiled-coil structure.
The Ankyrin Repeat Domain of XB3 Is Sufficient for in Vitro Interactions with XA21K
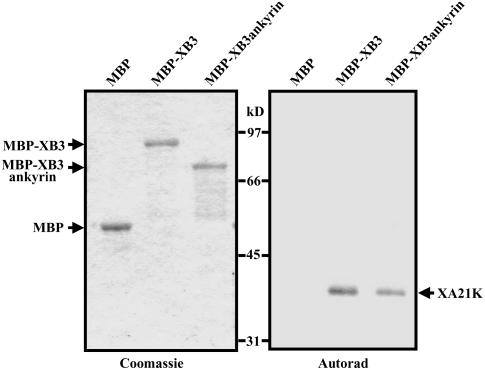
Because the C-terminal half of XB3, spanning the RF motif and the tail, did not interact with XA21K in the yeast two-hybrid system (data not shown), we tested whether the ankyrin domain physically binds to XA21K. XB3 and the ankyrin repeats of XB3 (not including the 65 bp 5′ untranslated region in the cDNA 98-2) were expressed as maltose binding protein (MBP) fusions, and the bacterially expressed proteins (MBP-XB3 and MBP-XB3ankyrin) were bound to amylose resin. MBP alone was used as a control. XA21K, produced and labeled with [35S]Met by an in vitro transcription and translation reaction, was mixed with the above resin-bound MBP fusion proteins. After extensive washing, proteins in the resin-bound fractions were eluted and resolved by SDS-PAGE. The 35S-labeled XA21K was only detected in the MBP-XB3 and MBP-XB3 ankyrin fractions but not in the MBP control (Figure 2), indicating that the ankyrin repeat domain of XB3 is sufficient for binding to XA21K.
Figure 2.
The Ankyrin Domain of XB3 Is Sufficient for in Vitro Interactions with XA21K.
35S-labeled XA21K was incubated with either resin-bound MBP, MBP-XB3, or MBP-XB3ankyrin (the ankyrin domain of XB3). After extensive washing, the resin-bound proteins were resolved by 8% SDS-PAGE. Both the Coomassie blue–stained gel (left) and its autoradiogram (right) are shown. XA21K visualized by autoradiogram is indicated. The experiment was repeated three times with similar results.
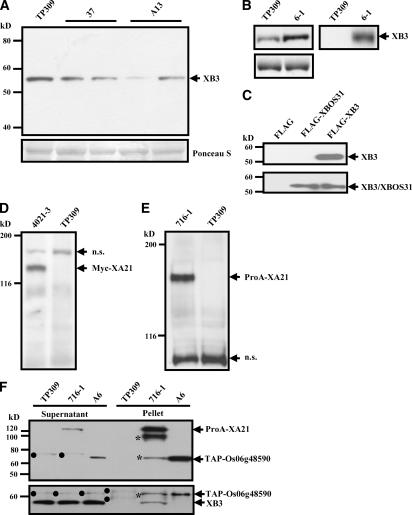
XB3 Interacts with XA21 in Vivo
We generated polyclonal antibodies against XB3 (anti-XB3M) and demonstrated their specificity against XB3. As shown in Figure 3A, anti-XB3M detected a major band at the position of 54 kD, which is slightly larger than the predicted XB3 (48 kD). The quantity of this product is reduced in the Xb3 RNA interference (RNAiXB3) lines when compared with the recipient line Taipei309 (TP309) but significantly enhanced in an XB3-FLAG overexpression line (Figures 3A and 3B). We then immunoprecipitated the XB3-FLAG from the overexpression line using anti-FLAG M2 agarose followed by detection with anti-XB3M. A single product of 54 kD was only found from the XB3-FLAG overexpression line (Figure 3B). To rule out the possibility that the 54-kD product could be XBOS31, we expressed XB3 and XBOS31 as FLAG-tagged fusion proteins in Eschericha coli. Protein gel blot analyses of bacterial extracts revealed that anti-XB3M only recognizes FLAG-XB3, not FLAG-XBOS31 (Figure 3C). As a control, the anti-FLAG M2 antibody detected both XB3 and XBOS31 in the same extracts. These results indicate that anti-XB3M is specific for XB3 and the 54-kD product in rice extracts is XB3.
Figure 3.
XB3 Interacts with XA21 in Vivo.
(A) Immunodetection of XB3 in the Xb3 RNAi lines. Equal amounts of total protein extracts isolated from the recipient line TP309 and four individuals of the T1 progeny of the RNAiXB3 lines 37 and A13 were immunoblotted with anti-XB3M (top). The XB3 band is indicated. The Ponceau S–stained blot is shown as the loading control (bottom).
(B) Immunodetection of XB3 in the Xb3 overexpression line. Left panels: Equal amounts of total protein extracts isolated from TP309 and the transgenic line 6-1 overexpressing a FLAG-tagged Xb3 were immunoblotted with anti-XB3M (top). Coomassie blue–stained gel of identical protein samples as a loading control (bottom). Right panel: Equal amounts of total protein extracts from TP309 and 6-1 were immunoprecipitated with anti-FLAG M2 agarose followed by detection with anti-XB3M.
(C) Anti-XB3M does not recognize bacterially expressed XBOS31. Protein extracts from the bacterial cultures expressing the indicated constructs were immunoblotted with anti-XB3M (top) or anti-FLAG M2 (bottom) antibodies.
(D) and (E) Immunodetection of Myc-XA21 and ProA-XA21, respectively. Equal amounts of total protein extracts from TP309 and the transgenic lines 4021-3 (carrying Myc-Xa21) or 716-1 (carrying ProA-Xa21) were immunoblotted with anti-c-Myc (D) or PAP (E) antibodies. The tagged XA21 proteins are indicated. n.s., nonspecific products.
(F) XB3 is coimmunoprecipitated with XA21. Twenty-five milliliters of total protein extracts from 5 g of leaf tissue of TP309, 716-1, and A6 (carrying TAP-Os06g48590) were immunoprecipitated with IgG sepharose beads. One-fifth of the precipitates were subjected to protein blot analyses using PAP (top) or anti-XB3M (bottom) antibodies. One microliter of total protein extracts from TP309, 716-1, and A6 was used as a control. XB3 is indicated. The XB3 band was not detectable in the A6 pellet even when 10 times more immunoprecipitates were used for the protein blot analyses (data not shown). The asterisks denote products degraded from XA21. Dots indicate nonspecific products. In the A6 line, the TAP-tagged kinase can be detected by anti-XB3M antibodies. The experiments were repeated three times with similar results.
We also generated transgenic plants expressing either a c-Myc–tagged or a double ProA-tagged XA21 for coimmunoprecipitation assays. Each of the tagged Xa21 variants was flanked by 2204-bp native 5′ and 3787-bp 3′ regulatory sequences that are sufficient for supporting Xa21-mediated resistance (Song et al., 1995). Transgenic plants carrying each of the constructs were resistant to Xoo Philippine race 6 (Xoo PR6), indicating that the tagged Xa21 variants are functional (data not shown). Anti-c-Myc antibody detected a 140-kD polypeptide in the resistant transgenic plants but not in the susceptible recipient line TP309 (Figure 3D). We concluded that the 140-kD polypeptide is Myc-XA21. A homozygous Myc-XA21 line (4021-3) was identified and used in this study. No segregation for the resistance was observed in the progeny of 4021-3 with >50 individuals (Xu et al., 2006). A similar strategy was used to specify ProA-XA21 in the rice protein extracts using the peroxidase-antiperoxidase (PAP) antibody (Figure 3E).
