Abstract
Plant mitochondrial genomes exist in a natural state of heteroplasmy, in which substoichiometric levels of alternative mitochondrial DNA (mtDNA) molecules coexist with the main genome. These subgenomes either replicate autonomously or are created by infrequent recombination events. We found that Arabidopsis thaliana OSB1 (for Organellar Single-stranded DNA Binding protein1) is required for correct stoichiometric mtDNA transmission. OSB1 is part of a family of plant-specific DNA binding proteins that are characterized by a novel motif that is required for single-stranded DNA binding. The OSB1 protein is targeted to mitochondria, and promoter–β-glucuronidase fusion showed that the gene is expressed in budding lateral roots, mature pollen, and the embryo sac of unfertilized ovules. OSB1 T-DNA insertion mutants accumulate mtDNA homologous recombination products and develop phenotypes of leaf variegation and distortion. The mtDNA rearrangements occur in two steps: first, homozygous mutants accumulate subgenomic levels of homologous recombination products; second, in subsequent generations, one of the recombination products becomes predominant. After the second step, the process is no longer reversible by backcrossing. Thus, OSB1 participates in controlling the stoichiometry of alternative mtDNA forms generated by recombination. This regulation could take place in gametophytic tissues to ensure the transmission of a functional mitochondrial genome.
INTRODUCTION
In higher plants, mitochondrial genomes are large (367 and 570 kb in Arabidopsis thaliana and maize [Zea mays], respectively) (Unseld et al., 1997; Clifton et al., 2004), and their complete genetic repertoire can theoretically assemble in a genome-size circular chromosome. However, electrophoresis and electron microscopy studies showed that plant mitochondrial DNA (mtDNA) is a heterogeneous population of circular, linear, and complex double- and single-stranded molecules (Oldenburg and Bendich, 1996; Backert et al., 1997; Backert and Börner, 2000). These structures exist in dynamic equilibrium and probably result from a rolling-circle mechanism of DNA replication, but also from a recombination-dependent mechanism of DNA replication, similar to the replication of phage T4 (Backert and Börner, 2000). In addition, numerous recombination events between large and small repeats result in a multipartite structure of the plant mtDNA (Adams and Daley, 2004). Because the plant mtDNA is rich in repeated sequences, strict control of homologous recombination (HR) is essential for its stability. If recombination regulation is relaxed, then alternative mtDNA structures generated by HR could be generated during replication. These structures could give rise to rapidly replicating selfish genomes, containing an incomplete set of genes or expressing deleterious chimeric genes. The Arabidopsis mtDNA (ecotype C24) contains 22 pairs of 100% identical repeats of >100 bp, but only the two largest ones (6.5 and 4.2 kb) are involved in frequent reciprocal HR (Unseld et al., 1997). In both prokaryotes and eukaryotes, several proteins are known to suppress HR (Pinto et al., 2005), but in plant mitochondria, the mechanisms that regulate HR have not yet been characterized.
Adding to the complexity of plant mitochondria, the mtDNA is essentially heteroplasmic (Kmiec et al., 2006)—that is, different genome forms can coexist and replicate differentially. The ratios of the different types of mtDNA may vary, but usually one mtDNA configuration is prevalent and alternative configurations are present at sublimon levels (Small et al., 1989). Heteroplasmy can also originate from selfish elements, such as those at the origin of cytoplasmic male sterility, an agronomically important trait used by breeders to create high-yielding hybrids. In plants, very rapid changes may occur in the relative proportions of mtDNA variants, a phenomenon called substoichiometric shifting. These changes can occur under natural conditions (Janska et al., 1998), but they can also be induced in cybrids, in specific cell culture conditions, and in certain nuclear backgrounds (Kanazawa et al., 1994; Bellaoui et al., 1998; Kuzmin et al., 2005). For example, in Phaseolus vulgaris pvs-orf239, a subgenomic molecule that undergoes substoichiometric shifting is amplified up to 2000-fold (Arrieta-Montiel et al., 2001), leading to cytoplasmic male sterility when the nuclear fertility-restorer Fr gene is inactive.
Mechanisms that regulate the stoichiometric transmission of the different mitotypes are still poorly understood. Substoichiometric shifting could result from increased HR, which continuously generates recombination products in somatic tissues, or from the favored replication of one of the mitotypes. It is also possible that increased HR activity creates a pool of sequences that, by strand invasion, prime the asymmetric replication of mtDNA chimeras. Whatever the mechanism regulating mtDNA heteroplasmy, it is expected to be active in rapidly dividing cells and in gametophyte cells. The nuclear control of substoichiometric shifting was shown to be more effective in undifferentiated meristem cells than in vegetative tissues (Arrieta-Montiel et al., 2001), and mtDNA reorganization was thus postulated to occur in transmitting tissues in which mtDNA replication is active. This hypothesis was recently corroborated by the work of Sheahan and colleagues (2005), who showed that massive mitochondrial fusion precedes fission and the dispersion of the organelles throughout the cytoplasm in newly prepared protoplasts. Massive mitochondrial fusion appears to be specific to the cell dedifferentiation process and therefore should facilitate the repackaging of mitochondrial genomes, thus ensuring the transmission of all subgenomic molecules. In animals, a sharp reduction in mitochondrial genome number (the so-called bottleneck effect) accompanies oogenesis, and it was suggested that this phenomenon is particularly relevant to understanding how differential mitochondrial segregation is achieved during mitotic divisions (Barr et al., 2005). As a corollary to this model, if the control of HR and illegitimate recombination is relaxed, then selfish mitochondrial genomes arising from rearrangements, deletions, and insertions could accumulate and be preferentially transmitted, leading to mitochondrial dysfunction.
In plants, a component of stoichiometric regulation was identified: the MSH1 gene (Abdelnoor et al., 2003), which encodes a protein similar to prokaryotic MutS and is responsible for the chm mutant phenotype (Martinez-Zapater et al., 1992) in Arabidopsis. In bacteria, MutS is involved in mismatch repair and the suppression of ectopic recombination. In yeast mitochondria, MSH1 is an essential gene required for mtDNA stability. However, recent evidence suggests that the mismatch repair function of yeast MSH1 is not essential but that other functions, such as recombination surveillance and heteroduplex rejection, are necessary for homoplasmic mtDNA sorting (Mookerjee and Sia, 2006). The precise mode of action of plant MSH1 in mtDNA substoichiometric shifting is still not known, but it could play a role similar to that of yeast MSH1. Another example of a gene involved in plant mtDNA instability by recombination was found in the maize P2 line (Kuzmin et al., 2005). In this line, a nuclear allele directly controls the amplification of several mitochondrial genome rearrangements. Several rearranged mtDNA forms could result from rare, reciprocal HR and could be generated continuously in vegetative tissues. However, other mtDNA forms could be created de novo by illegitimate recombination and amplified in P2 progeny lines.
Here, we describe the identification of a plant-specific OSB (for Organellar Single-stranded DNA Binding protein) family of mitochondrial and chloroplast proteins, several of which bind single-stranded DNA (ssDNA). Among these proteins, Arabidopsis OSB1 was shown to be required for mtDNA stability. In osb1 T-DNA insertion mutants, the accumulation of mtDNA molecules derived from HR leads to severe morphological phenotypes. OSB1 is expressed primarily in gametophytic cells, in correlation with the need for a nuclear control on gametophytic tissues to ensure the stoichiometric transmission of alternative mitochondrial genomes to the plant progeny.
RESULTS
The Family of OSB Proteins Is Characterized by a Plant-Specific Motif
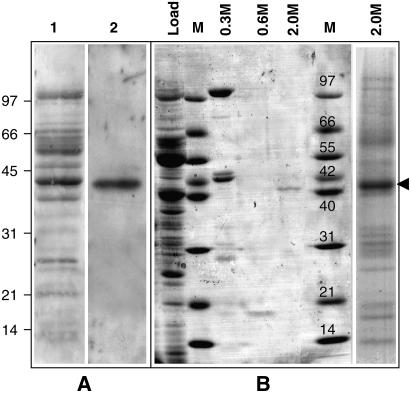
To identify plant mitochondrial proteins involved in mtDNA maintenance, potato (Solanum tuberosum) mitochondrial proteins with a high affinity for ssDNA were analyzed by protein/DNA gel blotting. To avoid sequence-specificity bias, a random sequence oligonucleotide was used as a probe. A 40-kD protein was the major mitochondrial ssDNA binding protein (SSB) detected by protein/DNA gel blotting (Figure 1A). A protein of the same size was purified from potato mitochondria by ssDNA-affinity chromatography (Vermel et al., 2002). This protein eluted from the column at high salt concentrations, denoting a high affinity for ssDNA (Figure 1B). It was thus named St OSB1, for Solanum tuberosum Organellar ssDNA Binding protein1. The complete St OSB1 cDNA was obtained by RT-PCR using degenerate oligonucleotides deduced from tryptic peptide sequences, followed by 5′ and 3′ rapid amplification of cDNA ends. The cDNA encodes a 43-kD protein predicted to be mitochondrial by Predotar, TargetP, and iPsort. Amplification was possible from total potato sprout RNA but not from leaf RNA, suggesting that the gene is expressed preferentially in rapidly dividing tissues.
Figure 1.
Purification of the Potato Mitochondrial OSB1 Protein.
(A) Soluble proteins (8 μg) from potato mitochondria were fractionated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and probed for DNA binding. Lane 1, Ponceau S staining of the proteins; lane 2, protein/DNA gel blot of proteins probed with a 5′ end-labeled random 37-nucleotide single-strand oligonucleotide.
(B) Purification of the 40-kD protein by ssDNA chromatography. Coomassie blue staining of the proteins eluted at 0.3, 0.6, and 2.0 M NaCl. The proteins eluting at 2.0 M were separated (right lane), and the 40-kD protein (arrowhead) was extracted and sequenced.
