Abstract
In plants, benzoic acid (BA) is believed to be synthesized from Phe through shortening of the propyl side chain by two carbons. It is hypothesized that this chain shortening occurs via either a β-oxidative or non-β-oxidative pathway. Previous in vivo isotope labeling and metabolic flux analysis of the benzenoid network in petunia (Petunia hybrida) flowers revealed that both pathways yield benzenoid compounds and that benzylbenzoate is an intermediate between l-Phe and BA. To test this hypothesis, we generated transgenic petunia plants in which the expression of BPBT, the gene encoding the enzyme that uses benzoyl-CoA and benzyl alcohol to make benzylbenzoate, was reduced or eliminated. Elimination of benzylbenzoate formation decreased the endogenous pool of BA and methylbenzoate emission but increased emission of benzyl alcohol and benzylaldehyde, confirming the contribution of benzylbenzoate to BA formation. Labeling experiments with 2H5-Phe revealed a dilution of isotopic abundance in most measured compounds in the dark, suggesting an alternative pathway from a precursor other than Phe, possibly phenylpyruvate. Suppression of BPBT activity also affected the overall morphology of petunia plants, resulting in larger flowers and leaves, thicker stems, and longer internodes, which was consistent with the increased auxin transport in transgenic plants. This suggests that BPBT is involved in metabolic processes in vegetative tissues as well.
INTRODUCTION
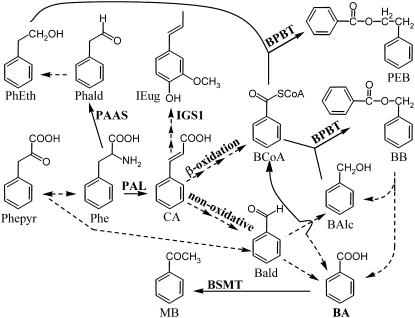
Benzenoid and phenylpropanoid volatile compounds, primarily derived from Phe, contribute to the aromas/scents of many plant species and play important roles in plant communication with surrounding environments (Dudareva and Pichersky, 2006; Knudsen and Gershenzon, 2006; Pichersky et al., 2006). Several enzymes that catalyze the final steps in the biosynthesis of these compounds have been isolated and characterized. However, the early steps leading to the formation of the benzenoid backbone still remain unclear (Beuerle and Pichersky, 2002; Schnepp and Dudareva, 2006; Wildermuth, 2006). In general, the biosynthesis of benzenoids from Phe requires the shortening of the carbon skeleton side chain by a C2 unit, which can potentially occur via either the β-oxidative pathway or nonoxidatively (Figure 1). Experiments with stable isotope-labeled precursors in tobacco (Nicotiana tabacum) leaves (Ribnicky et al., 1998) suggested that benzoic acid (BA) is produced from Phe-derived cinnamic acid (CA) via the β-oxidative pathway, yielding benzoyl-CoA first, which can then be hydrolyzed by a thioesterase to free BA. By contrast, labeling experiments along with initial enzyme characterization in Hypericum androsaemum cell cultures (Ahmed et al., 2002) supported the existence of the nonoxidative conversion of CA to benzaldehyde with subsequent formation of BA, which can be further converted to benzoyl-CoA (Beuerle and Pichersky, 2002; Figure 1). However, none of the genes or enzymes involved in any chain-shortening steps in BA biosynthesis have been identified or characterized in plants.
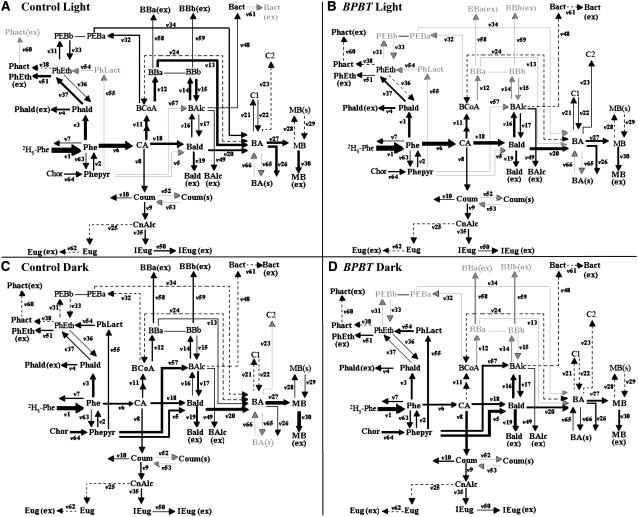
Figure 1.
Main Biochemical Reactions Leading to the Synthesis of Benzenoid Volatiles in Petunia Flowers.
Some reactions are hypothesized (dashed arrows) and have not been shown conclusively to occur in petunia. See text for details. BAlc, benzyl alcohol; Bald, benzylaldehyde; BB, benzylbenzoate; BCoA, benzoyl-CoA; CA, trans-cinnamic acid; IEug, isoeugenol; MB, methylbenzoate; PEB, phenylethyl benzoate; Phald, phenylacetaldehyde; Phe, l-Phe; PhEth, 2-phenylethanol; Phepyr, phenylpyruvic acid; BPBT, benzoyl-CoA:benzyl alcohol/2-phenylethanol benzoyltransferase; BSMT, benzoic acid/salicylic acid carboxyl methyltransferase; IGS1, isoeugenol synthase; PAAS, phenylacetaldehyde synthase.
Petunia (Petunia hybrida) flowers emit a diverse blend of floral scent phenylpropanoid and benzenoid compounds that are synthesized predominantly in the corolla with emission level peaking during the night (Kolosova et al., 2001; Verdonk et al., 2003; Boatright et al., 2004). These volatiles include benzyl alcohol, benzaldehyde, methylbenzoate and benzylbenzoate, as well as phenylacetaldehyde, 2-phenylethanol, phenylethylacetate, isoeugenol, and eugenol. Using petunia as a model system, we have recently identified enzymes responsible for the formation of some of these volatile compounds, including phenylacetaldehyde synthase (PAAS), which is responsible for the synthesis of phenylacetaldehyde from Phe (Kaminaga et al., 2006); benzoic acid/salicylic acid carboxyl methyltransferase, which is responsible for the synthesis of methylbenzoate from BA and S-adenosylmethionine (Negre et al., 2003); isoeugenol synthase (IGS1), which is responsible for the synthesis of isoeugenol from coniferyl acetate (Koeduka et al., 2006); and benzoyl-CoA:benzyl alcohol/phenylethanol benzoyltransferase (BPBT), which is responsible for the synthesis of benzylbenzoate and phenylethylbenzoate from benzoyl-CoA and benzyl alcohol or 2-phenylethanol, respectively (Boatright et al., 2004) (Figure 1).
Recently, we have also performed computer-assisted metabolic flux analysis of data obtained from in vivo stable isotope labeling of petunia petals to investigate the early steps of benzenoid biosynthesis (Boatright et al., 2004). This analysis revealed that both the β-oxidative and non-β-oxidative pathways contribute to the formation of benzenoid compounds in petunia flowers. The flux through the non-β-oxidative pathway with benzaldehyde as a key intermediate was estimated to be twofold higher than the flux through the β-oxidative pathway, which relies on the formation of CoA esters. Furthermore, the modeling of stable isotope labeling data uncovered the unexpected result that, in addition to benzaldehyde, benzylbenzoate is an alternative intermediate between l-Phe and BA.
To determine the role of benzylbenzoate within the benzenoid network in planta, we generated transgenic petunia plants in which this system was perturbed by suppression of BPBT gene expression via RNA interference–based (RNAi) technology. Analysis of transgenic plants not only confirmed that benzylbenzoate is indeed an intermediate in BA synthesis but also showed that a suppression of the flux to benzylbenzoate enhances the contribution of the non-β-oxidative route to some benzenoid compounds in the light. Moreover, the unexpected morphological and anatomical phenotypes of the transgenic plants indicated that BPBT plays a role in vegetative tissues as well.
RESULTS
Effect of RNAi-Mediated Repression of BPBT Expression on Volatile Benzenoid Production
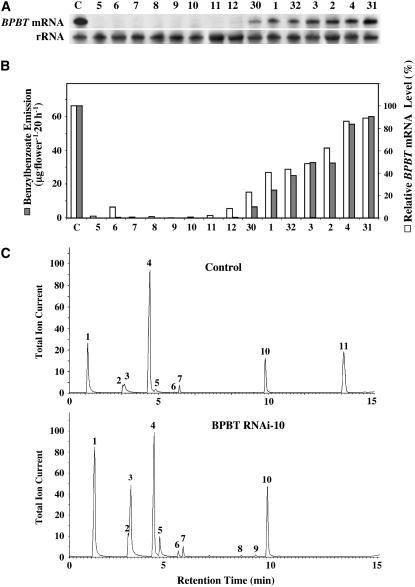
To investigate the specific role of benzylbenzoate within the benzenoid network, the expression of BPBT was downregulated in P. hybrida cv Mitchell using an RNAi strategy. An RNAi construct was generated using a 350-bp fragment within the 5′ end of the BPBT coding region and expressed under the control of the petal-specific LIS promoter (Cseke et al., 1998). Thirty independently transformed individuals were obtained, and RNA gel blot analysis showed that BPBT expression in petals was altered from complete elimination of transcripts (∼50% of transformants) to only slight (2%) reduction compared with untransformed control plants (Figures 2A and 2B). Quantitative analysis of the volatiles emitted from 2-d-old (2 d after fully opening) petunia flowers revealed that complete elimination (i.e., knockout) of BPBT expression eliminated benzylbenzoate emission, while a partial reduction (i.e., knockdown) in BPBT transcript levels resulted in a corresponding reduction of benzylbenzoate formation from 10 to 91% of that in control plants (Figures 2B, 2C, and 3), confirming that BPBT is responsible for benzylbenzoate formation in vivo. Consistent with expression data, BPBT activity was undetectable in transgenic flowers with eliminated BPBT expression, while in flowers with reduced BPBT expression (50%), BPBT activity was ∼50% of that in control flowers. Similar results were obtained for phenylethylbenzoate emission, indicating that BPBT is also responsible for its formation in vivo (Figure 3).