To demonstrate that XB3 interacts with XA21 in vivo, we used IgG sepharose beads to immunoprecipitate XA21 from the ProA-XA21 line (carrying only the native XB3). The ProA domains have proven to be a high-affinity tag for recovering protein complexes from a complex mixture (Rigaut et al., 1999; Rohila et al., 2004). Indeed, ProA-XA21 was strongly recognized by the PAP antibody in the precipitates, indicating that the ProA tag can efficiently recover the XA21 protein from plant extracts (Figure 3F). Anti-XB3M was then used to detect XB3 in the IgG precipitates. Figure 3F shows that a major band of 54 kD, identical to XB3, was detected in the ProA-XA21 line but not in the TP309 line. To exclude the possibility that XB3 could interact with the 128–amino acid ProA tag, we used the transgenic line A6 that expresses a tandem affinity purification (TAP)-tagged kinase (Os06g48590) unrelated to XA21. Similar to ProA-XA21, a ProA tag was placed at the N terminus of this kinase. No product of 54 kD was detected in the IgG precipitates prepared from the A6 plants. As a control, similar amounts of XB3 were present in the supernatant of these three reactions. These results are consistent with our in vitro observations and indicate that XB3 interacts with XA21 in vivo.
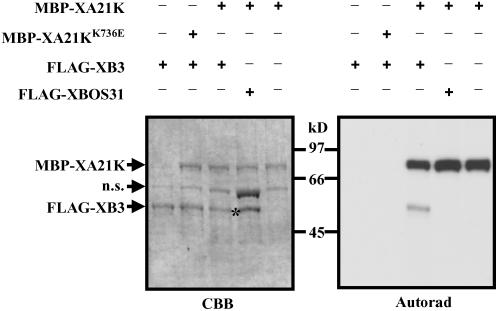
XB3 Is a Substrate of XA21K
The physical interaction between XA21 and XB3 suggests that XB3 may be a substrate of XA21K. To test this hypothesis, the purified FALG-XB3 protein was incubated with MBP-XA21K (a fusion of MBP and XA21K; Liu et al., 2002b) and [γ-32P]ATP. After incubation, the samples were resolved by SDS-PAGE. As indicated in Figure 4, MBP-XA21K can phosphorylate itself and FLAG-XB3 but not FLAG-XBOS31. As a control, MBP-XA21KK736E cannot autophosphorylate or phosphorylate FLAG-XB3. Moreover, we did not observe any kinase activity from FLAG-XB3. Since the FLAG tag does not contain any Ser or Thr residues, we therefore conclude that XA21K can specifically transphosphorylate XB3 in vitro.
Figure 4.
XA21K Can Specifically Phosphorylate XB3 in Vitro.
The purified MBP-XA21K was incubated either with purified FLAG-tagged XB3 or XBOS31 in the presence of [γ-32P]ATP. FLAG-XB3 alone and the kinase-deficient mutant MBP-XA21KK736E were used as negative controls. Samples were resolved with 8% SDS-PAGE. Coomassie blue staining (CBB) and autoradiogram (Autorad) of the same gel are shown. The asterisk denotes FLAG-XBOS31, which has been confirmed by protein blot analysis with anti-FLAG M2 antibodies (Figure 3C). The experiment was repeated three times with similar results. n.s., nonspecific bands.
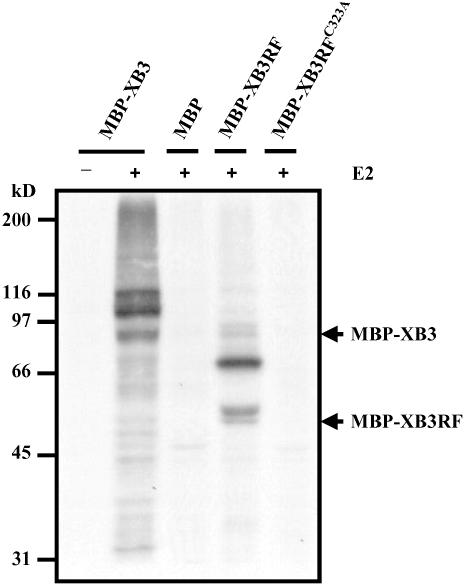
The RF Domain of XB3 Has E3 Activity
To determine whether XB3 has E3 activity, we examined its capability to mediate ubiquitination in vitro. Resin-bound MBP-XB3 was incubated with 32P-labeled ubiquitin (Ub), wheat (Triticum aestivum) E1, and ATP in the presence or absence of the E2 protein UbcH5B. The appearance of ubiquitinated products with high molecular weight was observed only in the presence of UbcH5B (Figure 5). The presence of multiple labeled bands larger than MBP-XB3 suggests a polyubiquitination of the protein, whereas the smaller labeled products could be due to degradation of the ubiquitinated MBP-XB3. The MBP control was not ubiquitinated. We then expressed the RF domain as an MBP fusion (MBP-XB3RF) and examined whether the RF domain alone can support ubiquitination using the above assays. As shown in Figure 5, MBP-XB3RF was ubiquitinated in the presence of UbcH5B. However, an MBP-XB3RF mutant in which the metal binding residue Cys-323 was replaced with Ala was not ubiquitinated (Saurin et al., 1996). Thus, the RF domain of XB3 is capable of catalyzing ubiquitination of MBP-XB3RF in an E2-dependent manner.
Figure 5.
Autoubiquitination of the XB3 and XB3 RF Domain (XB3RF) Fusion Proteins.
GST-ubiquitin was labeled with [γ-32P]ATP by protein kinase A and then digested with thrombin to release the 32P-ubiquitin. Resin-bound MBP and its fusion proteins were incubated with 32P-ubiquitin, ATP, and the wheat ubiquitin-activating enzyme E1 in the presence or absence of E2 (UbcH5B). MBP-XB3RFC323A is a mutant in which conserved Cys-323 is replaced with Ala. Samples were resolved with 8% SDS-PAGE, followed by autoradiography. MBP-XB3 and MBP-XB3RF migrate at ∼89 and 53 kD (indicated by arrows), respectively. The experiment was repeated three times with similar results.
Xb3 Is Required for Stability of the XA21 Protein and for Xa21-Mediated Resistance
We used RNAi to downregulate the expression of Xb3 in vivo. The RNAiXB3 construct, driven by the maize (Zea mays) ubiquitin promoter, contains both sense and antisense Xb3 sequences separated by a 979-bp uidA sequence. The Xb3 probe is generated using a 302-bp region derived from the 3′ end of the gene, including part of the last exon (see Supplemental Figure 2 online). This region shares <50% identity with the corresponding region of Xbos31. No sequence stretches of >11 bp are identical within the 302-bp region between Xb3 and Xbos31. Moreover, DNA gel blot analyses indicated that the 302-bp sequence only hybridizes to Xb3, not to Xbos31 (see Supplemental Figure 3 online). Therefore, RNAiXB3 should specifically downregulate the Xb3 gene.