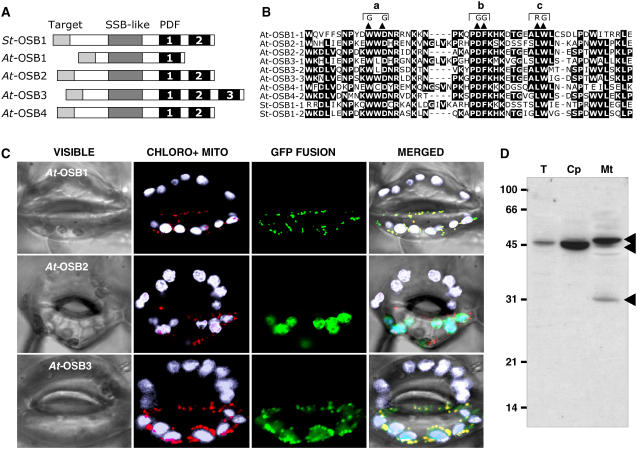
We found orthologs of the St OSB1 gene in the Arabidopsis thaliana, rice (Oryza sativa), and Zea mays genomes. The highest homology between these sequences (40 to 80% identity) was found in a 50–amino acid C-terminal motif that is present in one to three copies. This motif was named PDF, because these three residues are conserved in all proteins of the family. Two of the three putative OSB genes in the rice genome code for proteins with a single PDF motif, and one codes for a protein with two PDF motifs. PDF motifs were also found in numerous sequences deduced from ESTs of monocot and dicot plants. In the green alga Chlamydomonas reinhardtii, the chloroplast protein RB38 is constituted of four PDF motifs repeated in tandem (Barnes et al., 2004). RB38 is the only PDF motif protein in Chlamydomonas, according to the latest release of the C. reinhardtii genome (http://genome.jgi-psf.org/Chlre3/Chlre3.home.html). However, the PDF motif was not found in proteins of species outside of the plant kingdom. We focused our study on Arabidopsis, in which OSBs form a family of four putative proteins encoded by the nuclear genes At1g47720 (At OSB1), At4g20010 (At OSB2), At5g44785 (At OSB3), and At1g31010 (At OSB4). Complete cDNAs were obtained by RT-PCR for OSB1, OSB2, and OSB3. The OSB4 cDNA was not cloned, but the gene is apparently expressed, because there is a corresponding EST sequence. Figure 2A shows that OSB sequences can be divided into three domains: (1) a nonconserved sequence predicted to be an organellar targeting sequence; (2) a central region that has the signature of the prokaryotic SSB family (PFAM00436); and (3) a C-terminal region whose length depends on the number of PDF motifs: one in At OSB1, two in At OSB2, At OSB4, and St OSB1, and three in At OSB3 (Figure 2B).
Figure 2.
Characterization of the OSB Protein Family.
(A) Basic structure of OSB proteins from potato (St OSB1) and Arabidopsis (At OSB1 to OSB4). The PDF motifs are numbered.
(B) Alignment of PDF motifs of the proteins. The double substitution mutations (a, b, and c) analyzed in Figure 3 are indicated.
(C) Localization of protein-eGFP fusions in guard cells of N. benthamiana. Green, eGFP fluorescence; white, natural fluorescence of chloroplasts; red, fluorescence of the mitochondrial marker.
(D) Protein gel blot of protein fractions probed with antibodies raised against OSB1: total fraction (T)–, chloroplast (Cp)-, and mitochondria (Mt)- enriched fractions. Protein fractions were extracted from Arabidopsis cells in suspension culture as described (Laloi et al., 2001).
OSB Proteins Are Targeted to Mitochondria and Chloroplasts
OSB proteins are predicted to be organellar from sequence analysis. To localize At OSB1, two and three green fluorescent protein (GFP) fusion proteins, the N-terminal regions, up to the conserved SSB domain, were cloned in pCK-GFP3A (Menand et al., 1998) and transiently expressed in epidermal cells of Nicotiana benthamiana leaves. The N-terminal sequences of OSB1 and OSB3 target eGFP into mitochondria (Figure 2C). In all cells observed, the OSB3-eGFP fusion is also targeted to the chloroplast. By contrast, the OSB2 fusion protein was detected only in chloroplasts. To confirm these results, antibodies directed against At OSB1 were generated. Affinity-purified antibodies recognized both At OSB1 and At OSB2 recombinant proteins, respectively, expressed in Escherichia coli and in planta (see below; data not shown). Three OSB proteins were detected by these specific antibodies in fractions extracted from protoplasts of Arabidopsis cultured cells (Figure 2D). In the whole cell extract, a single 45-kD protein is detected. This protein is considerably enriched in the chloroplast fraction. In the mitochondrial extract, two other proteins of 48 and 30 kD are detected. The sizes of these three proteins are compatible with the sizes predicted for mature At OSB1 (31 kD), At OSB2 (42 kD), and At OSB3 (49 kD). These results suggest that OSB2 is targeted to chloroplasts, whereas OSB1 and OSB3 are targeted to mitochondria, in agreement with the localization of the corresponding GFP fusion proteins. The OSB3-GFP fusion protein was also detected in chloroplasts, and a faint band that could correspond to OSB3 was also immunodetected in the chloroplast fraction. However, this could result from mitochondrial contamination of the chloroplast fraction. Therefore, the dual targeting of At OSB3 should be confirmed by another approach. Given the limitations of the methods used, we cannot completely exclude dual targeting of a small proportion of other At OSB proteins.
OSB Proteins Preferentially Bind ssDNA
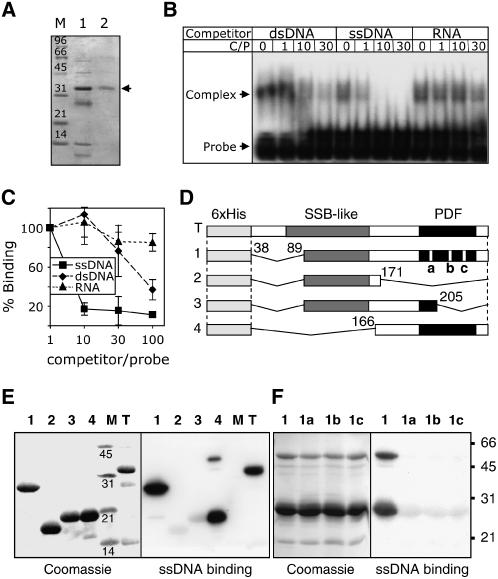
To investigate the nucleic acid binding capacity of Arabidopsis OSB proteins, recombinant OSB1 and OSB2 were expressed and purified (Figure 3A; see Supplemental Figure 1 online). The binding of soluble OSB1 protein to a labeled ssDNA probe was tested by electrophoretic mobility shift assay in the presence of cold competitors (double-stranded DNA [dsDNA], ssDNA, or RNA). To avoid sequence-specific effects and stable secondary structures, ACTG (or ACUG) repeats were used as probes. OSB1 binding was competed for by ssDNA, but it was poorly competed for by dsDNA or RNA (Figure 3B). Because it was difficult to obtain large quantities of soluble OSB1, additional binding assays were done on protein/DNA gel blots using renatured OSB1. The protein was purified under denaturing conditions, transferred to polyvinylidene difluoride membranes, and incubated with labeled ssDNA probe and variable amounts of cold competitor (ssDNA, dsDNA, or RNA). The renatured OSB1 has a much higher affinity for ssDNA than for dsDNA or RNA (Figure 3C), as shown above for the soluble protein. Both OSB1 (Figures 3B and 3C) and OSB2 (see Supplemental Figure 1 online) showed a higher affinity for ssDNA than for dsDNA, and neither bound to RNA. The affinity of OSB2 for ssDNA (Kd = 1.9 ± 0.5 nM; see Supplemental Figure 1 online) is comparable to that found for other high-affinity DNA binding proteins.
Figure 3.
ssDNA Binding Activity of At OSB1.
(A) Nickel-nitrilotriacetic acid agarose affinity chromatography purification of soluble At OSB1 expressed in E. coli. Lane 1, proteins eluting with 50 mM imidazole; lane 2, purified protein eluting with 150 mM imidazole.
(B) Electrophoretic mobility shift assay of nucleic acid binding. At OSB1 and 32P-labeled ssDNA probe were incubated with increasing quantities of cold competitor before electrophoresis. dsDNA, ssDNA, and RNA competitors were of the same size and sequence as the probe. The molar ratio of competitor to probe (C/P) is given. At OSB1/ssDNA complexes are shown by arrowheads.
(C) At OSB1 purified under denaturing conditions was tested on protein/DNA gel blots for binding to labeled ssDNA in the presence of increasing concentrations of cold competitor (ssDNA, dsDNA, or RNA). Results were quantified using a phosphor imager. Error bars indicate the sd of three independent experiments.
(D) Structure of At OSB1 mutants expressed in E. coli. SSB-like, PDF, and deleted regions are indicated. a, b, and c refer to the double substitution mutations described for Figure 2B.
(E) Analysis of ssDNA binding by At OSB1 mutant proteins. Expressed proteins described for (D) were fractionated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and probed with labeled ssDNA.
(F) Same as (E) using construct 1 with mutation a, b, or c.
The PDF Motif of OSB1 Is Involved in ssDNA Binding
The Arabidopsis OSB1 protein has a simple structure consisting of an SSB-like domain followed by a single PDF motif. To define the protein domains involved in ssDNA binding, deletion mutant constructs of OSB1 (Figure 3D) were expressed in E. coli and tested on protein/DNA gel blots for ssDNA binding. The results obtained showed that large portions of the protein (mutants 1 and 4) could be deleted without affecting its ability to bind ssDNA (Figure 3E). However, when the PDF motif was partially deleted (mutants 2 and 3), most of the ssDNA binding capacity was lost. To confirm that the PDF motif is directly involved in ssDNA binding, we constructed three additional mutants (a, b, and c; see Figure 2B) starting from deletion mutant 1 (Figure 3D). In each mutant, two amino acids were chosen among the most conserved residues in the PDF motif and were mutated to radically different residues (Figure 2B). Each of the mutations drastically reduced the ssDNA binding capacity of the protein (Figure 3F), thus suggesting that the PDF motif takes part in DNA binding. Additional bands were often observed by protein/DNA gel blots, corresponding to proteins twice the size of the protein monomers (mutants 1 and 4; Figures 3E and 3F). Because both prokaryotic and eukaryotic ssDNA binding proteins are known to form homomultimers or heteromultimers, it is possible that these bands are dimers that do not dissociate during SDS-PAGE.