Figure 2.
Effect of BPBT Silencing on Benzylbenzoate Emission.
(A) BPBT mRNA levels in corollas of control and different transgenic BPBT RNAi petunia lines. Representative RNA gel blot hybridization with total RNA (5 μg per lane) isolated from the limbs of corollas of 2-d-old control (C) petunia flowers and flowers from independent transgenic lines (numbers on top of the gel). A coding region of the BPBT genes was used as a probe. Autoradiography was performed overnight. The blot was rehybridized with an 18S rDNA probe (bottom gel) to standardize samples.
(B) Benzylbenzoate emission and the relative BPBT mRNA levels in control and transgenic BPBT RNAi petunia flowers. Floral volatiles were collected from petunia flowers using the closed-loop stripping method. The numbers represent independent transgenic lines (C, control). The relative BPBT mRNA levels were obtained by scanning RNA gel blots with a phosphor imager and corrected by standardizing for the amounts of 18S rRNA measured in the same runs. The transcript level in control plants was taken as 100%.
(C) Metabolic profiling of benzenoid compounds emitted from control (top chromatogram) and BPBT RNAi (bottom chromatogram) petunia flowers. The BPBT RNAi-10 transgenic line was used as an example. Floral scent collected from detached flowers of control and BPBT RNAi-10 transgenic plants was analyzed by electron ionization gas chromatography–mass spectrometry, and total ion currents are plotted. Compounds were identified based on their mass spectra and retention time: 1, benzaldehyde; 2, phenylacetaldehyde; 3, benzyl alcohol; 4, methylbenzoate; 5, 2-phenylethanol; 6, benzylacetate; 7, internal standard (naphthalene); 8, phenylethylacetate; 9, eugenol; 10, isoeugenol; 11, benzylbenzoate.
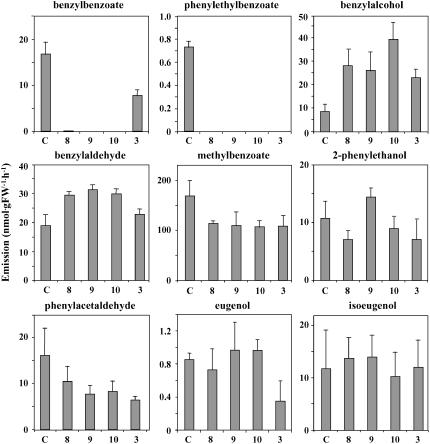
Figure 3.
Effect of BPBT Silencing on Emission of Benzenoid/Phenylpropanoid Compounds in Petunia Flowers.
Flowers from four independent BPBT RNAi lines, 8, 9, and 10 (knockouts) and 3 (knockdown), and control (C) were used for scent collection. Each graph represents the average of four to five independent experiments. Scent was collected for 12 h from detached flowers in the dark, and levels of volatiles were quantified by GC-MS. Emission rates are expressed on a per hour basis, assuming a constant emission rate over the 12-h period. Bars indicate sd.
Four independent BPBT RNAi lines, BPBTi-8, BPBTi-9, and BPBTi-10 (knockouts) and BPBTi-3 (a knockdown), were used for detailed metabolic profiling of other emitted floral volatiles (Figure 3). Knockout of the BPBT gene expression led to a significant increase (twofold to fourfold) of the emission of benzyl alcohol, which is a substrate for the BPBT enzyme (Figures 1 and 3). In addition, it resulted in increased emission of benzylaldehyde (∼1.7-fold) and reduced emission of methylbenzoate (∼1.6-fold). Interestingly, reduction/elimination of phenylethylbenzoate emission did not elevate the level of emitted 2-phenylethanol, and the amount of emitted phenylacetaldehyde was actually reduced in the BPBT RNAi transgenic plants. Silencing of BPBT had no effect on the emission of eugenol or isoeugenol (Figure 3) nor on emission of benzylacetate and phenylethylacetate (data not shown), two floral scent compounds that are usually present only in trace amounts during the night in petunia scent. Reduction of BPBT expression by 50% (BPBTi-3) had nearly the same effect on the volatile emission profile as complete elimination of BPBT transcripts (BPBTi-8, -9, and -10), with the exception of benzaldehyde, whose emission was only slightly elevated in the knockdown line when compared with control flowers (Figure 3).
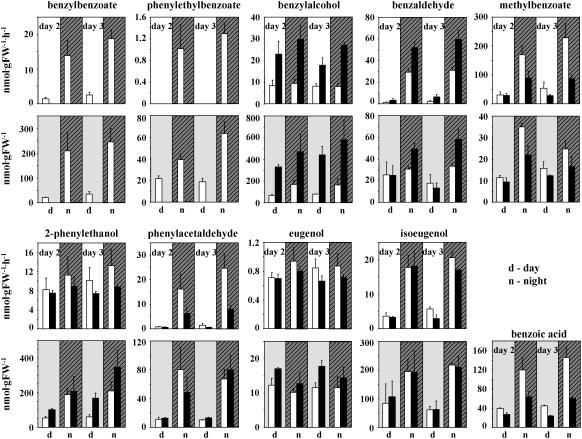
Complete or partial reduction of BPBT transcript level did not affect rhythmic emission of volatile compounds other than benzylbenzoate and phenylethylbenzoate (Figure 4). Rhythmic levels of emitted volatiles in transgenic and control plants positively correlated with their corresponding internal pools, with the exception of 2-phenylethanol, whose internal pool levels were higher in BPBT RNAi knockouts than in control plants in the light but showed no differences in emission between transgenics and controls (Figure 4, data shown for BPBTi-10).
Figure 4.
Rhythmic Changes in Emission and Internal Pools of Benzenoid/Phenylpropanoid Compounds in Petal Tissue of Control and BPBT Knockout Petunia Plants.
Data represent the average pools and emission rates from three independent experiments performed over a 48-h period beginning with 2-d-old control (white bars) and BPBT RNAi-10 transgenic (black bars) flowers. Headspace collections were performed for 12 h, and samples were taken at 8 am (night points) and 8 pm (day points). Tissue for internal pools was collected at 9 am (day points) and 9 pm (night points). Emission rates are shown in the top graphs with white backgrounds, while internal pools for the corresponding volatiles are shown in the bottom graphs with gray backgrounds. Striped areas indicate night period. Bars indicate sd.
Changes in Metabolite Levels and Fluxes in Transgenic BPBT RNAi Plants
To determine the effect of the reduction/elimination of benzylbenzoate synthesis on the benzenoid network in petunia flowers, we employed metabolic flux analysis with stable isotope labeling as described previously (Boatright et al., 2004). Since the emission rate of bezenoid volatiles in petunia flowers is highest at night (Kolosova et al., 2001; Verdonk et al., 2003), this analysis was initially conducted under dark (inverted photoperiod) conditions only (Boatright et al., 2004). Here, we examined the pathway under both natural dark and light conditions. Deuterium ring-labeled Phe (2H5-Phe) was supplied to excised petunia corolla limbs of both control and BPBT knockout (BPBTi-10) plants, and floral volatiles were collected at 30, 60, 120, and 240 min and analyzed by gas chromatography–mass spectrometry (GC-MS) for pool sizes and isotope abundances. The differences in scent profiles of control and transgenic plants during the 2H5-Phe feeding experiments were consistent with those found in scent collection from intact flowers. After scent collections, petal tissues were extracted with either methanol, to determine the labeling and pool sizes of endogenous nonvolatile metabolites, including organic acids, or with dichloromethane, to determine pool sizes and isotope abundances of endogenous intermediates and end products, including phenylacetaldehyde, 2-phenylethanol, phenylethylbenzoate, eugenol, isoeugenol, benzaldehyde, benzyl alcohol, benzylbenzoate, and methylbenzoate. Detection of additional endogenous compounds, including some organic acids, extended the previously described metabolic model for the benzenoid network in control flowers (Boatright et al., 2004) from 38 to 66 fluxes (Figure 5, Table 1). In addition, the rhythmicity in emission of volatile compounds from petunia flowers required the development of models showing the changes in flux between light and dark conditions. The fluxes generated by modeling these data from three independent labeling experiments are presented in Table 1.
Figure 5.
Four Models Representing the Benzenoid Network in the Light and Dark in Control and BPBT Knockout Petunia Flowers.
Computer-assisted metabolic modeling was performed using in vivo labeling kinetics and pool sizes of benzenoid and phenylpropanoid compounds in petunia petal tissue supplied with 2H5-Phe for up to 4 h in light ([A] and [B]) and dark ([C] and [D]) conditions in control ([A] and [C]) and BPBT RNAi-10 transgenic ([B] and [D]) plants. Thickness of lines correlates with strength of flux (see Table 1). Gray arrows indicate the absence of flux, and dashed lines indicate very low flux rate. Bact, benzylacetate; BBa, benzylbenzoate BA moiety; BBb, benzylbenzoate BAlc moiety; C1 and C2, unidentified nonvolatile BA conjugates (Boatright et al., 2004); Chor, chorismic acid; CnAlc, coniferyl alcohol; Coum, coumaric acid; Eug, eugenol; PEBa, phenylethyl benzoate BA moiety; PEBb, phenylethyl benzoate 2-phenylethanol moiety; Phact, phenylethylacetate; PhLact, phenyllactic acid. For remaining abbreviations, see Figure 1.
Table 1.