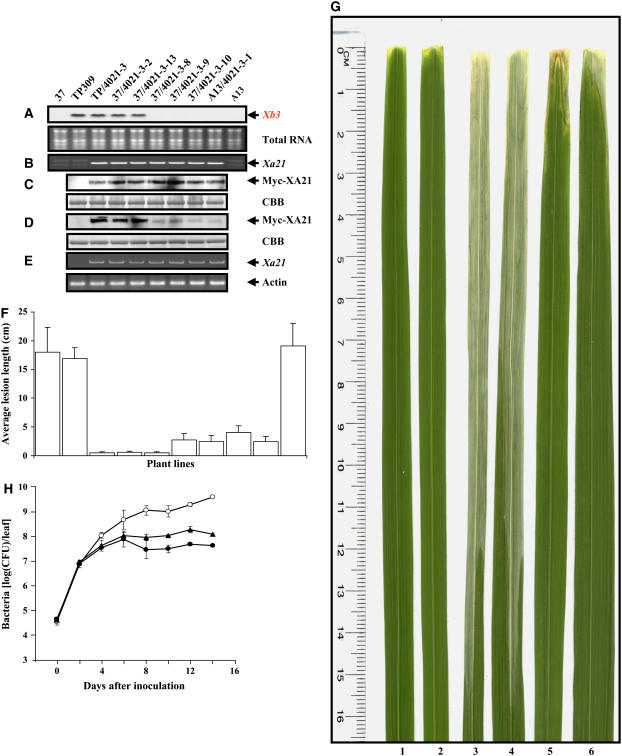
The RNAiXB3 construct was transformed into the susceptible cultivar TP309 using Agrobacterium tumefaciens–mediated transformation. More than 60 independent lines were generated and inoculated with Xoo PR6. All the plants were fully susceptible to Xoo PR6 (data not shown). Two RNAiXB3 lines (37 and A13) with drastically reduced levels of Xb3 RNA transcripts were chosen for further characterization (Figure 6A). Protein blot analyses confirmed that in comparison to TP309, XB3 was decreased in these two lines (Figure 3A).
Figure 6.
Reduction of the XA21 Protein Correlates with the Decrease of Xb3 RNA Transcripts in F1 Individuals.
The pollen recipient RNAiXB3 lines 37 and A13 were crossed with the Xa21-containing line 4021-3 (pollen donor). As a positive control, 4021-3 was crossed with the nontransgenic line TP309. Characterization of progeny at the 4-month-old stage is demonstrated.
(A) RNA gel blot analyses showing Xb3 RNA levels in the indicated parental lines and F1 progeny. Total RNA was probed with an Xb3-specific sequence. Both autoradiogram (top) and agarose gel (bottom) are shown. The Xb3 band can be visualized in all lines when the film was exposed for a longer time period (data not shown).
(B) PCR amplification showing the presence of the Xa21 gene in the indicated lines.
(C) and (D) Protein blot analyses showing the steady state levels of XA21 in the indicated lines at the 1-month-old (C) and 4-month-old (D) stages. Equal amounts of total protein extracts were immunoblotted with anti-c-Myc antibody (top). Coomassie blue–stained gel (CBB) of identical protein samples is shown as a loading control (bottom).
(E) Semiquantitative RT-PCR analyses of Xa21 transcripts in the indicated lines. Total RNA was used to amplify a Xa21 region (top) and the actin gene as control (bottom). The genomic contamination will result in a larger Xa21 fragment due to the presence of an intron in the amplified region (data not shown).
(F) Lesion length data of plants inoculated with Xoo PR6. Each data point represents three clonal individuals with two inoculations per individual. The standard error of the mean is indicated.
(G) Inoculated plants showing lesion development. Leaves 1 and 2, TP309/4021-3 expressing XA21; leaves 3 and 4, TP309; leaves 5 and 6, 37/4021-3-8 expressing a reduced XA21.
(H) Growth of Xoo PR6 in 37/4021-3-8 and control lines. For each time point, the bacterial populations were determined in three leaves separately. Open circles, TP309; triangles, 37/4021-3-8; closed circles, TP309/4021-3. CFU, colony-forming units. Bars indicate standard error.
To test the effects of reduced Xb3 on Xa21-mediated resistance, the RNAiXB3 lines 37 and A13 were used as the pollen recipient parents in crosses with the homozygous Myc-XA21 line 4021-3 (pollen donor). All the progeny tested contained the Xa21 gene, as shown by PCR analyses (Figure 6B, results from six representative F1 plants are shown). We have previously shown that the protein levels of XA21 are developmentally regulated with higher levels at the seedling stage (Xu et al., 2006), although the expression of the Xa21 gene at the RNA level is independent of developmental processes (Century et al., 1999). We then monitored the levels of XA21 at two developmental stages by protein blot analyses. XA21 accumulated at comparable levels at the seedling stage (Figure 6C), but in 4-month-old plants, XA21 accumulated at a significantly lower level in seven of the 12 F1 plants tested (Figure 6D, results from six representative F1 plants are shown). The observed reduction in the XA21 protein is not due to decreases in the Xa21 transcripts but strictly correlates with the decreases in the Xb3 transcripts (Figures 6A and 6E). Furthermore, this reduction also correlates with compromised resistance to Xoo PR6 (Figures 6F and 6G). Bacterial growth curve analyses confirmed that Xoo PR6 achieved a higher level in the F1 progeny with reduced XA21 levels than that generated from a cross of TP309 and 4021-3 (Figure 6H). Taken together, we conclude that Xb3 is required for an abundance of the XA21 protein and Xa21-mediated resistance.
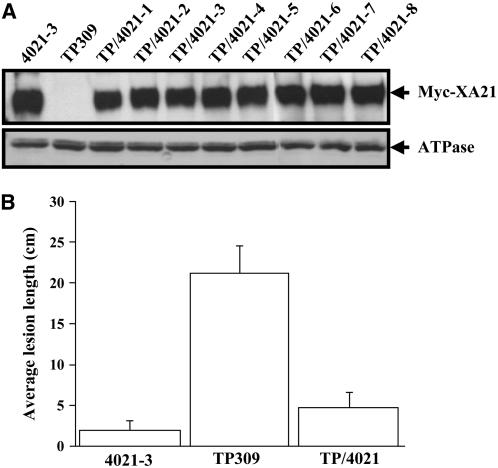
Xa21-Mediated Resistance Shows a Gene Dosage Effect
We crossed TP309 (pollen recipient) with 4021-3 (pollen donor) to determine whether Xa21-mediated resistance is gene dosage dependent. Eight F1 plants were characterized further. All the eight plants had similar levels of XA21, which were slightly lower than that of the homozygous parent 4021-3 (Figure 7A). These results exclude the possibility that the reduced XA21 protein in the above RNAiXB3/4021-3 plants was the result of a segregation of the functional copies of Xa21 and confirm that 4021-3 is homozygous for Xa21. To evaluate resistance, the eight F1 plants were inoculated with Xoo PR6. Although these plants and 4021-3 showed a typical resistance response, most of the F1 plants exhibited slightly longer lesions than 4021-3 (Figure 7B). Given that the F1 progeny only contain half the amount of the functional Xa21 gene, these results indicate that, over a certain range, Xa21 shows dosage-dependent resistance.
