Abstract
nifU of nitrogen-fixing bacteria is involved in the synthesis of the Fe–S cluster of nitrogenase. In a synthetic lethal screen with the mitochondrial heat shock protein (HSP)70, SSQ1, we identified a gene of Saccharomyces cerevisiae, NFU1, which encodes a protein with sequence identity to the C-terminal domain of NifU. Two other yeast genes were found to encode proteins related to the N-terminal domain of bacterial NifU. They have been designated ISU1 and ISU2. Isu1, Isu2, and Nfu1 are located in the mitochondrial matrix. ISU genes of yeast carry out an essential function, because a Δisu1Δisu2 strain is inviable. Growth of Δnfu1Δ isu1 cells is significantly compromised, allowing assessment of the physiological roles of Nfu and Isu proteins. Mitochondria from Δnfu1Δisu1 cells have decreased activity of several respiratory enzymes that contain Fe–S clusters. As a result, Δnfu1Δisu1 cells grow poorly on carbon sources requiring respiration. Δnfu1Δisu1 cells also accumulate abnormally high levels of iron in their mitochondria, similar to Δssq1 cells, indicating a role for these proteins in iron metabolism. We suggest that NFU1 and ISU1 gene products play a role in iron homeostasis, perhaps in assembly, insertion, and/or repair of mitochondrial Fe–S clusters. The conservation of these protein domains in many organisms suggests that this role has been conserved throughout evolution.
Mitochondria are essential organelles. Many critical metabolic pathways are known to occur partly or entirely within them, including the biosynthesis of many amino acids and nucleotides (1). In addition, the mitochondrial inner membrane contains the electron-transport chain responsible for the production of ATP by oxidative phosphorylation. Many electron-transport proteins, to perform redox reactions, contain iron in their active sites in the form of either Fe–S clusters or heme. Therefore, iron metabolism is also a critical, but poorly understood, aspect of mitochondrial function. The transport and internal metabolism of iron must be tightly regulated, because free iron can interact with oxygen to generate hydroxyl radicals, causing damage to a variety of cellular components. Advances have been made in the elucidation and regulation of iron transport across the plasma membrane in Saccharomyces cerevisiae, however, very little is known about how iron is carried and used within the cell (2).
Recently, Yfh1, a yeast homolog of mammalian frataxin, was found to be important in the efflux of iron from mitochondria (3). Δyfh1 cells accumulate ≈10-fold more iron in their mitochondria than wild-type (wt) cells, have reduced activity of the components of the electron-transport pathway and are sensitive to oxidative stress (4). A strain carrying a mutation in SSQ1, a mitochondrial heat shock protein (HSP)70 homolog, was found to be defective in the maturation of Yfh1 and also accumulated iron in mitochondria (5). In addition, an ssq1 mutant, which suppressed the metabolic defects associated with the absence of the copper/zinc superoxide dismutase (Sod1) was identified (6). This ssq1 mutant was found to have decreased activity of two Fe–S cluster-containing proteins: aconitase and succinate dehydrogenase (SDH).
To better understand the physiological role of Ssq1, we searched for genes that, when mutated, caused lethality in the presence of an SSQ1 deletion. This analysis led to the uncharacterized genes, NFU1, ISU1, and ISU2, which are related to domains of NifU, a protein from nitrogen-fixing bacteria. NifU is a modular protein containing three domains. The N-terminal domain of NifU, which bears homology to the Isu proteins discussed here, binds Fe via three conserved cysteines (7, 8). Nfu1 of yeast is related to the C-terminal domain of NifU. Although a function for this domain of NifU has not been established, it contains a highly conserved pair of cysteines, which may be involved in iron binding.
From the available genome sequences, it is now clear that many organisms, both nitrogen-fixing and non-nitrogen-fixing, possess ORFs that encode for proteins with homology to the individual domains of NifU. This homology suggests that different aspects of NifU function have been conserved throughout evolution of both prokaryotic and eukaryotic cells. Our analyses of the NFU1 and ISU yeast genes suggest the existence of a multicomponent system that functions in mitochondrial iron metabolism and is potentially involved in the synthesis, assembly, and or repair of Fe–S clusters.