Computer-Simulated Metabolic Fluxes of the Benzenoid Network in Control and BPBT RNAi-10 Petunia Flowers
| Flux | Description | Wild Type Dark | Trans Dark | Wild Type Light | Trans Light |
|---|---|---|---|---|---|
| v1 | Uptake rate of d5-Phe | 79.810 | 257.435 | 373.394 | 371.530 |
| v2 | Phepyr → Phe | 1.000 | 1.000 | 1.000 | 1.000 |
| v3 | Phe → Phald | 3.000 | 1.700 | 8.300 | 6.000 |
| v4 | Phald → Phald (ex) | 1.000 | 0.600 | 1.500 | 1.500 |
| v5 | Phepyr → Bald | 4.200 | 5.700 | 0.000 | 0.000 |
| v6 | Phe → CA | 4.810 | 4.735 | 14.794 | 15.230 |
| v7 | Phe → other | 1.000 | 1.000 | 1.000 | 1.000 |
| v8 | CA → coum | 0.780 | 1.735 | 0.745 | 0.750 |
| v9 | Coum → Cnalc | 0.650 | 0.735 | 0.635 | 0.550 |
| v10 | Coum → other | 0.100 | 1.000 | 0.100 | 0.200 |
| v11 | CA → BCoA | 1.130 | 0.100 | 7.450 | 4.400 |
| v12 | BCoA → BBa | 0.700 | 0.000 | 6.500 | 0.000 |
| v13 | BBa → BA | 0.200 | 0.000 | 6.000 | 0.000 |
| v14 | BAlc → BBb | 0.700 | 0.000 | 6.500 | 0.000 |
| v15 | BBb → BAlc (m) | 0.200 | 0.000 | 6.000 | 0.000 |
| v16 | Bald → BAlc | 0.430 | 4.000 | 1.549 | 4.100 |
| v17 | BAlc → Bald | 4.000 | 4.000 | 0.100 | 1.500 |
| v18 | CA → Bald | 2.900 | 2.900 | 6.599 | 10.080 |
| v19 | Bald → Bald (ex) | 3.800 | 5.200 | 2.500 | 2.300 |
| v20 | Bald → BA | 6.870 | 3.400 | 2.650 | 5.180 |
| v21 | BA → C1 | 0.100 | 0.100 | 0.600 | 0.600 |
| v22 | C1 → BA | 0.100 | 0.100 | 0.600 | 0.600 |
| v23 | BA → C2 | 0.000 | 0.100 | 0.060 | 0.100 |
| v24 | BCoA → BA | 0.050 | 0.050 | 0.050 | 0.050 |
| v25 | Cnalc → Eug | 0.050 | 0.035 | 0.035 | 0.050 |
| v26 | BA → other | 0.100 | 1.000 | 5.600 | 3.000 |
| v27 | BA → MB (m) | 6.000 | 2.200 | 3.800 | 2.080 |
| v28 | MB (m) → MB (s) | 0.200 | 0.008 | 0.000 | 0.010 |
| v29 | MB (s) → MB (m) | 0.200 | 0.008 | 0.000 | 0.010 |
| v30 | MB (m) → MB (ex) | 6.000 | 2.200 | 3.800 | 2.000 |
| v31 | PhEth → PEBb | 0.030 | 0.000 | 0.900 | 0.000 |
| v32 | BCoA → PEBa | 0.030 | 0.000 | 0.900 | 0.000 |
| v33 | PEBb → PhEth | 0.030 | 0.000 | 0.900 | 0.000 |
| v34 | PEBa → BA | 0.030 | 0.000 | 0.900 | 0.000 |
| v35 | Cnalc → Ieug | 0.600 | 0.700 | 0.600 | 0.500 |
| v36 | PhEth → Phald | 0.100 | 0.100 | 0.100 | 0.010 |
| v37 | Phald → PhEth | 0.600 | 0.400 | 4.190 | 2.500 |
| v38 | Pheth → Phact | 0.020 | 0.020 | 0.010 | 0.020 |
| v39 | MB (m) expansion flux | 0.000 | 0.000 | 0.000 | 0.080 |
| v40 | BA expansion flux | 1.000 | 0.100 | 0.090 | 0.000 |
| v41 | Coum expansion flux | 0.030 | 0.000 | 0.010 | 0.000 |
| v42 | Phe expansion flux | 70.000 | 250.000 | 350.000 | 350.000 |
| v43 | Phald expansion flux | 1.380 | 0.800 | 2.710 | 2.010 |
| v44 | PEB expansion flux | 0.010 | 0.000 | 0.000 | 0.010 |
| v45 | BB expansion flux | 0.000 | 0.000 | 0.250 | 0.000 |
| v46 | BCoA expansion flux | 0.400 | 0.100 | 0.050 | 4.400 |
| v47 | PhEth expansion flux | 0.500 | 0.580 | 2.280 | 2.270 |
| v48 | BAlc → Bact | 0.030 | 0.045 | 0.100 | 0.045 |
| v49 | BAlc → Balc (ex) | 0.500 | 0.700 | 0.500 | 1.000 |
| v50 | IEug → IEug (ex) | 0.350 | 0.200 | 0.550 | 0.250 |
| v51 | PhEth → PhEth (ex) | 0.600 | 0.400 | 1.800 | 0.200 |
| v52 | Coum (m) → coum (s) | 0.000 | 0.000 | 0.000 | 0.000 |
| v53 | Coum (s) → coum (m) | 0.000 | 0.000 | 0.000 | 0.000 |
| v54 | Phelact → PhEth | 0.700 | 0.700 | 0.000 | 0.000 |
| v55 | Phepyr → Phelact | 0.700 | 0.700 | 0.000 | 0.000 |
| v56 | BAlc expansion flux | 0.400 | 0.955 | 0.349 | 1.555 |
| v57 | Phepyr → BAlc | 5.000 | 1.700 | 0.000 | 0.000 |
| v58 | BBa → BBa (ex) | 0.500 | 0.000 | 0.250 | 0.000 |
| v59 | BBb → BBb (ex) | 0.500 | 0.000 | 0.250 | 0.000 |
| v60 | Phact → Phact (ex) | 0.020 | 0.020 | 0.000 | 0.020 |
| v61 | Bact → Bact (ex) | 0.030 | 0.045 | 0.000 | 0.045 |
| v62 | Eug → Eug (ex) | 0.040 | 0.035 | 0.035 | 0.040 |
| v63 | Phe → Phepyr | 2.000 | 1.000 | 0.300 | 0.300 |
| v64 | Chor → Phepyr | 8.900 | 8.100 | 0.700 | 0.700 |
| v65 | BA (m) → BA (s) | 0.000 | 0.300 | 0.000 | 0.000 |
| v66 | BA (s) → BA (m) | 0.000 | 0.300 | 0.000 | 0.000 |
For abbreviations, see Figures 1 and 6. Vn refers to the fluxes shown in Figure 5. Values given are nmol min−1 gFW−1. Numbers in boldface show fluxes in the dark. m, metabolically active pool; s, storage pool; ex, exogenous compound; wild type, control, nontransgenic petunia flowers; Trans, BPBT RNAi-10 petunia flowers.
Benzenoid Network in Light Conditions
In the light, both the β-oxidative and non-β-oxidative pathways contributed to the formation of benzenoid compounds in petunia flowers (Figure 5A). The flux to BA through the β-oxidative pathway contributed twice as much as the non-β-oxidative pathway (v13 versus v20) (Table 1). BPBT RNAi silencing eliminated the contribution of benzylbenzoate and phenylethylbenzoate to BA biosynthesis and simultaneously increased the flux via the non-β-oxidative pathway (v18 = 10.08 and 6.6 nmol min−1 gFW−1 in transgenic and control plants, respectively) (Figures 5A and 5B, Table 1). Although flux to benzaldehyde (v18) increased by 1.5-fold and its subsequent flux to BA (v20) increased approximately twofold, the total flux to BA (a sum of v13, v20, v22, v24, and v34) in transgenic plants decreased 1.75-fold, resulting in a lower flux to methylbenzoate (v27 = 2.08 and 3.8 nmol min−1 gFW−1 in transgenic and control plants, respectively) and its subsequent emission (v30 = 2.0 and 3.8 nmol min−1 gFW−1 in transgenic and control plants, respectively) (Figures 5A and 5B, Table 1). Inhibition of benzylbenzoate formation also led to the expansion of the benzyl alcohol internal pool and doubled its emission (see Supplemental Figure 1B online).
Benzenoid Network in Dark Conditions
Similar to the situation in the light, both the β-oxidative and non-β-oxidative pathways contributed to the formation of benzenoid compounds in control petunia flowers. Despite the higher rate of benzylbenzoate biosynthesis at night, its actual contribution to BA formation (v13) decreased due to a greater emission flux (v58 and v59). Thus, the relative input of the β-oxidative pathway in the dark was diminished when compared with its contribution in the light (ratio v13/v20 = 2.26 versus 0.03 in the light and dark, respectively) (Figures 5A and 5C, Table 1). The flux from phenylethylbenzoate to BA also decreased in the dark. Based on labeling kinetics, it can be assumed that phenylacetaldehyde is the major precursor of 2-phenylethanol in the light (Figure 5A), which was consistent with our recent report that an RNAi suppression of PAAS gene expression resulted in elimination of both phenylacetaldehyde and 2-phenylethanol emission in petunia flowers (Kaminaga et al., 2006). In the dark (Figure 5C), labeling of both phenylacetaldehyde and 2-phenylethanol was diluted, possibly due to a contribution from phenyllactic acid, as was recently hypothesized in rose (Rosa damascena) (Watanabe et al., 2002).
Analysis of the labeling patterns of internal pools and emitted volatiles revealed a dilution of isotopic abundance in most measured compounds in the dark versus light (Figure 6; see Supplemental Figures 1A and 1B online). While CA had similar labeling in the light and dark over a 4-h period (83 and 94% in the light and dark, respectively), endogenous benzaldehyde over the same time period was labeled to only 43% in the dark compared with 92% in the light (see Supplemental Figure 1 online). To simultaneously account for the divergent labeling patterns of CA and benzaldehyde in the dark, it was necessary to invoke an alternate biosynthetic pathway for benzaldehyde from a precursor other than Phe.