Figure 7.
Gene Dosage Effects on Xa21-Mediated Disease Resistance.
The Xa21 homozygous line 4021-3 was used as a pollen donor to cross with TP309. Eight F1 progeny were characterized by protein blot analyses using the anti-c-Myc antibody ([A], top) or anti-ATPase ([A], bottom) as a loading control and by bacterial inoculation using Xoo PR6 at the 6-week-old stage (B). Each data point of lesion length represents >16 leaves. The standard error of the mean is indicated.
DISCUSSION
We characterized a ubiquitin ligase XB3 containing eight ankyrin repeats and an RF motif for its role in Xa21-mediated disease resistance. The ankyrin domain is sufficient for binding to XA21K in vitro. Ankyrin repeats exist in a large number of proteins and interact with diverse partners, including Ser and Thr kinases (Mosavi et al., 2004). Coimmunoprecipitation experiments confirmed that XB3 interacts with XA21 in vivo. Because the leaf tissues used for the coimmunoprecipitation experiments were from uninoculated, healthy plants, the data strongly suggest that the ubiquitin ligase XB3 forms a protein complex with XA21 in planta.
The demonstration of the XA21–XB3 interaction in vivo may be generally important for understanding RLK complexes in rice and in other plants. In a large-scale yeast two-hybrid analysis, we found that nine of 50 randomly chosen rice RLKs interact with four putative E3 ubiquitin ligases (X. Ding and W.-Y. Song, unpublished data). In Brassica napus, the PUB-ARM protein ARC1 interacts with the kinase domain of the S receptor kinase in the yeast two-hybrid system and in vitro (Gu et al., 1998). ARC1 is positively involved in the self-incompatibility system (Stone et al., 1999). It has been proposed that ARC1 promotes the ubiquitination and proteasomal degradation of compatibility factors in the pistil (Stone et al., 2003). In tobacco (Nicotiana tabacum), the Nt PUB4 protein is also a member of the PUB-ARM family. Yeast two-hybrid analysis has linked Nt PUB4 to the kinase domain of the chitinase-related RLK CHRK1 that may be involved in development and cytokinin homeostasis (Kim et al., 2000; Lee et al., 2003). Thus, many plant RLKs may interact with E3 ubiquitin ligases.
One feature of XA21 in the protein complex is the requirement of XB3 for its accumulation, whereas XB3 does not require XA21 for its stability. In the Xb3 silencing lines, the steady state level of the XA21 protein is reduced at the adult stage; however, the XB3 protein accumulates to a similar level in the plants with or without the Xa21 gene (Figures 3F and 6D). These observations are consistent with our recent hypothesis that XA21 is intrinsically unstable owing to the presence of a putative proteolytic cleavage motif (XA21CS1) in the JM domain of this RLK (Xu et al., 2006). Mutation of three residues (Ser-686, Thr-688, and Ser-689) within XA21CS1 destabilizes XA21 only at the adult stage. It has been hypothesized that autophosphorylation of these residues directly or indirectly protects the resistance protein from cleavage by a developmentally regulated protease (Xu et al., 2006). However, Ser-686, Thr-688, and Ser-689 of XA21 appear to be unnecessary for the binding of XB3. The kinase domain of XA21 used for our original two-hybrid screening does not contain these three residues (Figure 1A). Moreover, mutation of these three residues did not abolish the XA21–XB3 interactions in yeast (Figure 1B). Sequence analysis revealed that XA21 possesses at least two additional stretches inside the kinase domain that fulfill the criteria of the P/GX 5-7 P/G motif (G. Cory and W.-Y. Song, unpublished data). Similar to XA21CS1, there are Ser and Thr residues within these two sequence stretches. Notably, the dead kinase mutant XA21K736E accumulates at a lower level than the XA21S686A/T688A/S689A mutant, in which autophosphorylation of other sites is not significantly affected (Xu et al., 2006). It is possible that multiple autophosphorylation and cleavage sites affect the stability of XA21 and that XB3 is recruited by a remaining identified autophosphorylation site(s) and influences the cleavage of this R protein. Although molecular details remain to be elucidated, our data suggest that protein–protein interactions are a second mechanism by which to stabilize XA21.
Consistent with this hypothesis, the XA21 protein appears to be sensitive to IgG precipitation. In total protein extracts, ProA-XA21 was stable even after incubation at 4°C for 2 h (Figure 3E; data not shown). By contrast, a significant portion of XA21 was cleaved into two products of ∼110 and ∼65 kD after precipitation with IgG beads (Figure 3F). The 110-kD band was present in almost all coimmunoprecipitation experiments performed, while the 65-kD product was detected only occasionally. Because the predicted mass of the ProA tag is 13 kD larger than that of Myc, the predominate 110-kD product is consistent in size with the 100-kD band that was previously observed in the microsomal fractions of Myc-XA21 and its mutants (Xu et al., 2006). It has been shown that protein cross-linking can efficiently increase the recovery of less stable protein complexes during TAP purification (Rohila et al., 2004). The enhanced degradation of ProA-XA21 in the immunoprecipitation experiments may be due to the dissociation of some components of the complex during IgG precipitation, which in turn leads to the cleavage of XA21 by a protease in the protein extracts.
The steady state levels of several NB-LRR proteins are regulated by their interacting partners. The Arabidopsis gene RIN4 encodes a novel protein that interacts with the R protein RPM1 (Mackey et al., 2002). RIN4 is required for RPM1-mediated resistance and for accumulation of the RPM1 protein. Interestingly, RIN4 is phosphorylated by an unknown kinase in response to infection with avirulent pathogens. Two additional proteins acting on RPM1 stability are At RAR1, encoded by the Arabidopsis ortholog of barley (Hordeum vulgare) RAR1, and HSP90 (Tornero et al., 2002; Hubert et al., 2003). At RAR1 is also required for the accumulation of the NB-LRR protein RPS5, and this regulation is negatively affected by SGT1b (Holt et al., 2005). In barley, RAR1 is important for accumulation of the NB-LRR proteins MLA1 and MLA6 (Bieri et al., 2004). Our results demonstrate that the steady state accumulation of RLK XA21 is also influenced by its binding protein. Therefore, the stability of R proteins may generally rely on the presence of their partners, and the measurement of the steady state levels of R proteins has become a common tool to dissect R gene–mediated signaling pathways.