At OSB1 Is Expressed in Roots and in Gametes
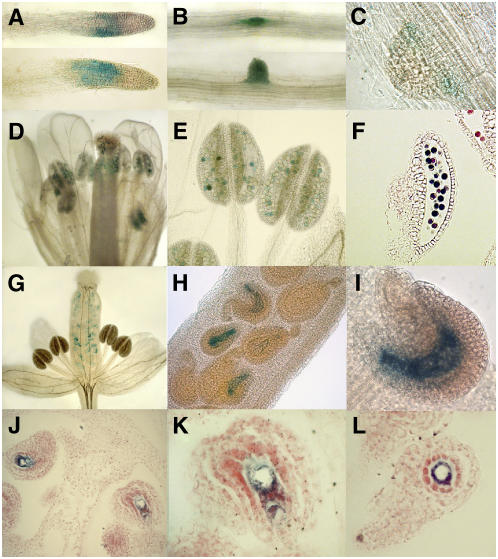
The OSB1 transcript is not detectable in total plant RNA by RNA gel blot hybridization. Microarray results, analyzed with GENEVESTIGATOR (https://www.genevestigator.ethz.ch/at/), showed a higher expression level in cell suspension cultures but did not reveal significant developmental regulation. To investigate expression in different tissues, promoter–β-glucuronidase (GUS) fusions were constructed. The sequence between the start codon of OSB1 and the stop codon of the upstream gene was cloned fused to the GUS gene (ProOSB1:GUS construct) and introduced in Arabidopsis by agroinfection. In young seedlings of six independent ProOSB1:GUS plant lines, gene expression was visible primarily in budding secondary roots (Figures 4B and 4C) but was no longer visible in developed lateral roots. In some plantlets, a short region of the root apex was also stained (Figure 4A), corresponding to the root elongation zone. In mature flowers (stage 14 according to Smyth et al. [1990]), anthers were fully colored by GUS staining of the pollen grains (Figures 4D to 4F). In the same flower (Figure 4E), anthers containing stained pollen were observed (from 0 to 100%), indicating that gene expression was independently induced in each maturing pollen grain. In young flowers (stage 11), strong GUS staining was also visible in the embryo sac of unfertilized ovules (Figures 4G to 4I), where the central cell was clearly stained (Figures 4J to 4L). The expression of At OSB1 thus appears to be temporally and spatially restricted primarily to gametophytic cells. Similarly, promoter-GUS fusions of the At OSB2 and At OSB3 genes showed that the mitochondria-targeted OSB3 protein is expressed primarily in the female gametophyte, but the chloroplast-targeted OSB2 protein is not (data not shown).
Figure 4.
Histochemical Localization of GUS Activity in Arabidopsis Plants Transformed with the Intergenic Region Upstream At OSB1 Gene Fused to the GUS Reporter Gene.
(A) to (C) Roots in an 18-d-old plantlet.
(D) to (F) Expression is detected in pollen grains of mature flowers. Expression in unfertilized ovules is visible in immature flowers.
(G) Whole flower.
(H) and (I) Closer views.
(J) to (L) Sections of unfertilized ovules showing that expression is restricted to the embryo sac.
The expression pattern we detected is consistent with the idea that mitochondrial OSB proteins are ssDNA binding proteins that could play a major role in mtDNA function during gametogenesis.
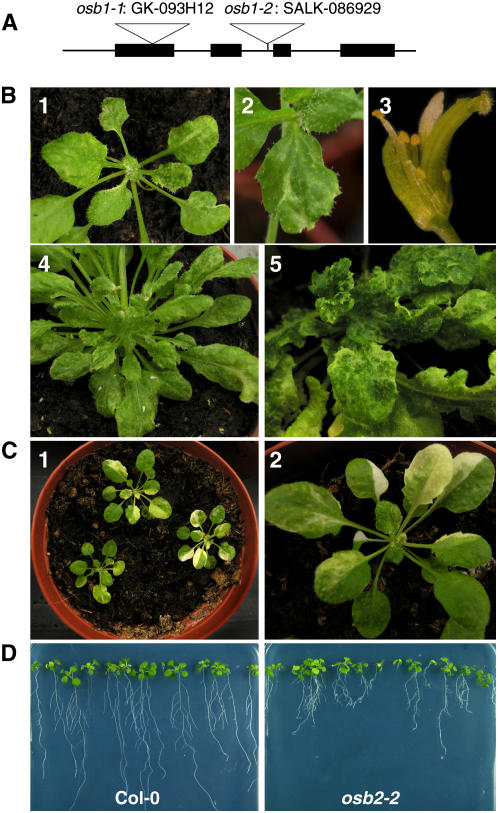
OSB1 T-DNA Insertion Mutants Can Develop Severe Leaf Variegation and Distortion Phenotypes
To identify the possible roles of OSB proteins in organellar genome maintenance, T-DNA insertion mutants were obtained for each of the At OSB1, OSB2, and OSB3 genes. Visible phenotypes were observed only for mutants affected in OSB1 expression. Two mutant alleles in the Columbia (Col-0) background were obtained, osb1-1 (GK-093H12 from GABI-Kat) and osb1-2 (SALK_086929 from the SALK collection). T-DNA insertions were in the first exon (position 173) and in the second intron (position 870) of OSB1, respectively (Figure 5A). In the homozygous lines, no OSB1 mRNA was detected by RT-PCR, suggesting that the insertion lines are authentic knockout OSB1 mutants. DNA gel blot hybridization of osb1-1 plants with T-DNA–specific probes indicated that there is a single T-DNA insertion. No particular phenotype was observed in the T3 homozygous plants that were analyzed. However, at generations T4 and T5, several osb1-1 and osb1-2 plants showed anatomical and/or developmental defects (Figure 5B): variegation, growth retardation, distortion of leaves and flowers, partial sterility, and production of unviable seeds. The severity of the phenotype varied from plant to plant and became more pronounced in later generations. Retarded growth was most evident in the roots. No major differences were observed between osb1 plants and wild-type plants at 7 d after germination. However, after 21 d, root growth was delayed in osb1 plants and the development of secondary roots was abnormal, giving a bushy appearance to the osb1 roots (Figure 5D).
Figure 5.
Variegated and Distorted Phenotypes of At OSB1 T-DNA Insertion Mutants.
(A) Physical map of the At OSB1 gene. The positions of the T-DNA insertions in osb1-1 and osb1-2 mutants are indicated.
(B) Examples of variegation and distortion phenotypes in leaves and flowers of osb1-1 (panels 1, 2, and 4) and osb1-2 (panels 3 and 5) mutants.
(C) Examples of variegation in leaves of an msh1 T-DNA insertion mutant.
(D) Comparison of the roots from Col-0 and osb2-2 plants at 21 d after germination on Murashige and Skoog agar plates.
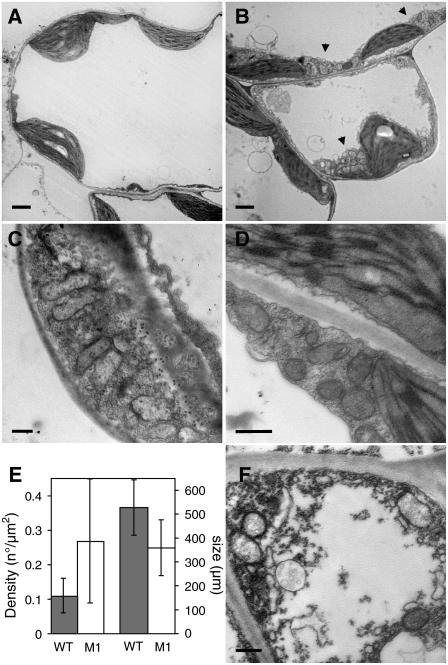
The leaf variegation and distortion phenotypes in osb1 are similar to the phenotypes described for chm mutants, which are affected in the expression of the MSH1 gene. This gene encodes a MutS homolog that is targeted to both mitochondria and chloroplasts and is involved in mtDNA substoichiometric shifting (Martinez-Zapater et al., 1992; Sakamoto et al., 1996; Abdelnoor et al., 2003; Christensen et al., 2005). However, the phenotypes conferred by osb1 and msh1 are not identical, because white sectors (attributable to chloroplast deficiency) frequently observed in msh1 plants (T-DNA insertion line SALK_046763; Figure 5C) have never been observed in osb1-1 or osb1-2 mutant plants. Furthermore, sections of variegated leaf sectors of osb1-1 plants were observed by transmission electron microscopy (Figure 6) and revealed no abnormal chloroplast morphology, unlike msh1 plants (Sakamoto et al., 1996). In most sections observed, mitochondria of mesophyll cells also had normal morphology but were smaller. In some cases, abnormal mesophyll cells were also observed (Figure 6F). Clusters of mitochondria were frequently visible in the mutant (Figures 6B to 6D) but not in control plants (Figure 6A). A statistical analysis of mitochondrion size and number showed that mitochondria were ∼30% smaller in osb1-1. The reduction in mitochondrion size appeared to be counterbalanced by an increase in density, with 0.28 mitochondria/μm2 in osb1-1 compared with 0.11/μm2 in wild-type Col-0 (Figure 6E). Variations in mitochondrion number and size may reflect an increase in mitochondrion division necessary to counterbalance a reduction in the respiratory performance of mutant mitochondria (Barr et al., 2005). Mitochondria of low electron density were also often observed (data not shown), suggesting that, in the affected tissues, there is a heterogeneous population of normal and deficient mitochondria.
Figure 6.
Clustering of Mitochondria in osb1 Mesophyll Cells.
(A) and (B) Transmission electron microscopy of sections from variegated leaves of osb1-1 plants. Mesophyll cells from wild-type (A) and osb1-1 (B) plants, showing clusters of mitochondria in osb1-1 (arrowheads).
(C) and (D) Details of mesophyll cells from osb1-1, showing clusters of mitochondria and the accumulation of mitochondria in regions of cell wall ingrowths.
(E) Increase in the number and decrease in the size of mitochondria in osb1-1. The mean surface of mitochondria cross sections was obtained from the analysis of 20 electron microscopy images of wild-type and osb1-1 (M1) leaves. Error bars indicate sd.
(F) Abnormal mesophyll cell in osb1-1.
Bars = 2 μm in (A) and (B) and 500 nm in (C), (D), and (F).