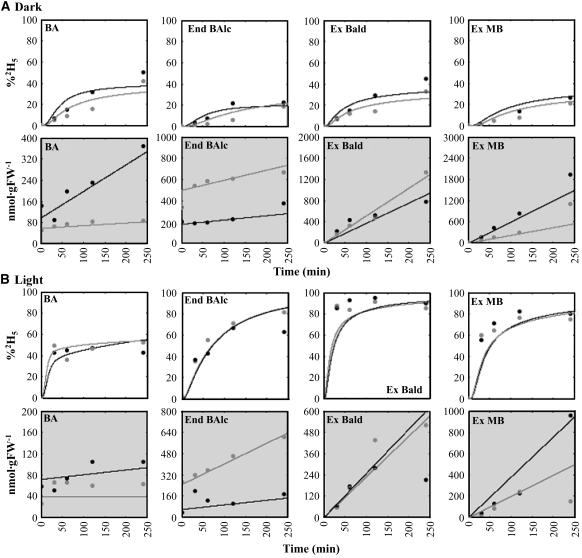
Figure 6.
Pool Sizes and Isotopic Abundances of Representative Compounds within the Benzenoid Network in Control and BPBT RNAi-10 Flowers.
Pool sizes and isotopic abundances are shown for BA, endogenous benzyl alcohol (End Balc), exogenous benzaldehyde (Ex Bald), and exogenous methylbenzoate (Ex MB) in the dark (A) and in the light (B). Charts with white background show experimentally determined and model-simulated isotopic labeling of key intermediates and end products. Charts with gray backgrounds show pool sizes of the various metabolites over a 4-h time course of feeding with 2H5-Phe. Simulated labeling curves and pool sizes (lines) were generated using the flux rates and initial pool sizes specified in Table 1 and are superimposed upon observed values (symbols). Controls are shown in black, and BPBT RNAi transgenics are shown in gray.
Several reports have shown that phenylpyruvic acid can be nonenzymatically converted to benzaldehyde in vivo in the presence of oxygen and metal cofactors (Nierop-Groot and De Bont, 1998; Rijnen et al., 2000). Thus, we investigated whether the experimentally obtained pool sizes and labeling pattern (see below) of phenylpyruvic acid over a 4-h period could account for the reduced labeling of compounds downstream of trans-cinnamic acid, including benzaldehyde, benzyl alcohol, BA, and their immediate products (Figure 6; see Supplemental Figure 1 online). Indeed, incorporation of these data in the model(s) suggested that phenylpyruvate is a possible candidate for the observed dilution of isotopic abundance. Unfortunately, the unavailability of labeled phenylpyruvate and its observed instability in solution caused by pH-dependent conversion to benzaldehyde and benzyl alcohol (data not shown) did not allow us to perform feeding experiments and test our hypothesis experimentally.
It is well established that in many species phenylpyruvate is synthesized directly from chorismate, although this reaction has not been conclusively demonstrated in plants (Jung et al., 1986; Warpeha et al., 2006). In our labeling experiments, phenylpyruvate was labeled up to 25% over a 4-h period when 2H5-Phe was supplied, suggesting that Phe can be converted back to phenylpyruvate in petunia petals, presumably by transamination. Although biosynthesis of phenylpyruvate from Phe is a one-step conversion, like formation of CA from Phe, labeling of phenylpyruvate was 70% less than labeling of CA, indicating that Phe is not likely the primary source of phenylpyruvate in petunia petals.
BPBT RNAi silencing did not affect the flux from Phe to CA to benzaldehyde (see fluxes v6 and v18) but slightly increased the relative contribution of the hypothesized alternative pathway from phenylpyruvate to benzaldehyde (v5 = 5.7 versus 4.2 nmol min−1 gFW−1 in transgenic and control plants, respectively) and simultaneously decreased its contribution to benzyl alcohol biosynthesis (v57 = 1.7 versus 5 nmol min−1 gFW−1 in transgenic and control plants, respectively) (Figures 5C and 5D, Table 1). This change in flux led to a twofold expansion of the endogenous benzaldehyde pool (see Supplemental Figure 1 online) as well as a 1.7-fold increase in its emission over the 4-h labeling period (Figure 6A). Similar to benzaldehyde, the endogenous benzyl alcohol pool (Figure 6A) and its emission rate (see Supplemental Figure 1 online) were increased in transgenics. This was achieved due to (1) elimination of flux to benzylbenzoate, for which benzyl alcohol is an immediate precursor, and (2) redirection of the flux from phenylpyruvate to benzyl alcohol via benzaldehyde.
The elimination of benzylbenzoate flux to free BA in transgenic plants also led to a twofold decrease in the flux from benzaldehyde to BA (v20 = 3.4 versus 6.87 nmol min−1 gFW−1 in transgenic and control plants, respectively) (Figures 5C and 5D, Table 1). As a result, the endogenous pool of BA and methylbenzoate emission were decreased approximately threefold and twofold, respectively (Figure 6A). Elimination of phenylethylbenzoate in BPBT RNAi plants resulted in only slight expansion of the endogenous 2-phenylethanol pool, which did not lead to an increase in its emission. The lack of expansion was a result of a reduction in flux from phenylacetaldehyde to 2-phenylethanol in transgenics (v37 = 0.4 versus 0.6 nmol min−1 gFW−1 in transgenic and control plants, respectively). There was also a decreased level of phenylacetaldehyde in BPBT RNAi plants as a result of the direct reduction in flux from Phe (Table 1, v3).
Elimination of BPBT transcripts also caused an increase in the flux from CA to coumaric acid (v8 = 1.74 versus 0.78 nmol min−1 gFW−1 in transgenic and control plants, respectively) but did not change the flux downstream toward eugenol and isoeugenol (Figures 5C and 5D). Rather, modeling suggested the possibility that knockout of BPBT may increase flux to flavonoids (v10 = 1.0 versus 0.1 nmol min−1 gFW−1 in transgenic and control plants, respectively), consistent with the accumulation of 3′,4′ flavondiols and flavonols in the transgenic plants (see below).
Effect of BPBT Gene Silencing on Morphology of Petunia Plants
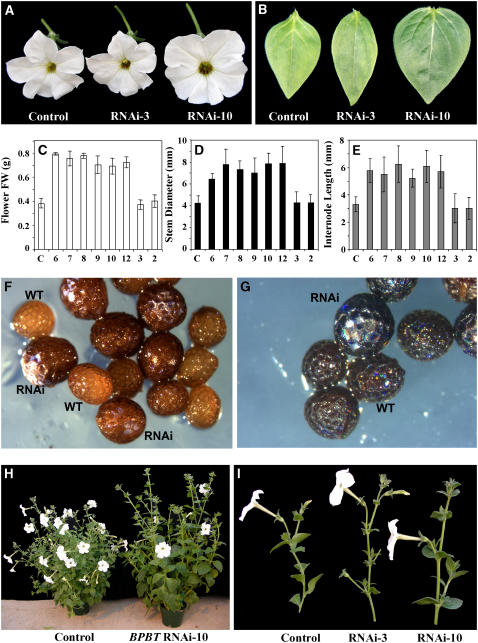
Complete elimination of BPBT transcripts resulted in phenotypic changes in growth and development when compared with control nontransgenic plants and plants with reduced BPBT expression (Figure 7). Flowers from BPBT knockouts were twofold larger based on fresh weight (Figures 7A and 7C) and bloomed at least 1 week later than flowers of control and BPBT knockdown plants. Flowers of BPBT knockouts also had larger anthers, which displayed a delay in pollen release. In addition, seeds of BPBT knockout plants were larger and darker than seeds from controls (Figure 7F). Staining of these seeds with p-dimethylaminocinnamaldehyde (DMACA), which changes to a dark color after binding to 3′,4′ flavon diols, proanthocyanindins, and its precursors, revealed that silencing of the BPBT gene resulted in qualitatively more 3′,4′ flavon diols, flavonols, and soluble flavonoids than in control seeds (Figure 7G). Levels of other flavonoids (primarily quercetin and kaempferol rhamnopyranosides and diglycosides) were slightly increased. However, levels of aglycone flavonols, which function as auxin transport and kinase/phosphatase inhibitors, were not different from nontransformed plants. These phenotypes were heritable and correlated with BPBT suppression in the T1 generation.
Figure 7.
Phenotypical Changes in BPBT RNAi Knockout Petunia Plants.
BPBT RNAi-10 knockout transgenic plants show increased flower size (A) and larger, more rounded leaves (B) than control and BPBT RNAi-3 knockdown plants. Increased flower size is manifested in increased flower weight (C), and this consistently caused a significant increase in all knockouts (6, 7, 8, 9, 10, and 12) in comparison with controls (C) and knockdowns (2 and 3). Knockouts also showed significantly increased stem diameter (D) and internode lengths (E). Each point in (C) to (E) represents the average of 7, 15, and 13 measurements, respectively. Bars indicate sd. These morphological changes are further illustrated in panels (H) and (I). BPBT RNAi-10 seeds are also larger and darker than controls (WT) (F). DMACA staining of both control and RNAi seeds is shown in (G).
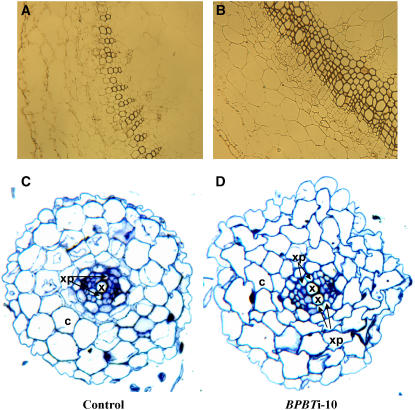
Comparisons of 6-week-old control and transgenic plants revealed that BPBT knockout plants exhibited longer internodes, thicker stems, and larger, more rounded leaves, while transgenic plants with reduced BPBT expression were not different from untransformed control plants (Figures 7B, 7D, 7E, 7H, and 7I). The average internode length and stem diameter were both 1.75-fold greater in BPBT knockouts than in control or BPBTi-2 and -3 plants (Figures 7D and 7E). Histochemical analysis of cross sections of the middle part of the first-order internodes of 3-week-old nonflowering BPBTi-10 plants stained with the Mäule reagent revealed that the increased thickness of knockouts' stems was due to larger cell sizes, increased secondary growth, and a proliferation of the vascular tissue, particularly the xylem (Figures 8A and 8B). Cross sections of the root revealed a doubling of central xylem and xylem pole cells and a loss of uniformity in cortical cells of knockout plants (Figures 8C and 8D).