XB3 can be phosphorylated by XA21K in vitro. Although this finding remains to be confirmed in planta, the direct interaction between XA21 and XB3 in vivo supports such an extrapolation. In previous studies, we have found that XA21K is capable of phosphorylating a 22–amino acid peptide derived from the glutathione S-transferase (GST) vector when the peptide is fused in frame with XA21K (Liu et al., 2002b). XA21K, however, cannot phosphorylate the free GST protein that contains the same peptide (G.-Z. Liu and W.-Y. Song, unpublished data), suggesting that a physical connection between XA21 and the artificial substrate is required for the phosphorylation. The two phosphorylated residues on the 22–amino acid peptide have been mapped (Liu et al., 2002a). Sequence comparisons reveal that the flanking sequences of these two residues share similarities with those of other identified autophosphorylation sites inside XA21K (G.-Z. Liu and W.-Y. Song, unpublished data). These observations suggest that XA21-mediated transphosphorylation requires the presence of a phosphorylation site and the physical interaction of the substrate with the kinase. XB3 fulfills these criteria. In line with this idea, XA21K was unable to phosphorylate XBOS31, which does not interact with XA21K in yeast. Therefore, protein–protein interactions may contribute to the specificity of XA21 phosphorylation.
When Xb3 is reduced, resistance to Xoo PR6 is compromised in the Xa21 lines. The compromised resistance can be attributed to a decrease in the level of the XA21 protein. The dosage effects have been suggested by several previous studies (Song et al., 1995; Zhang et al., 1998; Xu et al., 2006). Additionally, the heterozygous Xa21 plants generated by a cross between 4021-3 and TP309 are slightly less resistant than the homozygous plant 4021-3. Alternatively, but not mutually exclusive, the reduction of Xb3 may directly contribute to the compromised resistance as well. In this case, the XB3 level is a rate-limiting step for Xa21-mediated resistance.
Based on our data, we propose that the physical interaction of XB3 stabilizes the XA21 protein, thereby maintaining the level of the R protein. Upon pathogen infection, the XA21-XB3 complex is required for activation of XB3, presumably through transphosphorylation. The activated XB3 may ubiquitinate a third protein and target it for degradation. In this scenario, the degraded protein would be a negative regulator of the defense signaling. Alternatively, XB3 may autoubiquitinate, triggering activation of a downstream signaling protein(s). Support for the second model comes from studies in animal innate immunity. Recognition of IL-1 by IL-1R, for example, leads to the formation of a receptor/kinase complex, including IL-1R, MyD88, and the Ser and Thr kinase IRAK (Wu and Arron, 2003). Following this recognition is the activation of TRAF6, an RF-containing ubiquitin ligase. Polyubiquitination of TRAF6 has been suggested to activate the downstream kinase TAK1 (Wang et al., 2001). Our model therefore predicts an interesting parallel between plant and animal defense pathways mediated by a receptor kinase and a receptor/kinase complex. Such a parallel has been recognized based on the fact that a number of plant R proteins share the TIR domain (Baker et al., 1997).
METHODS
Yeast Two-Hybrid Screening
To make the BD-XA21K690 construct for yeast two-hybrid screening, XA21K690 was PCR amplified using the primer pair 5′-AGGTCGACCCCGGGAATGAAAGGCCACCCATT-3′/5′-GGGGTACCGGATCCTCAAAATTCAAGGCTCCCACCTTCAA-3′ and cloned into the two-hybrid vector pPC97 carrying the GAL4 BD domain (Chevray and Nathans, 1992; Chern et al., 2001). All the PCR products in this study were confirmed by DNA sequencing. A yeast two-hybrid library constructed from poly(A)+ mRNA harvested from 2-week-old seedlings of the rice (Oryza sativa) line TP309 (Yin et al., 1997) was kindly provided by R. Beachy. Yeast cells carrying BD-XA21 were transformed with the rice library. Transformation efficiency was estimated by plating an aliquot of transformation mixtures onto SD/-Leu-Trp medium. Candidates selected onto SD/-Leu-Trp-His medium were subjected to β-galactosidase assays as described in the manufacturer's procedure for the Matchmaker GAL4 system (Clontech). Plasmids were recovered from the cells showing the His+ and Lac+ phenotypes and sequenced to determine identity.
To make BD constructs for yeast two-hybrid analyses, XA21K, XA21KK736E, and XA21KS686A/T688A/S689A were subcloned from their GST versions of constructs into the pPC97 vector (Liu et al., 2002b; Xu et al., 2006).
In Vitro Binding Assays
XA21K was PCR amplified with the primer pair 5′-GGATCCGTCGACCACAAGAGAACTAAAAAGGGAGC-3′/5′-GGATCCGTCGACCCCGGGCAGAAGTCGATCTGAAGTGTGGCA-3′ and cloned into pET-28a (Novagen). The resulting plasmid was subjected to the expression of XA21K and labeling using the in vitro transcription and translation kit (Promega). Because the full-length XB3 was poorly expressed as an MBP fusion protein, the cDNA encoding amino acids 1 to 428 of XB3 (missing the last 22 amino acids) was cloned in frame into pMAL-c2X (New England Biolabs) to make MBP-XB3. The ankyrin domain of Xb3 was PCR amplified with the primer pair 5′-GGATCCATGATATCCATGGGTCACGGTGTC-3′/5′-CGGGATCCGATATCAGATGCAGCAAAGCTCC-3′ and cloned into pMAL-c2X. The 35S-labeled XA21K was incubated with resin-bound MBP or MBP fusion proteins in binding buffer (20 mM HEPES, pH 7.4, 1 mM EDTA, 5 mM MgCl2, 1 mM DTT, 0.1% Triton X-100, 1 mg/mL BSA, and 1× complete protease inhibitors [Roche]) for 120 min at 4°C with gentle shaking. After five washes using buffer (20 mM HEPES, pH 7.4, 1 mM EDTA, 5 mM MgCl2, 1 mM DTT, and 0.1% Triton X-100), the bound proteins were eluted with the same buffer supplemented with 10 mM maltose and resolved by 8% SDS-PAGE. The gel was stained with Coomassie Brilliant Blue, dried, and exposed to x-ray film.
Immunodetection
For the anti-XB3M antibodies, a region located in the middle of Xb3 (amino acids 230 to 365) was PCR amplified with the primer pair 5′-GGATCCATGATATCGAGAATTCCCTATTCTGTTGC-3′/5′-GGATCCGATATCTCATGAGGGCGGTGTCAGGGTC-3′ and cloned into the expression vectors pGTK (Liu et al., 2002b) and pMAL-c2X, respectively. The fusion proteins MBP-XB3M and GST-XB3M were expressed in the Escherichia coli strain ER2566 and affinity purified (Liu et al., 2002b). MBP-XB3M was used to immunize rabbits (Cocalico Biologicals). Antisera were subjected to affinity purification using GST-XB3M according to the method described by Lin et al. (1996).
To create the FLAG-tagged XB3 and XBOS31 for bacterial expression, the full-length coding regions of these two genes were PCR amplified with the primer pairs 5′-GTGTGATATCATGGGTCACGGTGTC-3′/5′-GGATCCATGATATCGAGGATGATGCGGCGA-3′ and 5′-GTGTAGATCTCCGAATTCCTACCTATGGGGCACGGCCTGAG-3′/5′-GTGTGCGGCCGCCCCAAAGCTCTAGTGCTAGTAAGGTG-3′ and cloned into pFLAG-N, an expression vector derived from pFLAG-MAC (Sigma-Aldrich). The fusion proteins FLAG-XB3 and FLAG-XBOS31 were expressed in the E. coli strain ER2566 (Liu et al., 2002b).