Arabidopsis OSB1 Plays a Role in Regulating mtDNA Recombination
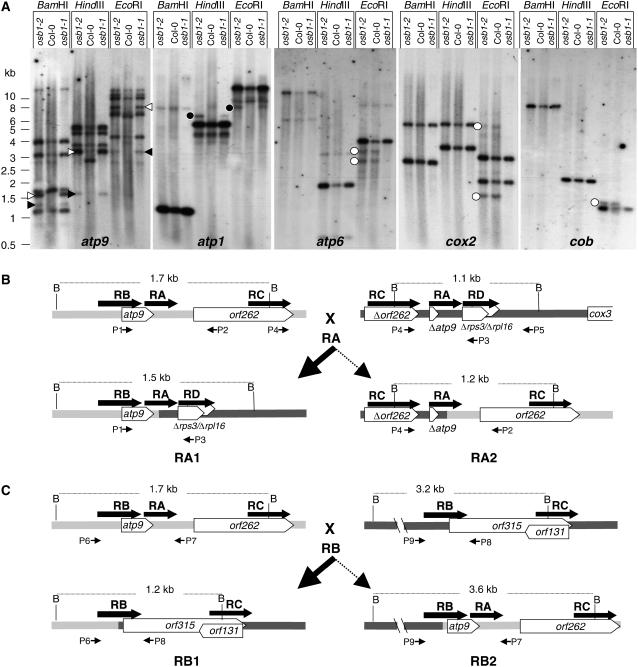
Because OSB1 binds ssDNA and is localized in gametophytic tissues, OSB1 could be involved in mtDNA maintenance. To investigate whether mtDNA structure was altered in osb1 mutants, several osb1 mutant plants were tested by PCR, using primers designed to amplify mtDNA regions of ∼500 nucleotides corresponding to mitochondrial genes and their flanking sequences (see Methods). In mutant plants, several specific pairs of primers failed to amplify the mtDNA in regions surrounding the nad5, ccmFe, rpl5, atp1, cox1, and cox2 genes, whereas these loci could be amplified from Col-0 DNA (see Supplemental Figure 2 online). These results suggested that mtDNA was rearranged or that changes had occurred in gene copy numbers. To test this hypothesis, total DNA from Col-0 wild-type plants and from one plant each of the osb1-1 and osb1-2 mutant lines having morphological phenotypes was analyzed by DNA gel blot hybridization using several mtDNA gene-specific probes. The patterns of mtDNA from mutants differed from wild-type mtDNA around several genes, including the atp9, atp1, atp6, cox2, and cob genes (Figure 7A): stoichiometry was altered, additional fragments were detected, and other fragments were no longer visible. Additional restriction fragments were detected in both osb1-1 and osb1-2 plants (atp9 and atp1 hybridization). However, some minor bands were absent only in the osb1-1 plant (atp6, cox2, and cob hybridization). In the wild-type plants, the level of these fragments was low compared with the gene-containing fragments. These fragments do not correspond to the published mtDNA sequence of Arabidopsis ecotype C24 and could be rearranged mtDNAs that exist as sublimons in wild-type mitochondria. These results suggest that, in the osb1 mutant background, there is segregation of plants with rearranged mtDNA, as a result of enhanced recombination activity and/or changes in the relative copy number of substoichiometric molecules.
Figure 7.
Recombination of mtDNA in osb1 Mutants.
(A) DNA gel blots of total flower DNA (3 μg/lane) from Col-0 and osb1-1 and osb1-2 mutants hybridized with atp9, atp1, atp6, cox2, and cob probes. Open arrowheads, RA1 recombination product; closed arrowheads, RA2 recombination product; closed circles, fragments that appear only in the mutant lines; open circles, fragments that disappear in one of the mutant plants.
(B) and (C) Analysis of the recombination process affecting the atp9 gene locus. RA, RB, RC, and RD indicate repeated sequences. BamHI fragments and their sizes are indicated. Triangles indicate partial gene sequences. P1 to P9 are primers used for PCR amplification.
(B) Recombination mediated by repeat RA present in the atp9 locus and upstream of cox3. The structures predicted for the most abundant heteroduplex (RA1) and the unfavored reciprocal heteroduplex (RA2) are shown. The corresponding fragments are indicated by open arrowheads in the atp9 DNA gel blot.
(C) Recombination mediated by repeat RB present in the atp9 and orf315 mtDNA environments. The structures of the most abundant heteroduplex (RB1) and the unfavored reciprocal heteroduplex (RB2) are shown. The corresponding fragments are indicated by closed arrowheads in the atp9 DNA gel blot.
Substoichiometric Shifting in osb1 Mutants Results from Asymmetric Amplification of HR Products
To investigate the mechanism that leads to substoichiometric shifting in osb1 plants, we focused on the atp9 gene locus, which undergoes substoichiometric shifting in the msh1 and msh1-derived MDL mutants (Sakamoto et al., 1996; Abdelnoor et al., 2003). As shown in Figure 7A, four major BamHI bands hybridize with a 748-bp fragment containing the whole atp9 gene and the 5′ region of orf262 (fragment amplified using primers P1 and P2 [Figure 7B], positions 278,908 to 279,656 of the mtDNA genome [ecotype C24]) (Unseld et al., 1997). The 1.7-, 3.2-, and 4.0-kb fragments contain the atp9 gene locus (Figure 7B), a 334-bp repeat (RB) that overlaps the 5′ end of orf315 (Figure 7B; positions 17,635 to 17,968 of the genome sequence), and a 259-bp repeat containing the 5′ end of exon e of nad5 (positions 142,922 to 143,180), respectively. The expected HindIII and EcoRI fragments were also identified, confirming our interpretation of the hybridization results (Figure 7A).
In Col-0, an additional 1.1-kb BamHI fragment hybridizes to the atp9 probe, which cannot be accounted for by the C24 mtDNA sequence. We mapped this fragment and showed that it differs in the mitochondrial genomes of ecotypes C24 and Col-0. By PCR and sequence analysis, the 1.1-kb BamHI fragment mapped to the region upstream of cox3. Recently, significant differences were reported in the cox3 region of ecotypes C24, Landsberg erecta (Ler), and Col-0 (Forner et al., 2005). In Col-0, upstream of cox3 there are three sequences repeated elsewhere in the mtDNA. We named these sequences repeats RA, RC, and RD (Figure 7B). Repeat RA (248 bp) is constituted of the atp9 3′ coding region and untranslated region sequences (sequence Δatp9 in Figure 7B). RA is the sequence responsible for the hybridization of the 1.1-kb fragment with the atp9 probe. Repeat RC (407 bp) corresponds to the 3′ end of orf262, an open reading frame that maps downstream of atp9 (sequence Δorf262 in Figure 7B). Repeat RD (251 bp) overlaps the rps3 and rpl16 genes (sequence Δrps3/Δrpl16 in Figure 7B), containing the 3′ end of rps3 and the 5′ end of rpl16.
As a result of substoichiometric shifting in the osb1 mutant lines, two additional BamHI fragments of 1.5 and 1.2 kb hybridizing with the atp9 probe (Figure 7A) were amplified. According to our PCR and sequence analysis, the 1.5-kb fragment corresponds to one of the products (RA1) obtained from the reciprocal HR between the two copies of the RA repeat (Figure 7B). As a result, the atp9 gene is relocated upstream of cox3. Similarly, the 1.2-kb fragment is the RB1 product resulting from HR between the two copies of the RB repeat (Figure 7C). In this rearrangement, the atp9-orf262 genes are replaced by the overlapping orf315-orf131 sequences (Figure 7C). Our interpretation of these hybridization results is supported by the results of the HindIII and EcoRI digests (Figure 7A). Fragments of the sizes expected for the RA1 and RB1 recombination products undergo the same changes in relative copy number as the BamHI fragments. The expected HR products RA1 and RB1 were amplified by PCR from the osb1 mutants and sequenced. Interestingly, the reciprocal products RA2 and RB2 resulting from the reciprocal HR processes were not detected by hybridization. This feature is discussed below.
In msh1 mutants, the same BamHI 1.5- and 1.2-kb fragments were associated with substoichiometric shifting in the Col-0 background (Martinez-Zapater et al., 1992; Abdelnoor et al., 2003). However, when an msh1 mutation was backcrossed into a Ler cytoplasm, giving rise to the MDL mutant lines (Sakamoto et al., 1996), different sequences were involved. In Ler, a copy of atp9 maps upstream of cox3. The product of the RA-mediated HR amplified by substoichiometric shifting in Col-0 appears to be the predominant wild-type sequence in Ler (Forner et al., 2005). The substoichiometric shifting events described by Sakamoto et al. apparently resulted from HR involving the pair of repeats RD (Figure 7B), leading to the relocation of atp9 upstream of rpl16 and the concomitant deletion of most of rps3. In the reciprocal recombination product, rps3 is found upstream of cox3 (fragments LMB56 and LMB23, respectively, in Sakamoto et al., 1996). This interpretation is consistent with the sequences reported by those authors and with the sizes of the restriction fragments on DNA gel blots. An important conclusion from this analysis is that substoichiometric shifting in osb1, as well as in the msh1 and msh1-derived MDL mutants, involves the products of reciprocal HR between rather large repeats (>200 bp) and not illegitimate recombination between short repeated sequences, as was assumed previously.
Analysis of Substoichiometric Shifting at the atp9 Gene Locus
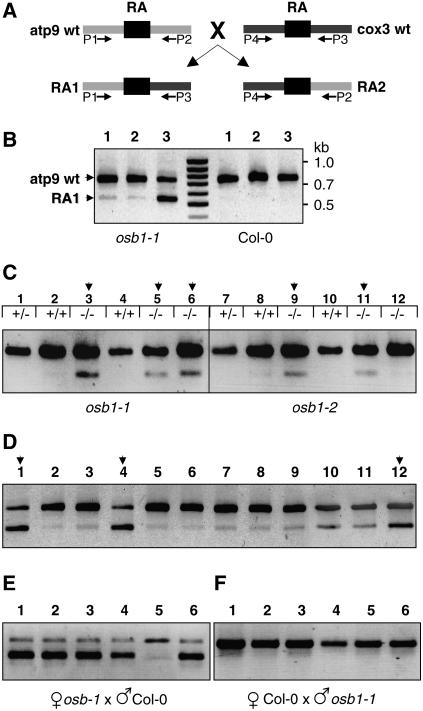
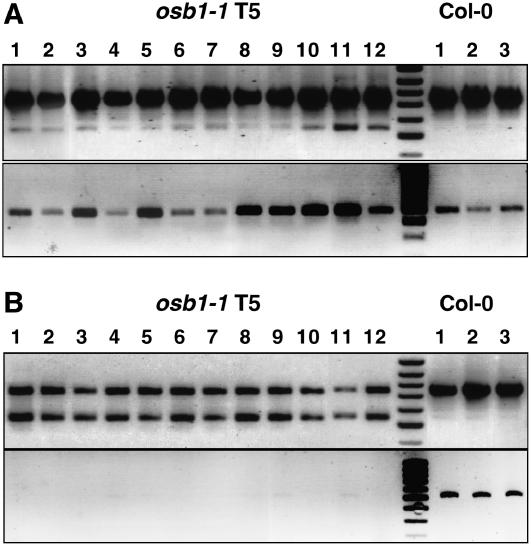
The distinctive morphological phenotypes of leaf distortion and variegation of osb1 mutants were observed only at generations T4 and T5, after selfing of homozygous plants. The substoichiometric shifting events described above at the atp9 gene locus were analyzed in these plants. To determine the effect of the mutation on mtDNA rearrangement, the appearance of recombination products was followed in successive plant generations. We developed a competitive PCR protocol, using three primers (P1 + P2 + P3; Figures 7B and 8A), to determine the proportion of the wild-type atp9 sequence and the RA1 recombination product. A similar competitive PCR method was used to analyze substoichiometric shifting in msh1 mutants (Sakamoto et al., 1996; Abdelnoor et al., 2003). It must be emphasized that dramatic differences in amplification of the two fragments were obtained with small differences in the primer ratio. Under these conditions, the osb1 RA1 sequence was amplified but no product was detected in the wild type (Figure 8B). The RA1 sequence was also amplified in the msh1 T-DNA insertion mutant using this technique (data not shown). The profiles obtained with osb1 homozygous mutant DNA (Figure 8B) show that the proportion of the atp9 wild-type (748 nucleotides) and RA1 (546 nucleotides) products varied according to the plant. In all Col-0 plants, only the atp9 sequence was detected. However, RA1 and RA2 sequences were present at low levels in wild-type plants, because both could be detected by a conventional two-primer PCR (Figure 9).