Figure 8.
Effects of BPBT Gene Knockout on the Histological Structure of Petunia Stems and Roots.
Cross sections of the middle part of the first-order internodes of 3-week-old nonflowering plants, stained with Mäule reagent, showed increased secondary growth and a proliferation of vascular tissue, particularly the xylem in BPBT RNAi-10 transgenic plants (B) in comparison with the control (A). Root cross sections of control (C) and BPBT RNAi-10 (D) transgenic plants showed a doubling of central xylem (x) and xylem pole (xp) cells and a loss of uniformity in cortical cells (c) of knockout plants.
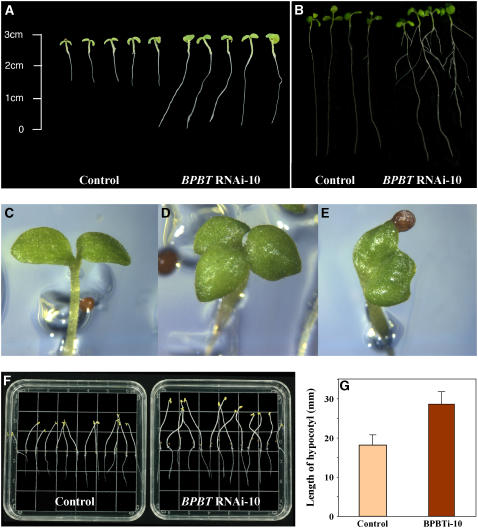
BPBT knockout plants also exhibited altered vegetative growth and development. Under greenhouse conditions, BPBT knockouts produced fewer first-order lateral branches (on average three to four) in contrast with the control plants of the same age with seven to eight branches. Transgenic 5-d-old seedlings grown under normal light/dark (12 h/12 h) conditions on half-strength Murashige and Skoog (MS) medium had longer primary roots (Figure 9A), and 10-d-old seedlings had on average approximately three lateral roots per seedling, while controls had one or none (2.8 ± 0.97 and 0.5 ± 0.62, P < 0.001, respectively; Figure 9B). BPBT knockout seedlings exhibited epinastic cotyledons, and ∼25% of BPBT knockout seedlings exhibited fused or supernumerary cotyledons (Figures 9C to 9E). Moreover, hypocotyl length in etiolated BPBT knockout seedlings was 1.6-fold greater than in controls (Figures 9F and 9G).
Figure 9.
Effect of BPBT Gene Silencing on Morphology of Petunia Seedlings.
Five-day-old (A) and 10-d-old (B) control and BPBT RNAi-10 seedlings grown under normal light/dark (12 h/12 h) conditions on half-strength MS medium. Five-day-old knockout seedlings exhibited fused (E) or supernumerary (D) cotyledons more frequently than controls (C). Etiolated 10-d-old seedlings of BPBT RNAi-10 grown on half-strength MS medium show elongated hypocotyls ([F] and [G]) in comparison with the control. Values are the average of 13 measurements. Bars indicate sd.
As altered flavonoid production was observed in seeds of BPBT knockouts and increased phenylpropanoid intermediates observed in BPBT flowers, total flavonols in methanolic extracts from seedling shoot tissues were analyzed as described previously (Murphy et al., 2000; Peer et al., 2001). Flavonol levels in seedling shoot tissues of BPBT knockouts were not different from the wild type.
Tissue-Specific Activity of Native BPBT and Lis Promoters
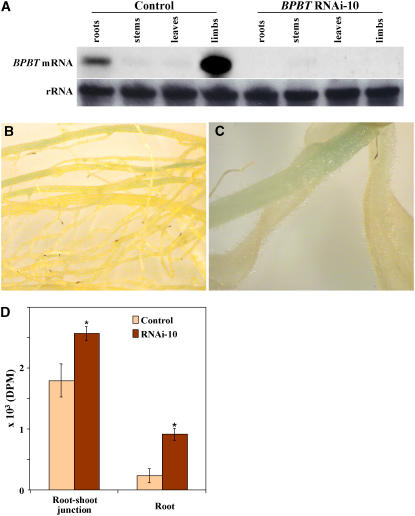
Differences in growth and development between BPBT RNAi knockouts and control plants were unexpected for at least two reasons. First, it was previously shown that BPBT has petal-specific expression, and second, the petal-specific (in Clarkia breweri) Lis promoter was used to silence the BPBT gene. However, expression of neither gene, BPBT or LIS, was previously analyzed in root or stem tissues (Dudareva et al., 1996; Boatright et al., 2004). To examine whether BPBT is expressed in these tissues, RNA gel blot analysis was performed with mRNA isolated from petunia roots, stems, and leaves as well as corolla limbs for comparison. Hybridization with a BPBT probe revealed that in addition to strong expression in corolla limbs, as previously reported (Boatright et al., 2004), BPBT was also expressed in roots but at a relatively low level (Figure 10A). The absence of BPBT expression in roots of BPBT RNAi knockouts (Figure 10A) suggested that the Lis promoter might be active in petunia roots. To analyze the spatial activity of the C. breweri Lis gene promoter, transgenic petunia plants containing the Lis promoter fused with a β-glucuronidase (GUS) reporter gene were generated and analyzed for Lis-driven GUS activity in roots and stems. Histochemical analysis of four independent Lis-GUS transgenic plants revealed the presence of low levels of GUS activity in roots (Figure 10B) and basal parts of the stems (Figure 10C) of these plants.
Figure 10.
BPBT Expression, Spatial Activity of the C. breweri Lis Gene Promoter, and Auxin Transport in Petunia.
(A) Representative RNA gel blot hybridization with total RNA (7 μg per lane) isolated from roots, stems, leaves, and corollas of 2-d-old flowers of control and BPBT RNAi-10 petunia plants. A coding region of the BPBT genes was used as a probe. Autoradiography was performed overnight. The blot was rehybridized with an 18S rDNA probe (bottom gel) to standardize samples.
(B) and (C) Histochemical analysis of GUS activity in four independent Lis-GUS transgenic plants revealed the presence of low levels of Lis-GUS expression in roots (B) and basal parts of the stems (C) of these plants.
(D) Auxin transport in roots and root-shoot junctions of 5-d-old control and BPBT RNAi-10 seedlings. Radiolabeled 3H-auxin was applied to the shoot tip, and its movement to the root-shoot transition zone and root tip over a 4-h period was monitored. Asterisks indicate significance determined by Student's t test (P < 0.02; n = 10). Bars indicate sd. DPM, disintegrations per minute.
Effect of BPBT Gene Silencing on Auxin Transport in Petunia Plants
Increased internode length and the anatomical changes observed in BPBT RNAi knockout plants were consistent with altered auxin transport or synthesis (Liu et al., 1993; Bennett et al., 1995; Al-Hammadi et al., 2003; Multani et al., 2003; Dai et al., 2006). To determine whether auxin transport is altered in BPBT knockouts, radiolabeled auxin (3H-indole-3-acetic acid) was applied to the shoot tip, and its movement to the root-shoot transition zone and root tip over a 5-h period was monitored. Significantly more auxin from the shoot apex reached the root-shoot transition zone and the root tip in BPBT seedlings than in the wild type (P < 0.02; Figure 10D).
DISCUSSION
The Benzenoid Network in Petunia Petals and the Role of BPBT
The biosynthesis of benzenoid compounds from a phenylpropanoid precursor requires the shortening of the propyl side chain by two carbons; however, the specific chain-shortening reactions have not yet been elucidated. We previously used stable isotope labeling in combination with metabolic flux analysis to investigate the possible routes leading to benzenoids in petunia flowers (Boatright et al., 2004). The model constructed based on experimental data obtained in the dark under an inverted photoperiod revealed that two routes, β-oxidative and non-β-oxidative, operate in petunia flowers. Under these conditions, the flux through the non-β-oxidative pathway with benzaldehyde as a key intermediate was estimated to be twofold higher than the flux through the β-oxidative pathway with benzoyl-CoA as an intermediate. Another conclusion from the analysis of these data was that benzylbenzoate is a precursor of BA in petunia petals. Although benzoyl-CoA can presumably be directly hydrolyzed to BA by a thiosterase, it appears that in the petunia petals, benzoyl-CoA is mostly converted to benzylbenzoate by BPBT (Boatright et al., 2004) and a portion of this ester is then hydrolyzed to contribute to the BA pool. Due to this unexpected role of benzylbenzoate and, therefore, BPBT in BA biosynthesis, we generated transgenic petunia plants with decreased BPBT activity. Flowers of transgenic and control plants were subjected to stable isotope labeling with 2H5-Phe and metabolic flux analysis. Since formation and emission of phenylpropanoid and benzenoid compounds from petunia flowers, including benzylbenzoate, occurs rhythmically and reaches a maximum during the night, it is likely that the contribution of benzylbenzoate to BA biosynthesis depends on the light/dark cycle. Thus, labeling experiments were performed under both natural light and dark conditions, resulting in the generation of two models of the benzenoid network for both control and transgenic species. The detection of more endogenous compounds and our recent identification of two additional scent biosynthetic enzymes (PAAS and IGS1) (Kaminaga et al., 2006; Koeduka et al., 2006) extended the original model and clarified previously unknown biochemical steps. Moreover, the generation of both light and dark models allowed us to confirm our prediction that benzylbenzoate contribution to BA biosynthesis is light-dependent.
The general conclusions drawn from the new models were consistent with those drawn from the previous model. However, the newly generated models showed that in control nontransgenic plants, the flux ratio between the oxidative and nonoxidative pathways was significantly higher in the light than in the dark (v13/v20 = 2.26 versus 0.03 in the light and dark, respectively) (Figures 5A and 5C, Table 1), indicating that there is a greater contribution of benzylbenzoate to BA biosynthesis in the light. Although contribution of the β-oxidative pathway to BA biosynthesis appeared to be lower in the dark than in the light, it was still required to account for the observed labeling patterns of BA and endogenous and exogenous benzyl alcohol (Figure 6; see Supplemental Figure 1 online). Comparison of the light and dark models in nontransgenic plants also showed that in the light there was a rapid turnover of benzylbenzoate (v12-v15, v58, and v59) in contrast with a significant expansion of its pool in the dark, despite a higher emission rate (v58 and v59).