Both the c-Myc and ProA tags were inserted in domain B of the Xa21 gene. The DNA sequences, encoding the 13–amino acid c-Myc and 128–amino acid double ProA epitope tags, were PCR amplified from the plasmids pATGMyc and pUbi.nc1300.ntapintron.new and cloned into the unique DraIII site of the plasmid pC822 to create pC822cMyc and pC822ProA, respectively. The KpnI fragment containing Myc-Xa21 or ProA-Xa21 was cloned into pCAMBIA1300 for rice transformation. The construction of TAP-Os06g48590 was as described by Rohila et al. (2006).
Total rice protein extracts were isolated by grinding leaf tissue in liquid nitrogen and thawing in an equal volume of extraction buffer [50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 5% (v/v) β-mercaptoethanol, 1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride (Sigma-Aldrich), 2 μg/mL antipain (Sigma-Aldrich), 2 μg/mL leupeptin (Sigma-Aldrich), and 2 μg/mL aprotinin (Sigma-Aldrich)]. Cell debris were removed by centrifugation at 12,000g for 10 min at 4°C. Protein concentration was measured with Bio-Rad protein assays.
To coimmunoprecipitate ProA-XA21 and XB3, protein extracts were prepared from 5 g of leaf tissue in 25 mL of ice-cold extraction buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% Triton X-100, 2.5 mM EDTA, 2 mM benzamidine [Sigma-Aldrich], 10 mM β-mercaptoethanol, 20 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 1% Protease Cocktail [Sigma-Aldrich], 10 μM leupeptin, and 10% glycerol). After filtering through double layers of Miracloth (Calbiochem) followed by centrifugation twice at 13,000g for 10 min at 4°C, the supernatant was mixed with 400 μL of IgG Sepharose beads (Amersham Biosciences) and incubated at 4°C for 1 h. The beads were then washed four times in 1 mL of protein extraction buffer lacking protease inhibitors and twice in 0.4 mL of 5 mM ammonium acetate, pH 5.0. The protein was eluted with 2 mL of 0.5 M HOAC, pH 3.4, neutralized with one-tenth volume of 1 M Tris-HCl, pH 8.0, and concentrated by acetone precipitation.
Protein blot analyses were performed as described previously (Xu et al., 2006).
Transphosphorylation Assays
Transphosphorylation of FLAG-XB3 by MBP-XA21K was essentially as previously described (Liu et al., 2002b).
Ubiquitination Assays
pMBP-XB3RING was made as follows: the RF domain and C-terminal tail of Xb3 were PCR amplified with the primer pair 5′-GGATCCATGATATCCGATGCATGCTCAGAG-3′/5′-GGATCCATGATATCGAGGATGATGCGGCGA-3′ and cloned into pMAL-c2X. To remove the C-terminal tail, the resulting construct pMALXB3C was digested with PstI and self-ligated. To create a stop codon after the RF domain, the self-ligated construct was digested with HindIII, blunted by filling-in, and self-ligated again. Site-directed mutagenesis was performed to create pMBP-XB3RINGC323A as described previously (Liu et al., 2002b). The fusion proteins MBP-XB3, MBP-XB3RING, and MBP-XB3RINGC323A were expressed in the E. coli strain ER2566 and affinity purified (Liu et al., 2002b). Ubiquitination assays were performed as described previously (Nodzon et al., 2004).
RNAiXB3 and Overexpression of the Xb3 Gene
The 3′ end of Xb3 was PCR amplified with the following primer pairs: 5′-GAATTCTCTAGACCGGGGCAGCATCTCA-3′/5′-ACTAGTGGATCCTTTCTGATACCAACGGA-3′ and 5′-GAATTCAGATCTCCGGGGCAGCATCTCA-3′/5′-ACTAGTGATATCTTTCTGATACCAACGGA-3′. The PCR products were ligated to the uidA fragment spanning nucleotides 815 to 1793 in both antisense and sense orientations. The resulting construct was then cloned into the overexpression vector pBHU-1, which contains the hph gene, whose product confers resistance to hygromycin B, at the site between the maize (Zea mays) ubiquitin promoter and the nos 3′ terminator. Rice transformation was performed as described previously (Xu et al., 2006).
A full-length Xb3 (lacking the stop codon) was PCR amplified with the primer pair 5′-GGATCCACTAGTATGGGTCACGGTGTCAG-3′/5′-GGATCCTAGATCGTGCTCAGGCT-3′ and in-frame fused to the FLAG tag (containing a stop codon at the 3′ end). The tagged Xb3 was cloned into pCmHU-1, an overexpression vector derived from pBHU-1, for rice transformation.
Characterization of Transgenic Plants
DNA and RNA gel blot analyses were performed using standard procedures. Plant inoculation and growth curve analyses were performed as described by Song et al. (1995), except that a bacterial suspension of OD ∼1.0 was used.
Semiquantitative RT-PCR analyses were performed with primer pairs 5′-CAGAAGTCGATCTGAAGTGTGGCA-3′/5′-GCACAAGAGAACTAAAAAGGGAGCCC-3′ (for Xa21 transcripts) and 5′-TGGCGCCCGAGGAGCACC-3′/5′-GTAACCCCTCTCAGTCAG-3′ (for actin transcripts). Three micrograms of total RNA were converted into cDNA using the SuperScript First-Stand Synthesis system (Invitrogen) followed by PCR amplification for 20, 25, 30, and 35 cycles. The amplified products were then resolved by gel electrophoresis.
The transgenic lines 37 and A13 were used as the pollen recipient parents to cross with pollen donor 4021-3. Eleven seeds were recovered from the 37/4021-3 cross, whereas only one seed was obtained from the A13/4021-3 cross. The nature of the F1 hybrids was confirmed by PCR amplification of a 690-bp fragment spanning part of the Xa21 and 35S promoters in the pCAMBIA1300-Myc-Xa21 construct using the primer pair 5′-ATTCATTAATGCAGCTGGCACGACA-3′/5′-GGTAATGGATGTACACTGCAGAACGA-3′.
Sequence Analyses
The PAUP (version 4.0b10) software package (Swofford, 2002) was used to perform phylogenetic analyses. Neighbor joining was used to reconstruct the phylogenetic relationships of these amino acids. Bootstrap values were derived from 1000 replicates to quantify the relative support for branches of the inferred phylogenetic tree. The identity and similarity between proteins were calculated using the GAP tool within the Genetics Computer Group program.
Accession Numbers
The rice cDNA accession numbers (KOME; http://cdna01.dna.affrc.go.jp/cDNA/) for the Xbos genes are as follows: Xbos31, AK106014; Xbos32, AK120632; Xbos33, AK065223; Xbos34, AK059792; and Xbos35, AK067289. The GenBank accession number for Xbos36 is DQ088999. The accession numbers for the XBAT genes were described previously (Nodzon et al., 2004). The GenBank accession number for Xb3 cDNA sequence is AF272860.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Comparisons of Amino Acid Sequences between the XB3-Related Proteins from Rice and Arabidopsis.