Figure 8.
Accumulation of the RA1 Recombination Product in osb1 Mutants Affected by mtDNA Substoichiometric Shifting.
(A) Simplified scheme showing the RA repeats in wild-type Col-0 atp9 and cox3 contexts and the resulting recombination products RA1 and RA2. Primers P1 to P4 (Figure 7B) were used to amplify the four mtDNA configurations.
(B) Analysis by optimized competitive, three-primer PCR of three osb1-1 plants affected by shifting and of three Col-0 plants. The top band (P1 + P2) corresponds to wild-type atp9 (748 nucleotides), and the smaller fragment (P1 + P3) corresponds to the RA1 recombination product (546 nucleotides).
(C) Analysis of RA1 accumulation in the first generation of osb1 homozygous mutants. The OSB1 genotype of each plant is indicated. Homozygous plants 3, 5, 9, and 11 have increased levels of RA1 (stage I shift; as described in Results and in Supplemental Table 2 online).
(D) Progeny (generation T3) of homozygous plant 3 from (C): plants 1, 4, and 12 show much higher levels of RA1 than their siblings (stage II shift).
(E) Analysis of plants resulting from the backcrossing, as pollen receptor, of plant 4 from (D).
(F) Analysis of plants resulting from the reciprocal cross.
Figure 9.
Substoichiometric Shifting Is Accompanied by the Loss of Reciprocal Recombination Products.
(A) Detection of the reciprocal recombination product RA2 (bottom panel) in the progeny of a T4 plant that shows partial mtDNA shifting (stage I; top panel).
(B) The same as (A), in the progeny of a plant that has a complete mtDNA shift (stage II).
Plants issued from mixed seeds of the osb1-1 insertion mutant obtained from GABI-Kat (line GK-093H12, generation T2) were genotyped and tested (Figure 8C). Fourteen of the 20 plants analyzed were either wild type (Col-0) or heterozygous for the T-DNA insertion. In the heterozygous plants, the atp9 wild-type gene locus was amplified predominantly, whereas the recombination product RA1 was barely visible (Figure 8C; see Supplemental Table 2 online). In Supplemental Table 2 online, this PCR pattern is referred to as shifting stage 0 (plants 1, 2, and 4 in Figure 8C). In pattern stage I, the amount of RA1 is higher, but it is still at a substoichiometric level (see Supplemental Table 2A online). The stage I pattern was found for three of the six homozygous mutant plants analyzed (plants 3, 5, and 6 in Figure 8C). Similar results were obtained for plants issued from the self-fertilization of a T3 osb1-2 plant that is heterozygous for the T-DNA insertion (line SALK_086929). Two of the three homozygous plants analyzed (plants 9 and 11 in Figure 8C) showed a significant increase in the RA1 copy number (see Supplemental Table 2A online). Because osb1-1 and osb1-2 mutants are both in the Col-0 background, the molecular phenotype is not specific to the plant line used for T-DNA insertion mutagenesis. From these results, we can conclude that the loss of OSB1 function led to an increase in the copy number of mtDNA molecules that arose from HR. This effect was already detected in the first generation of the mutants.
In Arabidopsis, the published mtDNA sequence of ecotype C24 contains >20 pairs of repeats of >100 bp. Only the two largest repeats (6.5 and 4.2 kb) are described as contributing to the multipartite structure of the genome (Unseld et al., 1997). Therefore, we tested whether recombination events could be mediated by short repeats of several hundred base pairs. A 435-bp sequence situated in exon 1 of the cox2 gene is repeated elsewhere in the genome (at positions 319,174 to 319,608 of the published sequence). A 556-bp sequence covering the first exon of rps3 and the tRNA-Lys gene is repeated at positions 204,102 to 204,657 in the genome sequence. By PCR, we found an increase in the copy number of sequences that result from reciprocal HR between these repeats (data not shown). As for the RA1 sequence, these differences were observed in the first generation of osb1 mutants. Hence, the increase in HR products in osb1 mutants is not restricted to the atp9 locus. It is likely that many HR products that are present in wild-type mitochondria at sublimon levels accumulate in osb1. The increased pool of HR-derived sequences could then trigger additional HR events.
The Level of Recombination Products Increases with Successive Generations
The progeny of the five homozygous osb1 plants described above were analyzed. The results shown in Supplemental Table 2B online demonstrate that the majority of the plants (138 of 194), like their parents, had higher levels of RA1 (stage I). However, 32 plants seemed to revert to wild type stage 0 RA1 levels. Interestingly, several (24) had a new pattern (stage II of stoichiometric shifting) characterized by a level of RA1 PCR product equal to or greater than the level of the PCR product corresponding to unrecombined DNA (plants 1 and 4 in Figure 8D). The segregation of shifted plants differed in the mutant lines: one-quarter of the progeny of homozygous plant 3 was affected, but none of the progeny of homozygous plant 9 showed shifting (see Supplemental Table 2B online). At generation T3, the shifted plants had no visible morphological phenotype.
After Complete Stoichiometric Shifting, Reversion to the Wild Type Is No Longer Possible
We tested whether the molecular phenotype of shifted mtDNA can be reverted by reintroduction of the wild-type OSB1 allele. Individual T3 osb1-1 plants with stage I and stage II mtDNA were selfed and backcrossed with wild-type Col-0. The progeny were then tested for mtDNA shifting by competitive PCR, as described above. Fifteen T4 plants obtained by selfing a stage I T3 plant were analyzed. Nine maintained the same molecular profile, whereas three reverted to the wild-type stage 0 pattern and three others shifted to stage II (see Supplemental Table 2C online). The backcross with wild-type plants revealed that, at generation T3, the shifting process can be reverted: of the 15 F1 daughter plants tested, 13 reverted to stage 0 wild-type patterns and only 1 plant evolved to stage II. As expected, all of the plants analyzed from the reciprocal cross displayed a wild-type profile.
When the same experiment was done with a plant of the same generation (T3) with a stage II shifted pattern, no revertants were found among the 15 T4 plants analyzed (see Supplemental Table 2D online). From the backcross with wild-type Col-0, of 21 F1 daughter plants, 11 conserved the stage II shifted pattern, whereas the others reverted to the wild-type or stage I pattern (Figure 8E). However, the analysis of progeny of the reciprocal cross showed that pollen of the same plant was unable to induce the stage I or stage II pattern in the F1 hybrids obtained from crossing with wild-type plants (Figure 8F). These results establish the non-Mendelian inheritance of the shifted phenotype.
In subsequent generations, reversion from the shifted phenotype was no longer detectable. This was clearly illustrated by results obtained by selfing or backcrossing T4 osb-1 plants with shifted stage II patterns, severe morphological distortion, and reduced fertility. As shown in Supplemental Table 2D online, the T5 plants obtained by selfing had the shifted stage II pattern (except for one plant that had the shifted stage I pattern). Most of these plants had distorted, variegated leaves. From the backcross with Col-0 (see Supplemental Table 2D online), the majority of the plants (15 of 18 analyzed) maintained shifted mtDNA. One plant had wild-type stage 0 mtDNA. However, further PCR analysis of this plant showed that its mtDNA had undergone numerous recombination events that could be responsible for its spectacular growth delay, leaf distortion, and early death. The reciprocal cross with Col-0 gave F1 plants with wild-type mtDNA, as expected from the maternal inheritance of the phenomenon.
Together, these results suggest that, above a threshold level of alternative structures obtained by infrequent HR, reversion to the original mtDNA structure is infrequent and independent of the OSB1 allele.
Shifting of the mtDNA from Stage I to Stage II Is Accompanied by the Loss of Reciprocal Recombination Products
As shown above (Figure 7), osb1 mutants have clear morphological phenotypes, and recombination products expected from reciprocal HR (RA2 and RB2; Figures 7B and 7C) are not detectable by DNA gel blot analysis. Therefore, we tested for the presence of these molecules using conventional two-primer PCR. Primers P2 + P4 (Figure 7B) were used to specifically amplify reciprocal recombination product RA2. As clearly shown in Figure 9A, plants in the first stage of mtDNA shifting (stage I, as shown by the level of RA1 in the top panel) contained levels of RA2 (bottom panel) comparable to those of wild-type Col-0. The relative copy number of RA2 was even higher in a few T2, T4, and T5 osb1 plants analyzed (data not shown). On the other hand, the RA2 product was absent, or detected very faintly, in all plants from subsequent generations with a shifted stage II pattern (Figure 9B), even after >35 PCR cycles. Similarly, the recombination product RB2 was not detectable in these plants after amplification using primers P8 + P9 (data not shown). Thus, the transition from partial and reversible mtDNA shifting to irreversible shifting correlates with the loss of reciprocal recombination products. Whether it is a cause or a consequence of the transition is discussed below.
DISCUSSION
The OSB Family Is a Novel Plant-Specific Protein Family
We have shown that the Arabidopsis OSB genes constitute a small family of organelle-targeted proteins. An OSB protein was initially identified in potato by ssDNA-affinity chromatography (Vermel et al., 2002). Putative OSB sequences were also detected in the O. sativa and Z. mays genomes, but no OSB sequences were found in nonplant genomes, suggesting that either the OSB proteins evolved to fulfill plant organelle-specific functions or that in nonplant species the same function is accomplished by other proteins.