Generation of transgenic plants with reduced/eliminated benzylbenzoate formation confirmed the model prediction that benzylbenzoate is a precursor of BA. The level of free BA in transgenic plants was lower than in control plants (Figures 4 and 6) in both the light and the dark, and a substantial decrease in emission of methylbenzoate was observed. Interestingly, this decrease was achieved differently in the light and dark. In the light, although the flux from benzaldehyde to BA (v20) was increased in transgenics, the total flux to BA decreased due to elimination of fluxes from benzylbenzoate (v13) and phenylethylbenzoate (v34) to BA biosynthesis. By contrast, during the dark period, in addition to the elimination of fluxes from benzylbenzoate (v13) and phenylethylbenzoate (v34) to BA biosynthesis, the decrease in BA pool and methylbenzoate emission was also the result of a decrease in flux from benzylaldehyde to BA (v20).
A quantitative analysis of the 2H5-Phe labeling kinetics of internal pools and emitted volatiles in nontransgenic plants also revealed that there is a dilution of their isotopic abundances in the dark compared with the light (Figure 6; see Supplemental Figure 1 online), suggesting the contribution of flux from a precursor with low or no labeling. The compounds with closest proximity to Phe, which exhibited the reduced isotopic abundance, were benzaldehyde and benzyl alcohol, followed by their immediate products. A search of the literature revealed that benzaldehyde could be nonenzymatically formed from phenylpyruvate (Nierop-Groot and De Bont, 1998; Lapadatescu et al., 2000; Rijnen et al., 2000; Lomascolo et al., 2001). Feeding of 2H5-Phe to petunia petals led to a 25% labeling of phenylpyruvate over a 4-h period, indicating that Phe can be converted to phenylpyruvate, as was recently demonstrated in etiolated Arabidopsis thaliana seedlings (Warpeha et al., 2006). Incorporation of flux from phenylpyruvic acid to benzaldehyde and benzyl alcohol in the model accounted for the reduced labeling of these two compounds as well as that of downstream compounds in the dark.
Knockout of BPBT expression led to the redistribution of flux from the non-Phe source, presumably phenylpyruvic acid, to benzaldehyde and benzyl alcohol in the dark (Figures 5C and 5D), causing an expansion of their endogenous pools and an increase in their emission (Figure 6; see Supplemental Figure 1 online). This redistribution in flux could be the result of possible feedback regulation of benzyl alcohol biosynthesis from phenylpyruvate by benzyl alcohol, whose internal pool was expanded due to the elimination of flux to benzylbenzoate. The reason for the contribution of phenylpyruvic acid only in the dark remains unknown; however, we do not exclude the possibility of the involvement of pH and oxygen in influencing benzaldehyde synthesis from phenylpyruvate, as has been demonstrated in Lactococcus lactis (Rijnen et al., 2000), nor can we exclude the possibility of the existence of alternative pathways independent of Phe and phenylpyruvate. A similar dilution of 2-phenylethanol labeling occurred in the dark, possibly due to the contribution of flux from phenylpyruvic acid via phenyllactic acid (Watanabe et al., 2002) leading to a subsequent dilution of phenylacetaldehyde, which was more rapidly labeled in the light (see Supplemental Figure 1 online).
It should be noted that in these feeding experiments, we are likely measuring maximum velocities, which may not correspond to fluxes in vivo. A comparison of emission rate on a plant with that in feeding experiments with an exogenous precursor can provide useful information about potential rate-limiting steps. Indeed, the overall level of volatile emission was higher in Phe-fed, excised corollas, especially in the light, indicating that the level of Phe in the cell plays a crucial role in the regulation of the formation of benzenoid compounds in petunia flowers (data not shown).
The analyses presented here and earlier (Boatright et al., 2004) illustrate the concept of an integrative cycle of metabolic flux analysis followed by metabolic engineering, which together can lead to increased understanding of the complexity of metabolic networks. Additional genetic perturbations based on the data presented here are likely to reveal new information that will in turn lead to further refinements in the flux models.
The Role of BPBT in Vegetative Tissues
The altered growth phenotypes observed in multiple independent BPBT knockout lines were consistent and more pronounced than in knockdown lines, which suggests that BPBT is involved in metabolic processes in vegetative and floral tissues. Elongated internodes (Figures 7E, 7H, and 7I) and increased lateral root number (Figure 9B) found in knockout lines both correlate with increased auxin transport (Noh et al., 2001; Li et al., 2005) and are consistent with the observed increase in auxin transport capacity of BPBT knockout plants (Figure 10D). Altered vascular tissue structure and increased cell proliferation seen in BPBT knockouts (Figure 8) were similar to what is seen in the br2/pgp1 mutant in maize (Zea mays) (Multani et al., 2003). Moreover, similar morphological changes have been found to correlate with accumulation of auxin in localized regions in br2 tissues (A. Knoeller, J.J. Blakeslee, and A.S. Murphy, unpublished data).
Epinastic, fused, and supernumerary cotyledons similar to those seen in BPBT knockouts (Figures 9C to 9E) are often associated with defects in auxin transport. Epinastic cotyledons observed in the pgp19 auxin transport mutant of Arabidopsis result from increased auxin accumulation in cotyledons and can be phenocopied by application of exogenous auxin (Noh et al., 2001; W.A. Peer and A.S. Murphy, unpublished data). Fused and supernumerary cotyledons have been associated with auxin accumulation resulting from localized increases or decreases in auxin transport as is seen in multiple Arabidopsis mutants (Bennett et al., 1995; Swarup et al., 2004; Dai et al., 2006; Kuroha et al., 2006) and the polycotyledon mutant of tomato (Solanum lycopersicum) (Al-Hammadi et al., 2003). Alterations in filament length and anther size seen in BPBT knockouts are also seen in auxin transport mutants (Noh et al., 2001; Al-Hammadi et al., 2003). Loss of BPBT function might be expected to modulate auxin-dependent growth by altering accumulations of phenylpropanoid compounds, which are shown to inhibit transport protein activity, interfere with auxin action, enhance apoplastic acidification, and/or contribute to cell wall structure.
Suppression of BPBT activity increases phenylpropanoid intermediate levels in flowers and results in high flavonoid content in the BPBT transgenic testa (Figure 7G), which could be sufficient to alter auxin-dependent development in the embryonic cotyledons (Kakiuchi et al., 2006). However, BPBT suppression did not affect the concentration or distribution of flavonols in seedlings, except in the root-shoot transition zone (see Supplemental Figure 2 online), where changes in flavonol levels do not appear to alter polar auxin transport (Peer et al., 2004).
While trans-cinnamic acid has no apparent auxin-like activity (Thimann, 1938), it has been shown to competitively interfere with auxin signaling at low micromolar concentrations (van Overbeek et al., 1951). We found an ∼1.4-fold increase in trans-cinnamic acid levels in flowers of BPBT knockout plants under both light and dark conditions (see Supplemental Figure 1 online). However, we were unable to detect differences in CA levels between embryos of knockout and control plants. As such, more detailed studies will be needed to determine whether trans-cinnamic acid accumulations directly regulate development and auxin transport in BPBT knockouts.
Another possibility for increased auxin transport in BPBT knockouts is apoplastic acidification by increased organic acid production. According to chemiosmotic models, auxin transport depends on the pH gradient across the plasma membrane (Goldsmith and Goldsmith, 1977; Li et al., 2005). Analysis of apoplastic root pH using bromocresol purple dye suggested that there was no obvious increase in root apoplastic pH (data not shown).
Altered auxin transport has been seen in Arabidopsis mutants exhibiting aberrant vascular structure (Zhong and Ye, 2001; Surpin et al., 2003; Blakeslee et al., 2005). BPBT knockout shoots and roots showed increased size and number of auxin-conducting vascular parenchyma cells as well as altered cortical cell shape (Figure 8). Staining of BPBT knockout roots with Toluidine Blue O did not reveal any obvious differences in the distribution of lignin or pectin, but BPBT knockout roots were more permeable to bromocresol purple dye than controls (data not shown), suggesting altered cell wall composition or structure. Cinnamic and p-coumaric acids have been shown to affect membrane permeability and fluidity (Castelli et al., 1999). Both characteristics would affect rates of indole-3-acetic acid partitioning into membranes and interactions with PGP and PIN transporters that are thought to bind their substrates within the inner membrane leaflet (Blakeslee et al., 2005). Thus, the developmental phenotypes and increased auxin transport observed in BPBT knockouts are more likely a consequence of vascular tissue expansion resulting from modified cell wall structure than a direct modulation of cellular auxin transport processes by loss of BPBT. However, it is possible that altered metabolic fluxes in BPBT knockouts could result in increased auxin production and consequent developmental changes. We also cannot exclude the possibility that BPBT RNAi silencing affects another acyltransferase, which may in some unknown way impact auxin metabolism. Further analysis of BPBT activity in vegetative phenylpropanoid metabolism is required to determine its exact function in these tissues.
METHODS
Plant Material and Transformation
Petunia hybrida cv Mitchell (Ball Seed) and the transgenic petunia plants were grown under normal greenhouse conditions and in vitro to provide material of the same age and physiological conditions for experiments. Transgenic petunias were obtained via Agrobacterium tumefaciens (strain GV3101 carrying plasmid pEF1.LIS-BEBTi) leaf disk transformation using the standard leaf disk transformation method (Horsch et al., 1985). Rooted plants were screened for the presence of the LIS promoter by PCR with the specific primers LIS-F (5′-GGCACCCACTTCTTAATGATC-3′) and LIS-R (5′-CTGGGATATGATAGGATGTGG-3′) and for the hygromycin resistance ntpII gene with the primers Hyg-F (5′-AAGCCTGAACTCACCGCGAC-3′) and Hyg-R (5′-TCCACTATCGGCGAGTACTTC-3′). PCR-positive plants were transferred to the greenhouse. T0 ant T1 transformants were self-pollinated manually, and obtained seeds were analyzed for segregation by germinating on MS medium supplement with hygromicin (20 mg/L).