Supplemental Figure 1. Phylogenetic Tree Based on the Predicted Amino Acid Sequences of XB3 and XB3-Related Proteins from Rice (XBOS) and Arabidopsis (XBAT).
Supplemental Figure 2. Schematic Representation of the Xb3 and Xbos31 Genomic Gene Structures.
Supplemental Figure 3. DNA Gel Blot Analyses Showing Specificity of the RNAiXB3 Probe.
Supplementary Material
Acknowledgments
We thank R. Beachy for the rice yeast two-hybrid library; M. Fromm for pUbi.nc1300.ntapintron.new; M. Boutry for anti-ATPase; O. da Costa e Silva for pATGmyc; S. Dai, Y. Chen, Y. Xu, P. Tinjuangjun, Y.-R. Chen, A. Snyder, and H. Xiao for technical assistance; Lisa Nodzon for cloning Xbos36; M.-S. Shern for providing BD-XA21 for use in these studies; and Fahong Yu for phylogenetic analyses. We also thank Jeff Chang, J.B. Jones, H. Klee, Terry Davoli, and Margaret Joyner for critical reading of the manuscript and their invaluable comments. This research was supported primarily by the National Science Foundation under Grant 0080155 to W.-Y.S. and by a grant from National Institutes of Health to P.C.R.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Wen-Yuan Song (wsong@ifas.ufl.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Austin, M.J., Muskett, P., Kahn, K., Feys, B.J., Jones, J.D., and Parker, J.E. (2002). Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295 2077–2080. [DOI] [PubMed] [Google Scholar]
- Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., and Schulze-Lefert, P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295 2073–2076. [DOI] [PubMed] [Google Scholar]
- Baker, B., Zambryski, P., Staskawicz, B., and Dinesh-Kumar, S.P. (1997). Signaling in plant-microbe interactions. Science 276 726–733. [DOI] [PubMed] [Google Scholar]
- Ben-Neriah, Y. (2002). Regulatory functions of ubiquitination in the immune system. Nat. Immunol. 3 20–26. [DOI] [PubMed] [Google Scholar]
- Bieri, S., Mauch, S., Shen, Q.H., Peart, J., Devoto, A., Casais, C., Ceron, F., Schulze, S., Steinbiss, H.H., Shirasu, K., and Schulze-Lefert, P. (2004). RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell 16 3480–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueggeman, R., Rostoks, N., Kudrna, D., Kilian, A., Han, F., Chen, J., Druka, A., Steffenson, B., and Kleinhofs, A. (2002). The barley stem rust-resistance gene Rpg1 is a novel disease-resistance gene with homology to receptor kinases. Proc. Natl. Acad. Sci. USA 99 9328–9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century, K.S., Lagman, R.A., Adkisson, M., Morlan, J., Tobias, R., Schwartz, K., Smith, A., Love, J., Ronald, P.C., and Whalen, M.C. (1999). Developmental control of Xa21-mediated disease resistance in rice. Plant J. 20 231–236. [DOI] [PubMed] [Google Scholar]
- Chen, X., et al. (2006). A B-lectin receptor kinase gene conferring rice blast resistance. Plant J. 46 794–804. [DOI] [PubMed] [Google Scholar]
- Chern, M.S., Fitzgerald, H.A., Yadav, R.C., Canlas, P.E., Dong, X., and Ronald, P.C. (2001). Evidence for a disease-resistance pathway in rice similar to the NPR1-mediated signaling pathway in Arabidopsis. Plant J. 27 101–113. [DOI] [PubMed] [Google Scholar]
- Chevray, P.M., and Nathans, D. (1992). Protein interaction cloning in yeast: Identification of mammalian proteins that react with the leucine zipper of Jun. Proc. Natl. Acad. Sci. USA 89 5789–5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411 826–833. [DOI] [PubMed] [Google Scholar]
- Diener, A.C., and Ausubel, F.M. (2005). RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis resistance gene, is not race specific. Genetics 171 305–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godiard, L., Sauviac, L., Torii, K.U., Grenon, O., Mangin, B., Grimsley, N.H., and Marco, Y. (2003). ERECTA, an LRR receptor-like kinase protein controlling development pleiotropically affects resistance to bacterial wilt. Plant J. 36 353–365. [DOI] [PubMed] [Google Scholar]
- Goes da Silva, F., Shen, Y., Dardick, C., Burdman, S., Yadav, R., Sharma, P., and Ronald, P.C. (2004). Components of a type I secretion system and a sulfotransferase-like protein are required for the XA21 receptor kinase mediated defense response. Mol. Plant Microbe Interact. 17 593–601. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Lamothe, R., Tsitsigiannis, D.I., Ludwig, A.A., Panicot, M., Shirasu, K., and Jones, J.D. (2006). The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18 1067–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, T., Mazzurco, M., Sulaman, W., Matias, D.D., and Goring, D.R. (1998). Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc. Natl. Acad. Sci. USA 95 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, J.A., and Reichhart, J.M. (2002). Drosophila innate immunity: An evolutionary perspective. Nat. Immunol. 3 121–126. [DOI] [PubMed] [Google Scholar]
- Holt III, B.F., Belkhadir, Y., and Dangl, J.L. (2005). Antagonistic control of disease resistance protein stability in the plant immune system. Science 309 929–932. [DOI] [PubMed] [Google Scholar]
- Hubert, D.A., Tornero, P., Belkhadir, Y., Krishna, P., Takahashi, A., Shirasu, K., and Dangl, J.L. (2003). Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 22 5679–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro, C.A.P., and Weissman, A.M. (2000). RING finger proteins: Mediators of ubiquitin ligase activity. Cell 102 549–552. [DOI] [PubMed] [Google Scholar]
- Kawasaki, T., Nam, J., Boyes, D.C., Holt III, B.F., Hubert, D.A., Wiig, A., and Dangl, J.L. (2005). A duplicated pair of Arabidopsis RING-finger E3 ligases contribute to the RPM1- and RPS2-mediated hypersensitive response. Plant J. 44 258–270. [DOI] [PubMed] [Google Scholar]
- Kim, H.S., and Delaney, T.P. (2002). Arabidopsis SON1 is an F-box protein that regulates a novel induced defense response independent of both salicylic acid and systemic acquired resistance. Plant Cell 14 1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.S., Lee, J.H., Yoon, G.M., Cho, H.S., Park, S.W., Suh, M.C., Choi, D., Ha, H.J., Liu, J.R., and Pai, H.S. (2000). CHRK1, a chitinase-related receptor-like kinase in tobacco. Plant Physiol. 123 905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.H., Takei, K., Sakakibara, H., Sun Cho, H., Kim, D.M., Kim, Y.S., Min, S.R., Kim, W.T., Sohn, D.Y., Lim, Y.P., and Pai, H.S. (2003). CHRK1, a chitinase-related receptor-like kinase, plays a role in plant development and cytokinin homeostasis in tobacco. Plant Mol Biol. 53 877–890. [DOI] [PubMed] [Google Scholar]