We showed that GFP fusion proteins are targeted to mitochondria (At OSB1), to chloroplasts (At OSB2), or to both (At OSB3). From our results, we cannot exclude the possibility that a small proportion of At OSB1 and At OSB2 proteins could be dual-targeted. However, it is unlikely that At OSB1 is dual-targeted, because a protein of the corresponding size was detected only in the mitochondrial protein extract. In addition, osb1 mutants do not show the phenotypes associated with chloroplast deficiency observed in msh1 plants, supporting the idea that At OSB1 is targeted only to mitochondria. Although the dual targeting of At OSB3 should be confirmed by an additional approach, it is interesting that dual targeting is also observed for other proteins involved in organellar gene expression, such as aminoacyl-tRNA synthetases, RNA polymerases, DNA gyrases, and DNA polymerases (Hedtke et al., 2000; Wall et al., 2004; Christensen et al., 2005; Duchene et al., 2005). The redundancy of specifically targeted and dual-targeted proteins could reflect a high degree of flexibility in the regulation of organellar gene expression by the nucleus.
Evolution of OSB Proteins as ssDNA Binding Proteins
The OSB proteins are characterized by the presence of PDF motifs. Our results suggest that, at least for At OSB1, the PDF motif is required for the protein–DNA interaction. It is surprising that the intermediate region of At OSB1 (Figure 2), which is clearly related to the SSB protein family (30 to 50% similarity), is apparently not required for ssDNA binding. However, none of the residues, including Trp and Phe, required for high-affinity binding to ssDNA of E. coli SSB are conserved in the SSB-like domains of OSB proteins (Raghunathan et al., 1997). It is also possible that the protein/DNA gel blot analysis underestimates the influence of this domain if, for instance, the SSB domain is involved in protein oligomerization. Nevertheless, the PDF can bind ssDNA and thus constitutes a new ssDNA binding motif. We have shown that At OSB1 and At OSB2 have a preference for ssDNA, but we cannot exclude the possibility that other members of the family evolved to have different binding specificities. Multiple PDF motifs in different OSB proteins could lead to differences in their affinity for nucleic acids.
The phylogenetic origin of the PDF motif remains unknown. Apart from plant OSB proteins, this motif was found only in the RB38 protein of Chlamydomonas. RB38 binds the 5′ untranslated region of the chloroplast psbA mRNA (Barnes et al., 2004) and is composed of a chloroplast-targeting peptide directly followed by four PDF motifs responsible for RNA binding. The apparent different substrate specificities of At OSB1 (ssDNA) and RB38 (RNA) could be attributable to differences in the positions of the predicted α-helixes and β-sheets within the PDF motifs, to nonconserved amino acids, and to sequences surrounding the motif. We can speculate that the PDF motif evolved primarily as an RNA binding domain and that OSB proteins originated in higher plants by the fusion of a bacterium-type SSB and a PDF-containing protein. In this context, the PDF domain could have evolved to preferentially bind ssDNA.
OSB proteins have the common structure of an SSB-like domain followed by a PDF motif(s). Therefore, it is possible that they have overlapping functions, which could explain the absence of visible phenotypes of At OSB2 and At OSB3 T-DNA insertion mutants. In the case of At OSB1, our results show that its deficiency results in mtDNA instability, leading to an unbalanced transmission of alternative mtDNA configurations.
Accumulation of mtDNA Recombination Products in osb1 Plants
In osb1 mutant lines, changes in mtDNA profiles apparently result from reciprocal HR between small repeats, such as the RA and RB repeats, which are a few hundred nucleotides long. In wild-type plants, recombination mediated by these repeats seems infrequent, because the corresponding products usually are present at very low levels and can be detected only by PCR. The very strong HR activity, thought to be responsible for the multipartite structure of plant mitochondrial genomes, involves large repeats of several kilobases in length (Unseld et al., 1997).
The hot spot of HR that we followed as a marker of mtDNA substoichiometric shifting concerns the RA repeat, consisting of atp9 gene sequences. RA-mediated recombination is a good marker for recombination deregulation in Arabidopsis Col-0: the RA1 recombination product is amplified in osb1 and msh1 mutants and also in cell suspension cultures (Forner et al., 2005). In other ecotypes, such as Ler, the RA1 recombination product is the predominant mtDNA configuration. The repeats surrounding the atp9 gene are prone to recombination, which could be attributable to the very high transcription activity associated with the atp9 gene locus. Local melting of the DNA double strand could favor invasion by homologous sequences. However, the RA1 recombination product is not linked to the morphological phenotypes of leaf variegation and distortion, because the phenotype severity is not correlated with the ratio between wild-type and RA recombination products. Systematic analysis by PCR of mtDNA sequences surrounding expressed genes detected changes affecting several mtDNA loci in osb1 mutants. These changes probably result from additional recombination events and suggest that inactivation of OSB1 affects the maintenance of the mtDNA genome as a whole. Multiple mtDNA configurations probably segregate from the same original mutant line, explaining the variegated phenotype of the most affected osb1 plants. A comprehensive analysis of the mtDNA substoichiometric shifting process induced by the osb1 or msh1 mutations would require detailed mapping and sequencing of the different mtDNA genomes for several lines segregating from the same mutant plant.
mtDNA Shifting in osb1 Lines Is a Two-Step Process
Repression of HR is likely to be essential in ensuring mtDNA stability. The loss of OSB1 appears to affect nuclear control, which represses HR of the mtDNA. Although our analysis was restricted to the recombination events involving the atp9 locus, several conclusions can be made concerning the process of mtDNA shifting. (1) In the first mutant generation, there is already a patent increase in the accumulation of HR products. (2) In the next generation, individual lines segregate in which certain rearranged mtDNA configurations obtained by HR are preferentially replicated, compared with the parental sequences (what we called stage II). This process is accompanied by an almost complete disappearance of the reciprocal recombination product, suggesting that this product is no longer produced by reversible reciprocal HR. (3) In the first stages of mtDNA shifting (stage I), the process is reversible if OSB1 activity is restored. This is probably attributable to the low copy number of HR products, which are segregated out in the subsequent generations, if their continuous production by HR is repressed. However, after stage II, reversion to wild-type mtDNA no longer occurs, even in the presence of the osb1 allele. It is possible that the recombination product is now the major mtDNA sequence and can no longer be segregated out during gametogenesis.
The shift from stage I to stage II could result either from the stochastic segregation of the parental mtDNA molecules and HR products or from additional recombination events, leading to the preferential replication of the mtDNA molecules containing recombined sequences. The accumulation of HR products could contribute to the pool of sequences that prime the asymmetric replication of mtDNA chimeras by strand invasion. It is reasonable to assume that, after the stage II mtDNA shift, the accumulation of the predominant recombination product no longer depends on continuous production by HR. This could explain why reciprocal recombination products are no longer detected: if they are not replication-competent, their production in somatic tissues by HR requires the presence of the wild-type parental sequences. Because the wild-type sequences are minor in the shifted mtDNA, they should preferentially undergo HR with the new predominant mtDNA sequence, a process that can no longer generate the reciprocal recombination products.
In cybrid rapeseed (Brassica napus) plants, changes in mtDNA stoichiometry are associated with the selective amplification of low copy number molecules produced by recombination (Bellaoui et al., 1998). As for osb1 mutants, this process involves the amplification of only one of the recombination products, whereas the reciprocal sequence remains at low levels. Therefore, it seems that different causes (cybrid production and the osb1 mutation) can result in a similar mechanism of mtDNA substoichiometric shifting.
A Possible Role of OSB1 as a Repressor of HR Required for mtDNA Genome Stability
ssDNA binding proteins play a central role in HR, both as enhancers (by holding DNA strands open and melting ssDNA secondary structures that block recombinases) and as suppressors (by outcompeting the initial binding of recombinases to ssDNA). Recombination mediators, such as bacterial RecO and eukaryotic Rad52, overcome ssDNA binding protein inhibition by promoting the assembly of recombinases on ssDNA (Gasior et al., 2001). Bacterium-type SSB proteins also exist in plant mitochondria. Like E. coli SSB, plant mitochondria SSB promotes the RecA-dependent strand invasion of dsDNA by homologous ssDNA (Edmondson et al., 2005). At OSB1 is an ssDNA binding protein with no apparent sequence specificity, but its function is apparently not redundant with that of bacterium-type SSB, which is essential for plant viability (our unpublished data). At OSB1 could function as a suppressor of HR, a function that could explain its role in the mechanism of copy number suppression of subgenomes generated by HR during mtDNA replication.
Such a putative function is in agreement with the preferential expression of OSB1 in gametophytic tissues, postulated to be the locus where the stoichiometry of the alternative genomes is determined. Models of how mtDNA is maintained with minimal heteroplasmy, both in animals and in plants, suggest that mitochondria pass through a stringent genetic bottleneck in transmitting tissues (Shoubridge, 2000; Arrieta-Montiel et al., 2001). Thus, genes that directly affect the structure of the transmitted mtDNA are expected to be preferentially expressed in those tissues. According to our model, to replicate a functional, high copy number mitochondrial genome, the role of At OSB1 in meristematic and gametophytic cells is to reduce the mitochondrial genome complexity resulting from the production of aberrant subgenomes generated by infrequent recombination events between small repeats. In somatic tissues, recombination control is relaxed, and the ratio of normal to subgenomes is dictated by their inherent replication efficiency. In the absence of At OSB1, the predominance of these subgenomes during mtDNA replication in transmitting cells results in the preferential replication of selfish subgenomes and in the nonreversible imbalance observed in shifted plants.
We have identified At OSB1 as a component of the system regulating mtDNA stoichiometry. Our results suggest that At OSB1 represses the production of recombination products considered illegitimate and thus takes part in the nuclear control that prevents mtDNA instability. Determining the roles of proteins such as OSB1 in plant development will shed light on the mechanism of substoichiometric shifting in the mitochondrial genome and its contribution to plant evolution.