Generation of RNAi Silencing Construct
A modified pRNA69 was first constructed in which the 35S promoter was replaced with the LIS promoter (a SacI-XhoI fragment). Next, the RNAi construct for BPBT was generated by first splicing a BPBT fragment from nucleotide 159 to nucleotide 509 of the coding region in direct orientation into the polylinker site downstream of the intron in the pRNA69 vector. Next, the same BPBT fragment was spliced in the inverted orientation into the polylinker site upstream of the intron in this vector. Finally, the section of this constructed plasmid that contained the LIS promoter and the two BPBT fragments in opposite orientation separated by the intron was cut out using SacI-PstI and spliced into a modified pCAMBIA 1303 that had previously been digested with PmlI and HindIII, treated with Klenow, and self-ligated.
Morphological Analysis of Transgenic Plants
To estimate the phenotypic differences between the BPBT knockouts and control petunia plants, 6-week-old plants were measured to calculate average length of the internodes, stem diameters, and fresh weight of flowers. To measure the number and length of the lateral roots, the sterilized seeds were germinated on solid half-strength MS medium, pH 5.2, without antibiotics for 10 d. Three repetitions of 30 seedlings of each variant were analyzed, plates were scanned, and images were analyzed with Scion Image for Windows (http://www.scioncorp.com) to measure length of lateral roots.
Sampling Volatiles
Floral volatiles were collected from control and BPBT RNAi flowers, using a closed-loop stripping method (Donath and Boland, 1995; Dudareva et al., 2005) under growth chamber conditions (21°C, 50% relative humidity, 150 μmol·m−2.s−1 light intensity, and 12-h photoperiod). To determine the internal pools of volatiles, 1 g of 2-d-old corolla tissue was collected from each transgenic and control plant at the same time of day to minimize the effect of rhythmicity. Volatiles were extracted with 10 mL of dichloromethane, concentrated to 160 mL, and analyzed by GC-MS (Boatright et al., 2004).
BPBT Enzyme Assays
Crude protein extracts were prepared from corollas of 2-d-old control and transgenic petunia flowers and analyzed for BPBT activity as described previously (Boatright et al., 2004).
Labeling Experiment and Metabolic Flux Analysis
Feeding experiments were performed as described previously (Boatright et al., 2004). Corollas from 2-d-old control and BPBT knockout flowers were placed on moist filter paper supplied with deuterium-labeled Phe (l-Phe-ring-2H5; Cambridge Isotope Laboratories) (total of 10 corollas per each experiment), and emitted volatiles and internal pools were collected after 30, 60, 120, and 240 min. Labeling experiments under light conditions always started at 9 am and under dark conditions at 11 pm. Computer modeling of labeling data was performed as described by Boatright et al. (2004).
Organic and Amino Acid Analyses
For internal pool, organic acid, and amino acid analyses, flower petal tissue was collected after each feeding, frozen in liquid nitrogen and stored at −80°C. For organic and amino acids, 0.5 g of frozen tissue was extracted overnight with 10 mL of 100% methanol. Then, 5 mL of chloroform and 6 mL of deionized water were added to each vial, as well as 25 μL of each internal standard (10 mM Hyp for amino acid fraction and 10 mM 4-chlorobenzoic acid for organic acid fraction). After shaking, vials were stored at 4°C overnight to allow separation of layers. The aqueous layer was removed, dried, and then redissolved in 4 mL of deionized water. Three milliliters were used for organic acid analysis and 1 mL for amino acid analysis. For organic acid analysis, 20 μL of 1 n HCl and 3 mL of ethyl acetate were added, and the sample was vortexed for 1 min and centrifuged for 2 min at 2000 rpm. The ethyl acetate fraction was transferred to a clean vial and the above extraction was repeated. The ethyl acetate fraction was then dried, redissolved in 400 μL of 100% methanol, and dried again. For derivatization, 20 μL of pyridine and 250 μL of BSTFA were added to the sample and incubated at 75°C for 2 h. Samples were then analyzed by GC-MS.
For the amino acid analysis, the 1 mL of remaining extract underwent ion exchange chromatography using a Dowex 50-H+ × 8, 200 mesh column. Amino acids were eluted from the Dowex column with 6 mL of 6 M NH4OH, dried, dissolved in 0.4 mL of 60% methanol, and dried again. For derivatization, 100 μL of methylene chloride was added to a sample, which was then dried. After adding 0.2 mL of a 5:1 (v/v) isobutanol:acetyl chloride mixture, samples were heated for 20 min at 120°C and then dried again. An aliquot of 100 μL of heptafluorobutyric anhydride was added, and samples were incubated at 120°C for 10 min. Samples were then cooled to room temperature and brought to incipient dryness to minimize loss of the more volatile amino acid derivatives. Finally, 200 μL of a 1:1 (v/v) ethyl acetate:acetic anhydride mixture was added, and samples were analyzed by GC-MS as described previously (Boatright et al., 2004).
RNA Isolation and Analysis
Total RNA was isolated from petal limbs, leaves, stems, and roots of P. hybrida cv Mitchell and BPBT RNAi transgenics and analyzed as previously described (Boatright et al., 2004). A 1.9-kb EcoRI fragment containing the coding region of the BPBT gene was used as a probe in RNA gel blot analysis. Seven micrograms of total RNA were loaded in each lane. The blots were rehybridized with 18S rDNA for loading control.
Light Microscopy
Stem cross sections (10-μm thick) were prepared from paraffin-embedded samples. The sections were first deparaffinized followed by staining in 0.05% (w/v) aqueous solution of Toluidine Blue O or Mäule reagent (Chapple et al., 1992), rinsed, and examined using bright-field microscopy. Images were recorded using an Olympus Vanox-S AH-2 microscope equipped with a digital imaging system. Root samples from 10-d-old etiolated seedlings were fixed in formaldehyde (10% [v/v]) in phosphate buffer, pH 7.0 (Ricca Chemical Company), washed with 0.1 M potassium phosphate buffer, pH 6.8, dehydrated, and embedded in Spurr's resin using a low viscosity embedding kit (Electron Microscopy Sciences). Root cross sections (1-μm thick) were stained with TBO.
Auxin Transport Analysis
Plants were grown as previously described (Murphy and Taiz, 1995). Auxin transport assays were conducted as described by Geisler et al. (2005).
pH Determination
As previously described by Massoneau et al. (2001), the root systems of the different plants were spread between two layers of 1% agar gel containing bromocresol purple 0.01% (w/v) as a pH indicator. The pH of the gel was adjusted between 6 and 6.5 to visualize change of color: yellow for the acidification and purple of the alkalinization. Vacuolar pH was examined as by Li et al. (2005).
Flavonoid Staining
Diphenylboric acid 2-aminoethyl ester staining was performed as previously described (Murphy et al., 2000; Peer et al., 2001). DMACA staining was performed as described by Baxter et al. (2005). HPLC analysis of flavonoids in methanolic extracts was performed as described by Peer et al. (2001).
Statistical Analysis
Student's t test and analysis of variance followed by Newman-Keuls post-hoc analysis were preformed using SigmaStat (Jandel Scientific).
GUS Analysis
Transgenic petunia plants containing the Lis-GUS construct were generated via Agrobacterium (strain GV3101) transformation using the standard leaf disk transformation protocol (Horsch et al., 1985). GUS staining for morphogenic callus, primary regenerants, stems, leaves, roots, and flowers was performed as described by Jefferson et al. (1987).
Accession Numbers
Sequence data for BPBT cDNA and the LIS promoter from this article can be found in the GenBank nucleotide database under the accession numbers AY611496 and AF067601.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Metabolic Modeling of in Vivo Labeling Kinetics and Pool Sizes of Benzenoid and Phenylpropanoid Compounds in Petal Tissues of Control and BPBT RNAi-10 Petunia Flowers Supplied with 2H5-Phe for up to 4 h in the Dark and in the Light.
Supplemental Figure 2. Flavonoid Fluorescence in 5-d-Old Control and BPBT RNAi-10 Petunia Seedlings Stained with Diphenylboric Acid 2-Aminoethyl Ester.
Supplementary Material
Acknowledgments
We thank Yasuhisa Kaminaga for help with detection of enzyme activities. This work was supported by the National Science Foundation/USDA–National Research Initiative Interagency Metabolic Engineering Program, Grants MCB 0331333 (N.D.) and MCB 0331353 (E.P.), and by a grant from the Fred Gloeckner Foundation to N.D. This article is contribution No. 17981 from the Purdue University Agricultural Experiment Station.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Natalia Dudareva (dudareva@purdue.edu).
Online version contains Web-only data.