- Lin, Y., Wang, Y., Zhu, J.K., and Yang, Z. (1996). Localization of a Rho GTPase implies a role in tip growth and movement of the generative cell in pollen tubes. Plant Cell 8 293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, G.Z., Pi, L.-Y., Walker, J.C., Ronald, P.C., and Song, W.-Y. (2002. b). Biochemical characterization of the kinase domain of the rice disease resistance receptor-like kinase XA21. J. Biol. Chem. 277 20264–20269. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Schiff, M., Serino, G., Deng, X.W., and Dinesh-Kumar, S.P. (2002. a). Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to Tobacco mosaic virus. Plant Cell 14 1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente, F., Alonso-Blanco, C., Sanchez-Rodriguez, C., Jorda, L., and Molina, A. (2005). ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant J. 43 165–180. [DOI] [PubMed] [Google Scholar]
- Lorick, K.L., Jensen, J.P., Fang, S., Ong, A.M., Hatakeyama, S., and Weissman, A.M. (1999). RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 96 11364–11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey, D., Holt III, B.F., Wiig, A., and Dangl, J.L. (2002). RIN4 interacts with Pseudomonas syringae Type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108 743–754. [DOI] [PubMed] [Google Scholar]
- Mosavi, L.K., Cammett, T.J., Desrosiers, D.C., and Peng, Z.Y. (2004). The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 13 1435–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodzon, L.A., Xu, W.H., Wang, Y., Pi, L.-Y., Chakrabarty, P.K., and Song, W.-Y. (2004). The ubiquitin ligase XBAT32 regulates lateral root development in Arabidopsis. Plant J. 40 996–1006. [DOI] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405 462–466. [DOI] [PubMed] [Google Scholar]
- Resh, M.D. (1999). Fatty acylation of proteins: New insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451 1–16. [DOI] [PubMed] [Google Scholar]
- Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. (1999). A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17 1030–1032. [DOI] [PubMed] [Google Scholar]
- Rohila, J.S., Chen, M., Cerny, R., and Fromm, M.E. (2004). Improved tandem affinity purification tag and methods for isolation of protein heterocomplexes from plants. Plant J. 38 172–181. [DOI] [PubMed] [Google Scholar]
- Rohila, J.S., et al. (2006). Protein-protein interactions of TAP-tagged protein kinases in rice. Plant J. 46 1–13. [DOI] [PubMed] [Google Scholar]
- Rowland, O., Ludwig, A.A., Merrick, C.J., Baillieul, F., Tracy, F.E., Durrant, W.E., Fritz-Laylin, L., Nekrasov, V., Sjolander, K., Yoshioka, H., and Jones, J.D. (2005). Functional analysis of Avr9/Cf-9 rapidly elicited genes identifies a protein kinase, ACIK1, that is essential for full Cf-9-dependent disease resistance in tomato. Plant Cell 17 295–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin, A.J., Borden, K.L., Boddy, M.N., and Freemont, P.S. (1996). Does this have a family RING? Trends Biochem. Sci. 21 208–214. [PubMed] [Google Scholar]
- Scheer, J.M., and Ryan, C.A., Jr. (2002). The systemin receptor SR160 from Lycopersicon peruvianum is a member of the LRR receptor kinase family. Proc. Natl. Acad. Sci. USA 99 9585–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick, S.G., and Smerdon, S.J. (1999). The ankyrin repeat: A diversity of interactions on a common structural framework. Trends Biochem. Sci. 24 311–316. [DOI] [PubMed] [Google Scholar]
- Shirasu, K., Lahaye, T., Tan, M.W., Zhou, F., Azevedo, C., and Schulze-Lefert, P. (1999). A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 99 355–366. [DOI] [PubMed] [Google Scholar]
- Smalle, J., and Vierstra, R.D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55 555–590. [DOI] [PubMed] [Google Scholar]
- Song, W.-Y., Wang, G., Chen, L., Kim, H., Pi, L.-Y., Gardner, J., Wang, B., Holsten, T., Zhai, W., Zhu, L., Fauquet, C., and Ronald, P.C. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene Xa21. Science 270 661–667. [DOI] [PubMed] [Google Scholar]
- Staskawicz, B.J., Mudgett, M.B., Dangl, J.L., and Galan, J.E. (2001). Common and contrasting themes of plant and animal diseases. Science 292 2285–2289. [DOI] [PubMed] [Google Scholar]
- Stone, S.L., Anderson, E.M., Mullen, R.T., and Goring, D.R. (2003). ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15 885–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, S.L., Arnoldo, M., and Goring, D.R. (1999). A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science 286 1729–1731. [DOI] [PubMed] [Google Scholar]
- Sun, X., Cao, Y., Yang, Z., Xu, C., Li, X., Wang, S., and Zhang, Q. (2004). Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 37 517–527. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. (2002). PAUP: Phylogenetic Analysis Using Parsimony, Version 4.0b10. (Sunderland, MA: Sinauer Associates).
- Tor, M., Gordon, P., Cuzick, A., Eulgem, T., Sinapidou, E., Mert-Turk, F., Can, C., Dangl, J.L., and Holub, E.B. (2002). Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell 14 993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero, P., Merritt, P., Sadanandom, A., Shirasu, K., Innes, R.W., and Dangl, J.L. (2002). RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis, and their relative contributions are dependent on the R gene assayed. Plant Cell 14 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., Deng, L.M., Hong, M., Akkaraju, G.R., Inoue, J., and Chen, Z.J. (2001). TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412 346–351. [DOI] [PubMed] [Google Scholar]
- Wu, H., and Arron, J.R. (2003). TRAF6, a molecular bridge spanning adaptive immunity, innate immunity and osteoimmunology. Bioessays 25 1096–1105. [DOI] [PubMed] [Google Scholar]
- Xie, D.-X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xu, W.H., Wang, Y., Liu, G., Chen, X., Tinjuangjun, P., Pi, L.Y., Zhang, Y., and Song, W.Y. (2006). The autophosphorylated Ser686, Thr688 and Ser689 residues within a putative protease cleavage motif in the juxtamembrane domain of XA21 are implicated in the stability control of the rice receptor-like kinase. Plant J. 45 740–751. [DOI] [PubMed] [Google Scholar]
- Yang, C.W., Gonzalez-Lamothe, R., Ewan, R.A., Rowland, O., Yoshioka, H., Shenton, M., Ye, H., O'Donnell, E., Jones, J.D., and Sadanandom, A. (2006). The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18 1084–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y., Zhu, Q., Dai, S., Lamb, C., and Beachy, R.N. (1997). RF2a, a bZIP transcriptional activator of the phloem-specific rice tungro bacilliform virus promoter, functions in vascular development. EMBO J. 16 5247–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, L.R., Qu, S., Bordeos, A., Yang, C., Baraoidan, M., Yan, H., Xie, Q., Nahm, B.H., Leung, H., and Wang, G.L. (2004). Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16 2795–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S.P., Song, W.-Y., Chen, L.-L., Ruan, D.L., Ronald, P.C., Beachy, R., and Fauquet, C. (1998). Genetic engineering of agronomically important Indica rice varieties for resistance to bacterial blight disease. Mol. Breed. 4 551–558. [Google Scholar]
- Zipfel, C., Robatzek, S., Navarro, L., Oakeley, E.J., Jones, J.D., Felix, G., and Boller, T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.