METHODS
Identification, Purification, and Cloning of OSB Genes
For protein/DNA gel blot analysis of soluble mitochondrial proteins, potato (Solanum tuberosum) mitochondria were purified as described (Neuburger et al., 1982) and lysed by freeze-thaw (three cycles). Soluble proteins (3 mg) were recovered by centrifugation, adjusted to 250 mM NaCl, and run through a 1.5-mL DEAE anion-exchange column (DE52; Whatmann) to remove contaminating nucleic acids. The eluted proteins were fractionated by SDS-PAGE (8 μg/lane) and probed for ssDNA binding with an oligonucleotide containing a 25-nucleotide random sequence (5′-GGCCACTAGTCGGATCCC[N]25GGGCTGCAGGAATTCGACG-3′). The St OSB protein was purified by ssDNA-affinity purification as described previously (Vermel et al., 2002), starting from 200 mg of potato mitochondrial protein. The purified protein was excised from the gel, and tryptic peptides were N-terminal sequenced on an Applied Biosystems 473A apparatus. From two peptide sequences (Pep1, 5′-IEIYDEAEDVSSWPKPSEI-3′; Pep2, 5′-NFLLDENDNQHSDY-3′), two sets of degenerated primers were derived that allowed RT-PCR amplification of the St OSB1 cDNA from total potato germ RNA. The sequence of the 450-bp amplified fragment was used to derive new primers and amplify the complete St OSB1 cDNA by 5′ and 3′ rapid amplification of cDNA ends (5′ RACE system; Invitrogen).
At OSB cDNAs were amplified from total Arabidopsis thaliana cDNA primed with oligo(dT) using primers complementary to the 5′ and 3′ untranslated region sequences of genes At1g47720, At4g20010, and At5g44785. For in vivo intracellular localization, the sequences encoding the first 43, 85, and 68 amino acids of At OSB1, At OSB2, and At OSB3, respectively, were cloned in plasmid pCK-GFP3A to express protein-eGFP fusions under the control of a 35S promoter (Menand et al., 1998). The resulting plasmids were transfected in Nicotiana benthamiana leaves by bombardment (Sanford et al., 1993), and images were obtained at 24 h after transfection with a Zeiss LSM510 confocal microscope. The pCK-mRFP plasmid (Vermel et al., 2002) was cotransfected as an internal mitochondrial marker. Chloroplasts were visualized by the natural fluorescence of chlorophyll. Organelle-targeting predictions were determined with the programs Predotar, TargetP, and iPsort (genoplante-info.infobiogen.fr/tools/predotar/; www.cbs.dtu.dk/services/TargetP/; and biocaml.org/ipsort/iPSORT/, respectively).
Plants, DNA, and RNA
Arabidopsis plants, ecotype Col-0, were transformed by the floral dip method (Clough and Bent, 1998). T1 and T2 plants were sown on agar plates containing Murashige and Skoog salts, 0.5% sucrose, and 50 μg/mL kanamycin. After 2 to 3 weeks, kanamycin-resistant plants were transferred to soil and grown in the greenhouse. T-DNA insertion mutant lines were obtained from the GABI-Kat and SALK collections. Insertion sites were determined by PCR using gene- and T-DNA–specific primers and mapped to positions 17,565,117 to 17,565,126 (in Arabidopsis chromosome 1) with a 9-nucleotide deletion for mutant GK-93H12 and to positions 175,645,593 to 175,645,568 with a 25-nucleotide deletion for mutant SALK-086929, where insertion is composed of at least two T-DNA molecules head to head. Mutant seeds were genotyped by PCR, and homozygote plants were self-propagated. Plants were grown at 23°C under a 16-h-light/8-h-dark photoperiod. Genomic DNA was extracted with DNAzol (Invitrogen) and total RNA was extracted with TRIzol (Invitrogen) as described (Chomczynski and Sacchi, 1987).
Protein Expression
The At OSB1 cDNA sequence encoding the predicted mature protein (between amino acids 38 and 261) was cloned between HindIII and BamHI sites in expression vector pRSETc (Invitrogen) (construct T in Figure 3). Deletion mutant constructs 1 and 4 (Figure 3D) were obtained by PCR from construct T, using primers taken at the borders of the deleted region (residues 38 to 89 for construct 1 and residues 38 to 166 for construct 4) followed by clone religation. Similar deletion constructs 2 and 3 were obtained starting from construct 1 and resulting in the removal of residues 171 to 262 (construct 2) and residues 205 to 261 (construct 3). Recombinant proteins were expressed in Escherichia coli strain BL21(DE3) (Novagen). The cells were grown at 37°C in Luria-Bertani medium containing carbenicillin (50 μg/mL) and 0.2% glucose up to A600 of 0.6, and protein expression was then induced with 1 mM isopropyl β-d-thiogalactopyranoside. After 2 h at 37°C, cells were pelleted and vortexed in the presence of buffer A (10 mM Tris-HCl, pH 8.0, 100 mM NaH2PO4, and 8 M urea). Debris were removed by centrifugation, and the soluble fraction was incubated with nickel-nitrilotriacetic acid agarose beads (Qiagen) for 2 h at room temperature. After washing with buffer A containing 50 mM imidazole, the expressed proteins were eluted with 150 mM imidazole.
The At OSB2 cDNA sequence encoding the predicted mature protein (between amino acids 69 and 372) was cloned in the pBin+ vector (van Engelen et al., 1995) fused to a C-terminal tag sequence comprising the FLAG peptide and the calmodulin binding peptide under the control of the cauliflower mosaic virus 35S promoter and terminator sequences, as described (Perrin et al., 2004). These constructs were used to express the protein in N. benthamiana 3-week-old leaves by agroinfiltration, as described (Voinnet et al., 2003). After 4 d, infiltrated leaves were ground in liquid nitrogen and the tagged protein was purified by tandem affinity purification, as described previously (Perrin et al., 2004).
Antibody Production
Purified At OSB1 (amino acids 38 to 261) produced in E. coli using vector pRSETc was transferred to nitrocellulose membranes after SDS-PAGE. The protein was stained with Ponceau red, cut, dried for 12 h under vacuum, and dissolved in DMSO. A rabbit antiserum was prepared, and the antibodies were purified against the protein antigen immobilized on CNBr-activated Sepharose. For protein gel blot analysis, the purified antibodies were used at a concentration of 1:5000.
DNA Binding Assays
Electrophoretic mobility shift assays were performed as described in Promega Technical Bulletin 110. Briefly, purified protein (1 to 10 ng) and 5′ end radiolabeled oligonucleotide (0.1 pmol) purified on polyacrylamide gels were incubated for 20 min on ice in 10% glycerol, 30 mM Tris-HCl, pH 8.0, 3 mM β-mercaptoethanol, 10 mM MgCl2, 100 μg/mL BSA, 50 mM NaCl, the appropriate concentration of competitor, and a mixture of protease inhibitors (Complete-EDTA; Roche Applied Science). DNA–protein complexes were resolved on 8% polyacrylamide gels in 1× Tris-Borate-EDTA buffer at 4°C. The gels were then vacuum-dried and revealed using a BAS 1000 phosphor imager (Fujix). Quantification of the signals was done with MacBas version 2.2 software (Fuji Photo Film). Routinely, the (ACTG)x8 32-mer oligonucleotide was used as an ssDNA probe. The dsDNA probe was prepared by annealing with the complementary oligonucleotide, and the RNA probe was prepared by in vitro transcription using a primer of the same sequence containing a 5′ T7 promoter sequence. All probes were gel-purified before use. Protein/DNA gel blot analysis was done as described by Ghosh et al. (1994) for protein/RNA gel blot analysis. Briefly, purified proteins were fractionated by SDS-PAGE, transferred to polyvinylidene difluoride membranes (Immobilon P; Millipore), and renatured by four 30-min incubations at 4°C in 100 mM Tris-HCl, pH 7.5, 0.1% Nonidet P-40, and 100 mM NaCl. The membrane was blocked for 10 min at room temperature in binding buffer (10 mM Tris-HCl, pH 7.5, 5 mM Mg-acetate, 2 mM DTT, 50 mM NaCl, and 0.01% Triton X-100) containing 5% BSA, and then incubated for 10 min at room temperature in binding buffer added with 100 μg of poly(dCdI). Radiolabeled probe was added, and after 2 h of incubation, membranes were washed four times for 10 min each in binding buffer before autoradiography.
Promoter-GUS Fusion Analysis
The 5′ intergenic region of At OSB1 (1074 nucleotides between the stop codon of the upstream At1g47730 gene and the initiation codon of At1g47720) was cloned in the binary vector pBI101.2 (Clontech) upstream of the GUS gene. Transgenic Arabidopsis plants were produced, and tissues from promoter-GUS fusion plants issued from six independent lines were stained with 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (Biosynth) as described (Jefferson et al., 1987). For observation of thin sections, tissues were fixed in 3.7% formaldehyde, 50% ethanol, and 5% acetic acid, stained using eosine (0.002% final concentration), and embedded in Paraplast wax using conventional techniques.
Transmission Electron Microscopy
Leaf tissue samples were fixed overnight in 3% glutaraldehyde, then treated for 2 h with 10% (w/v) picric acid and for 2 h with 2% uranyl acetate, and stained with 0.1% (v/v) osmium tetroxide in 150 mM phosphate buffer, pH 7.2. Samples were dehydrated through an ethanol series and infiltrated with EPON812 medium-grade resin (Polysciences). Polymerization was done for 48 h at 60°C. Ultrathin sections (90 μm) were cut using an Ultracut E microtome (Reichert) and collected on grids coated with formvar (Electron Microscopy Sciences). Samples were visualized with a Hitachi H-600 electron microscope operating at 75 kV. The mitochondrial surface was measured with ImageJ software (http://rsb.info.nih.gov/ij/).
PCR Screening of mtDNA Structural Modifications
The DNA integrity of the Arabidopsis mitochondrial genome was checked by PCR using the primer collection described by C. Andrés (unpublished data; see Supplemental Table 1 online). PCR was performed on 384-well plates in a final volume of 10 μL using Taq DNA polymerase (M0267S; New England Biolabs). PCR conditions were as described in the product manual except that primer concentrations were 0.1 μM and that Cresol Red (0.2 mg/mL) and sucrose (60 mg/mL) were added. Genomic DNA concentration was 25 to 75 ng per reaction. The Thermocycler (Bio-Rad) was set with the following program: 5 min at 94°C, then 35 cycles of 20 s at 94°C, 20 s at 52°C, and 60 s at 72°C, and a final step of 5 min at 72°C. PCR products were analyzed on 2.0% agarose gels and visualized by ethidium bromide staining under UV light. Gel images of wild-type and mutant samples were processed using Genetools software (Syngene).
Accession Numbers
Sequence data for the genes mentioned in this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AY942642 (St OSB1), At1g47720 (At OSB1), At4g20010 (At OSB2), At5g44785 (At OSB3), At1g31010 (At OSB4), At3g24320 (MSH1), At4g11060 (mitochondrial SSB), and NC_001284 (Arabidopsis ecotype C24 mitochondrion complete genome).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Methods. Analysis of At OSB2 Binding Activities.
Supplemental Figure 1. ssDNA Binding Activity of At OSB2.