References
- Ahmed, M.A., El-Mawla, A., and Beerhues, L. (2002). Benzoic acid biosynthesis in cell cultures of Hypericum androsaemum. Planta 214 727–733. [DOI] [PubMed] [Google Scholar]
- Al-Hammadi, A.S., Sreelakshmi, Y., Negi, S., Siddiqi, I., and Sharma, R. (2003). The polycotyledon mutant of tomato shows enhanced polar auxin transport. Plant Physiol. 133 113–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter, I.R., Young, J.C., Armstrong, G., Foster, N., Bogenschutz, N., Cordova, T., Peer, W.A., Hazen, S.P., Murphy, A.S., and Harper, J.F. (2005). A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102 2649–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, S.R.M., Alvarez, J., Bossinger, G., and Smyth, D.R. (1995). Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 8 505–520. [Google Scholar]
- Beuerle, T., and Pichersky, E. (2002). Purification and characterization of benzoate:coenzyme A ligase from Clarkia breweri. Arch. Biochem. Biophys. 400 258–264. [DOI] [PubMed] [Google Scholar]
- Blakeslee, J.J., Peer, W.A., and Murphy, A.S. (2005). Auxin transport. Curr. Opin. Plant Biol. 8 494–500. [DOI] [PubMed] [Google Scholar]
- Boatright, J., Negre, F., Chen, X., Kish, C.M., Wood, B., Peel, G., Orlova, I., Gang, D., Rhodes, D., and Dudareva, N. (2004). Understanding in vivo benzenoid metabolism in petunia petal tissue. Plant Physiol. 135 1993–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelli, F., Uccella, N., Trombetta, D., and Saija, A. (1999). Differences between coumaric and cinnamic acids in membrane permeation as evidenced by time-dependent calorimetry. J. Agric. Food Chem. 47 991–995. [DOI] [PubMed] [Google Scholar]
- Chapple, C.C.S., Vogt, T., Ellis, B.E., and Somerville, C.R. (1992). An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 4 1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cseke, L., Dudareva, N., and Pichersky, E. (1998). Structure and evolution of linalool synthase. Mol. Biol. Evol. 15 1491–1498. [DOI] [PubMed] [Google Scholar]
- Dai, Y., Wang, H., Li, B., Huang, J., Liu, X., Zhou, Y., Mou, Z., and Li, J. (2006). Increased expression of MAP KINASE KINASE7 causes deficiency in polar auxin transport and leads to plant architectural abnormality in Arabidopsis. Plant Cell 18 308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donath, J., and Boland, W. (1995). Biosynthesis of acyclic homoterpenes: Enzyme selectivity and absolute configuration of the nerolidol precursor. Phytochemistry 39 785–790. [Google Scholar]
- Dudareva, N., Andersson, S., Orlova, I., Gatto, N., Reichelt, M., Rhodes, D., Boland, W., and Gershenzon, J. (2005). The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. Proc. Natl. Acad. Sci. USA 102 933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva, N., Cseke, L., Blanc, V.M., and Pichersky, E. (1996). Evolution of floral scent in Clarkia: Novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant Cell 8 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva, N., and Pichersky, E. (2006). Floral scent metabolic pathways: Their regulation and evolution. In Biology of Floral Scent, N. Dudareva and E. Pichersky, eds (Boca Raton, FL: CRC Press), pp. 55–78.
- Geisler, M., et al. (2005). Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 44 179–194. [DOI] [PubMed] [Google Scholar]
- Goldsmith, M.H.M., and Goldsmith, T.H. (1977). Chemiosmotic model for polar transport of auxin through plant tissue. Biophys. J. 17 A260. [Google Scholar]
- Horsch, R.B., Fry, J.E., Hofman, N.L., Elichholz, D., Rogers, S.G., and Fraley, R.T. (1985). A simple and general method for transferring genes into plants. Science 227 1229–1231. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, E., Zamir, L.O., and Jensen, R.A. (1986). Chloroplasts of higher plants synthesize L-phenylalanine via L-arogenate. Proc. Natl. Acad. Sci. USA 83 7231–7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakiuchi, Y., Gàlis, I., Tamogami, S., and Wabiko, H. (2006). Reduction of polar auxin transport in tobacco by the tumorigenic Agrobacterium tumefaciens AK-6b gene. Planta 223 237–247. [DOI] [PubMed] [Google Scholar]
- Kaminaga, Y., et al. (2006). Plant phenylacetaldehyde synthase is a bifunctional homotetrameric enzyme that catalyzes phenylalanine decarboxylation and oxidation. J. Biol. Chem. 281 23357–23366. [DOI] [PubMed] [Google Scholar]
- Knudsen, J.T., and Gershenzon, J. ( 2006). The chemical diversity of floral scent. In Biology of Floral Scent, N. Dudareva and E. Pichersky, eds (Boca Raton, FL: CRC Press), pp. 27–52.
- Koeduka, T., et al. (2006). Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a coniferyl alcohol ester. Proc. Natl. Acad. Sci. USA 103 10128–10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolosova, N., Gorenstein, N., Kish, C.M., and Dudareva, N. (2001). Regulation of circadian methylbenzoate emission in diurnally and nocturnally emitting plants. Plant Cell 13 2333–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroha, T., Ueguchi, C., Sakakibara, H., and Satoh, S. (2006). Cytokinin receptors are required for normal development of auxin-transporting vascular tissues in the hypocotyl but not in adventitious roots. Plant Cell Physiol. 47 234–243. [DOI] [PubMed] [Google Scholar]
- Lapadatescu, C., Ginies, C., Le Quere, J.-L., and Bonnarme, P. (2000). Novel scheme for biosynthesis of aryl metabolites from L-phenylalanine in the fungus Bjerkandera adusta. Appl. Environ. Microbiol. 66 1517–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., et al. (2005). Arabidopsis H+-PPase AVP1 regulates auxin mediated organ development. Science 310 121–125. [DOI] [PubMed] [Google Scholar]
- Liu, C.M., Xu, Z.H., and Chua, N.-H. (1993). Auxin polar transport is essential for the establishment of bilateral symmetry during early plant embryogenesis. Plant Cell 5 621–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomascolo, A., Asther, M., Navarro, D., Antona, C., Delattre, M., and Lesage-Meessen, L. (2001). Shifting the biotransformation pathways of L-phenylalanine into benzaldehyde by Trametes suaveolens CBS 334.85 using HP20 resin. Lett. Appl. Microbiol. 32 262–267. [DOI] [PubMed] [Google Scholar]
- Massonneau, A., Langlade, N., Leon, S., Smutny, J., Vogt, E., Neumann, G., and Martinoia, E. (2001). Metabolic changes associated with cluster root development in white lupin (Lupinus albus L.): Relationship between organic acid excretion, sucrose metabolism and energy status. Planta 213 534–542. [DOI] [PubMed] [Google Scholar]
- Multani, D.S., Briggs, S., Chamberlin, M.A., Blakeslee, J.J., Murphy, A.S., and Johal, G. (2003). Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 302 81–84. [DOI] [PubMed] [Google Scholar]
- Murphy, A., and Taiz, L. (1995). A new vertical mesh transfer technique for metal-tolerance studies in Arabidopsis (ecotypic variation and copper-sensitive mutants). Plant Physiol. 108 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, A.S., Peer, W.A., and Taiz, L. (2000). Regulation of auxin transport by aminopeptidases and endogenous flavonoids. Planta 211 315–324. [DOI] [PubMed] [Google Scholar]
- Negre, F., Kish, C.M., Boatright, J., Underwood, B., Shibuya, K., Wagner, C., Clark, D.G., and Dudareva, N. (2003). Regulation of methylbenzoate emission after pollination in snapdragon and petunia flowers. Plant Cell 15 2992–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierop-Groot, M.N., and De Bont, J.A.M. (1998). Conversion of phenylalanine to benzaldehyde initiated by an aminotransferase in Lactobacillus plantarum. Appl. Environ. Microbiol. 64 3009–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh, B., Murphy, A.S., and Spalding, E.P. (2001). Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13 2441–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer, W.A., Bandyopadhyay, A., Blakeslee, J.J., Makam, S.N., Chen, R.J., Masson, P.H., and Murphy, A.S. (2004). Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 16 1898–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer, W.A., Brown, D.E., Tague, B.W., Muday, G.K., Taiz, L., and Murphy, A.S. (2001). Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis. Plant Physiol. 126 536–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky, E., Noel, J.P., and Dudareva, N. (2006). Biosynthesis of plant volatiles: Nature's diversity and ingenuity. Science 311 808–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribnicky, D.M., Shulaev, V., and Raskin, I. (1998). Intermediates of salicylic acid biosynthesis in tobacco. Plant Physiol. 118 565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijnen, L., Courtin, P., Gripon, J.-C., and Yvon, M. (2000). Expression of a heterologous glutamate dehydrogenase gene in Lactococcus lactis highly improves the conversion of amino acids to aroma compounds. Appl. Environ. Microbiol. 66 1354–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnepp, J., and Dudareva, N. (2006). Floral scent: Biosynthesis, regulation, and genetic modifications. In Flowering and Its Manipulation, C. Ainsworth, ed (Oxford, UK: Blackwell Publishing), pp. 240–257.
- Surpin, M., Zheng, H., Morita, M.T., Saito, C., Avila-Teeguarden, E., Blakeslee, J.J., Bandyopadhyay, A., Kovaleva, V., Carter, D., Murphy, A.S., Tasaka, M., and Raikhel, N. (2003). The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 15 2885–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup, R., et al. (2004). Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell 16 3069–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimann, K.V. (1938). Hormones and the analysis of growth. (Inaugural Steven Hales address December 28, 1937). Plant Physiol. 13 437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Overbeek, J., Blondeau, R., and Horne, V. (1951). Trans-cinnamic acid as an anti-auxin. Am. J. Bot. 38 589–595. [Google Scholar]
- Verdonk, J.C., Ric de Vos, C.H., Verhoeven, H.A., Haring, M.A., van Tunen, A.J., and Schuurink, R.C. (2003). Regulation of floral scent production in petunia revealed by targeted metabolomics. Phytochemistry 62 997–1008. [DOI] [PubMed] [Google Scholar]
- Warpeha, K.M., Lateef, S.S., Lapik, Y., Anderson, M., Lee, B.-S., and Kaufman, L.S. (2006). G-protein-coupled receptor 1, G-protein Gα-subunit 1, and prephenate dehydratase 1 are required for blue light-induced production of phenylalanine in etiolated Arabidopsis. Plant Physiol. 140 844–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, S., Hayashi, K., Yagi, K., Asai, T., MacTavish, H., Picone, J., Turnbull, C., and Watanabe, N. (2002). Biogenesis of 2-2-phenylethanol in rose flowers: Incorporation of [2H8] L-phenylalanine into 2-2-phenylethanol and its β-D-glucopyranoside during the flower opening of Rosa ‘Hoh-Jun’ and Rosa damascena Mill. Biosci. Biotechnol. Biochem. 66 943–947. [DOI] [PubMed] [Google Scholar]
- Wildermuth, M.C. (2006). Variations on a theme: Synthesis and modification of plant benzoic acids. Curr. Opin. Plant Biol. 9 288–296. [DOI] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.H. (2001). Alteration of auxin polar transport in the Arabidopsis ifl1 mutants. Plant Physiol. 126 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.