Supplemental Figure 2. PCR Analysis of Col-0 and osb1 Mutant mtDNA.
Supplemental Table 1. Mitochondrial Genomic Positions of PCR Products.
Supplemental Table 2. Analysis of Shifting in the Progeny of osb1 Plants.
Supplemental Table 3. Oligonucleotides Used.
Supplementary Material
Acknowledgments
We thank Philippe Hammann, Malek Alioua, and Leo Baettig for technical assistance and the gardeners of the Institut de Biologie Moléculaire des Plantes for excellent plant care. We are grateful to Dominique Gagliardi and Philippe Giege for helpful discussions and critical reading of the manuscript. We also thank the ABRC, GABI-Kat, and the Salk Institute Genomic Analysis Laboratory for providing the T-DNA insertion mutant lines. V.Z. was supported by a PhD fellowship of the French government. This work was supported by the Centre National de la Recherche Scientifique and by the French plant genomic program Génoplante.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: José M. Gualberto (jose.gualberto@ibmp-ulp.u-strasbg.fr) and Patrice Imbault (patrice.imbault@ibmp-ulp.u-strasbg.fr).
Online version contains Web-only data.
References
- Abdelnoor, R.V., Yule, R., Elo, A., Christensen, A.C., Meyer-Gauen, G., and Mackenzie, S.A. (2003). Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc. Natl. Acad. Sci. USA 100 5968–5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, K.L., and Daley, D. (2004). Plant mitochondrial genome evolution and gene transfer to the nucleus. In Plant Mitochondria: From Genome to Function, D.A. Day, A.H. Millar, and J. Whelan, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 107–120.
- Arrieta-Montiel, M., Lyznik, A., Woloszynska, M., Janska, H., Tohme, J., and Mackenzie, S. (2001). Tracing evolutionary and developmental implications of mitochondrial stoichiometric shifting in the common bean. Genetics 158 851–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert, S., and Börner, T. (2000). Phage T4-like intermediates of DNA replication and recombination in the mitochondria of the higher plant Chenopodium album (L.). Curr. Genet. 37 304–314. [DOI] [PubMed] [Google Scholar]
- Backert, S., Nielsen, B.L., and Börner, T. (1997). The mystery of the rings: Structure and replication of mitochondrial genomes from higher plants. Trends Plant Sci. 2 477–483. [Google Scholar]
- Barnes, D., Cohen, A., Bruick, R.K., Kantardjieff, K., Fowler, S., Efuet, E., and Mayfield, S.P. (2004). Identification and characterization of a novel RNA binding protein that associates with the 5′-untranslated region of the chloroplast psbA mRNA. Biochemistry 43 8541–8550. [DOI] [PubMed] [Google Scholar]
- Barr, C.M., Neiman, M., and Taylor, D.R. (2005). Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 168 39–50. [DOI] [PubMed] [Google Scholar]
- Bellaoui, M., Martin-Canadell, A., Pelletier, G., and Budar, F. (1998). Low-copy-number molecules are produced by recombination, actively maintained and can be amplified in the mitochondrial genome of Brassicaceae: Relationship to reversion of the male sterile phenotype in some cybrids. Mol. Gen. Genet. 257 177–185. [DOI] [PubMed] [Google Scholar]
- Chomczynski, P., and Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162 156–159. [DOI] [PubMed] [Google Scholar]
- Christensen, A.C., Lyznik, A., Mohammed, S., Elowsky, C.G., Elo, A., Yule, R., and Mackenzie, S.A. (2005). Dual-domain, dual-targeting organellar protein presequences in Arabidopsis can use non-AUG start codons. Plant Cell 17 2805–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton, S.W., et al. (2004). Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol. 136 3486–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Duchene, A.M., Giritch, A., Hoffmann, B., Cognat, V., Lancelin, D., Peeters, N.M., Zaepfel, M., Marechal-Drouard, L., and Small, I.D. (2005). Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102 16484–16489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson, A.C., Song, D., Alvarez, L.A., Wall, M.K., Almond, D., McClellan, D.A., Maxwell, A., and Nielsen, B.L. (2005). Characterization of a mitochondrially targeted single-stranded DNA-binding protein in Arabidopsis thaliana. Mol. Genet. Genomics 273 115–122. [DOI] [PubMed] [Google Scholar]
- Forner, J., Weber, B., Wietholter, C., Meyer, R.C., and Binder, S. (2005). Distant sequences determine 5′ end formation of cox3 transcripts in Arabidopsis thaliana ecotype C24. Nucleic Acids Res. 33 4673–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior, S.L., Olivares, H., Ear, U., Hari, D.M., Weichselbaum, R., and Bishop, D.K. (2001). Assembly of RecA-like recombinases: Distinct roles for mediator proteins in mitosis and meiosis. Proc. Natl. Acad. Sci. USA 98 8411–8418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, A., Ghosh, T., Ghosh, S., Das, S., and Adhya, S. (1994). Interaction of small ribosomal and transfer RNAs with a protein from Leishmania donovani. Nucleic Acids Res. 22 1663–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke, B., Börner, T., and Weihe, A. (2000). One RNA polymerase serving two genomes. EMBO Rep. 1 435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janska, H., Sarria, R., Woloszynska, M., Arrieta-Montiel, M., and Mackenzie, S.A. (1998). Stoichiometric shifts in the common bean mitochondrial genome leading to male sterility and spontaneous reversion to fertility. Plant Cell 10 1163–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa, A., Tsutsumi, N., and Hirai, A. (1994). Reversible changes in the composition of the population of mtDNAs during dedifferentiation and regeneration in tobacco. Genetics 138 865–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec, B., Woloszynska, M., and Janska, H. (2006). Heteroplasmy as a common state of mitochondrial genetic information in plants and animals. Curr. Genet. 50 149–159. [DOI] [PubMed] [Google Scholar]
- Kuzmin, E.V., Duvick, D.N., and Newton, K.J. (2005). A mitochondrial mutator system in maize. Plant Physiol. 137 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi, C., Rayapuram, N., Chartier, Y., Grienenberger, J.M., Bonnard, G., and Meyer, Y. (2001). Identification and characterization of a mitochondrial thioredoxin system in plants. Proc. Natl. Acad. Sci. USA 98 14144–14149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Zapater, J.M., Gil, P., Capel, J., and Somerville, C.R. (1992). Mutations at the Arabidopsis CHM locus promote rearrangements of the mitochondrial genome. Plant Cell 4 889–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand, B., Marechal-Drouard, L., Sakamoto, W., Dietrich, A., and Wintz, H. (1998). A single gene of chloroplast origin codes for mitochondrial and chloroplastic methionyl-tRNA synthetase in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 95 11014–11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mookerjee, S.A., and Sia, E.A. (2006). Overlapping contributions of Msh1p and putative recombination proteins Cce1p, Din7p, and Mhr1p in large-scale recombination and genome sorting events in the mitochondrial genome of Saccharomyces cerevisiae. Mutat. Res. 595 91–106. [DOI] [PubMed] [Google Scholar]
- Neuburger, M., Journet, E.P., Bligny, R., Carde, J.P., and Douce, R. (1982). Purification of plant mitochondria by isopycnic centrifugation in density gradients of Percoll. Arch. Biochem. Biophys. 217 312–323. [DOI] [PubMed] [Google Scholar]
- Oldenburg, D.J., and Bendich, A.J. (1996). Size and structure of replicating mitochondrial DNA in cultured tobacco cells. Plant Cell 8 447–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin, R., Meyer, E.H., Zaepfel, M., Kim, Y.J., Mache, R., Grienenberger, J.M., Gualberto, J.M., and Gagliardi, D. (2004). Two exoribonucleases act sequentially to process mature 3′ ends of atp9 mRNAs in Arabidopsis mitochondria. J. Biol. Chem. 279 25440–25446. [DOI] [PubMed] [Google Scholar]
- Pinto, A.V., Mathieu, A., Marsin, S., Veaute, X., Ielpi, L., Labigne, A., and Radicella, J.P. (2005). Suppression of homologous and homeologous recombination by the bacterial MutS2 protein. Mol. Cell 17 113–120. [DOI] [PubMed] [Google Scholar]
- Raghunathan, S., Ricard, C.S., Lohman, T.M., and Waksman, G. (1997). Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-Å resolution. Proc. Natl. Acad. Sci. USA 94 6652–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, W., Kondo, H., Murata, M., and Motoyoshi, F. (1996). Altered mitochondrial gene expression in a maternal distorted leaf mutant of Arabidopsis induced by chloroplast mutator. Plant Cell 8 1377–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford, J.C., Smith, F.D., and Russell, J.A. (1993). Optimizing the biolistic process for different biological applications. Methods Enzymol. 217 483–509. [DOI] [PubMed] [Google Scholar]
- Sheahan, M.B., McCurdy, D.W., and Rose, R.J. (2005). Mitochondria as a connected population: Ensuring continuity of the mitochondrial genome during plant cell dedifferentiation through massive mitochondrial fusion. Plant J. 44 744–755. [DOI] [PubMed] [Google Scholar]
- Shoubridge, E.A. (2000). Mitochondrial DNA segregation in the developing embryo. Hum. Reprod. 15 (suppl. 2): 229–234. [DOI] [PubMed] [Google Scholar]
- Small, I.D., Suffolk, R., and Leaver, C.J. (1989). Evolution of plant mitochondrial genomes via substoichiometric intermediates. Cell 58 69–76. [DOI] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unseld, M., Marienfeld, J.R., Brandt, P., and Brennicke, A. (1997). The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 15 57–61. [DOI] [PubMed] [Google Scholar]
- van Engelen, F.A., Molthoff, J.W., Conner, A.J., Nap, J.P., Pereira, A., and Stiekema, W.J. (1995). pBINPLUS: An improved plant transformation vector based on pBIN19. Transgenic Res. 4 288–290. [DOI] [PubMed] [Google Scholar]
- Vermel, M., Guermann, B., Delage, L., Grienenberger, J.M., Marechal-Drouard, L., and Gualberto, J.M. (2002). A family of RRM-type RNA-binding proteins specific to plant mitochondria. Proc. Natl. Acad. Sci. USA 99 5866–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet, O., Rivas, S., Mestre, P., and Baulcombe, D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33 949–956. [DOI] [PubMed] [Google Scholar]
- Wall, M.K., Mitchenall, L.A., and Maxwell, A. (2004). Arabidopsis thaliana DNA gyrase is targeted to chloroplasts and mitochondria. Proc. Natl. Acad. Sci. USA 101 7821–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.