MATERIALS AND METHODS
Strains, Media, and Chemicals. PJ53 is isogenic to W303: trp1-1/trp1-1 ura3-1/ura3-1 leu2-3,112/leu2-3,112 his3-11,15/his3-11,15 ade2-1/ade2-1 can1-100/can1-100 GAL2+/GAL2+ met2-Δ1/met2-Δ1 lys2-Δ2/lys2-Δ2 (9). Δssq1, Δnfu1, Δisu1, and Δisu2 are haploid derivatives of PJ53 transformed with Δssq1∷LYS2 (10), Δnfu1∷TRP1, Δisu1∷LEU2, or Δisu2∷HIS3 respectively (see below). Δnfu1Δisu1 is a haploid derivative of PJ53 obtained by dissection of spores from a cross between Δnfu1 and Δisu1 strains. Mitochondria were purified from PK82 (11).
Yeast were grown on YPD, YPGal, YPGly, and YPLac [1% yeast extract, 2% peptone, and 2% glucose, 2% galactose, 3% glycerol, or 1% lactate (pH 5.0), respectively]. Synthetic media were prepared as described (12). For hydrogen peroxide sensitivity testing, 4 mM H2O2 was added to sterilized YPD media before pouring plates.
All chemicals, unless stated otherwise, were purchased from Sigma.
Synthetic Lethal Screen. To perform an ade2 ade3 colony-sectoring assay for synthetic lethality (13), a Δade2Δade3Δssq1 haploid strain was constructed and transformed with pRS316-ADE3-SSQ1, a plasmid carrying ADE3 (3.3 kilobase Bst1107I–NheI fragment), SSQ1 (3.8 kilobase EcoRI fragment), and URA3 (14). This strain was mutagenized by transformation with the yeast∷mTn3lacZLEU2 DNA library digested with NotI (15). Transformants possessing a mutation that requires the presence of the SSQ1 plasmid were identified as colonies that failed to produce white sectors in a red background, indicating that the plasmid carrying ADE3 and SSQ1 was not being lost at a detectable rate. To determine that the requirement for SSQ1 was independent of the plasmid pRS316-ADE3-SSQ1, candidates were transformed with a wt copy of SSQ1 on a TRP1 marked plasmid (pRS314-SSQ1) to show restored sectoring. Tetrad analysis of the candidates crossed with the wt parent were performed to demonstrate 2:2 segregation of the LEU2-marked insertional mutation, verifying that only one recombination event occurred.
The plasmid, pRSQ, was used to rescue the DNA sequence adjacent to the transposon insertion in mutagenized yeast strains as described (16). Yeast DNA flanking the inserted Tn3 element was sequenced by using dideoxy sequencing protocol (17) and the M13 −40 primer.
Construction of Chromosomal Deletions and Tagged Proteins. The genes for Nfu1, Isu1, and Isu2 were amplified from chromosomal DNA by PCR using Pfu polymerase (Stratagene) and cloned into the pRS series of vectors (14). The entirety of the NFU1 (+1 to 771), ISU1 (+1 to 499), and ISU2 (+1 to 471) coding sequences were replaced with the TRP1, LEU2, and HIS3 genes, respectively. The various constructs were transformed as linear DNA fragments into PJ53 to replace one wt copy of the gene by homologous recombination. Correct transformants, and haploids resulting from sporulation, were identified by PCR amplification by using primers homologous to sequences outside the inserted marker genes.
Nfu1, Isu1, and Isu2 were epitope-tagged at their C termini with 3× EQKLISEEDLN (MYC) by using the PCR technique as described (18) except that larger flanking regions of the genes were used. The tag-URA3-tag cassette was amplified by PCR and sewn to DNA sequences flanking the terminal amino acid codon of the desired genes. The final purified PCR product was transformed into a haploid derivative of PJ53. Correct Ura+ transformants were identified by using PCR. Cells having undergone the correct recombination event between the repeated epitopes, which removes the URA3 gene, were identified by patching cells on plates containing 5-fluoroorotic acid. Complete loss of the URA3 gene with the retention of the epitope tag was confirmed by using PCR. Immunoblot analysis of the three MYC-tagged protein strains by using mouse monoclonal c-myc antibodies (clone 9E10) from Boehringer Mannheim and the Renaissance detection kit from New England Nuclear only produced a detectable signal when proteins from purified mitochondria were isolated. To enhance a weak Nfu1-MYC signal, the NFU1-MYC fusion was cloned by PCR onto the high-copy plasmid, pRS426. The Nfu1–MYC fusion protein, expressed from the plasmid, was easily detected.
Translocation of Proteins Into and Fractionation of Mitochondria.
All methods for translocation experiments have been described (10, 11). For submitochondrial localization, mitochondrial protein isolated from a strain expressing myc-tagged Nfu1 (25 μg) encoded on a high-copy plasmid, myc-tagged chromosomal Isu1 (50 μg), or myc-tagged chromosomal Isu2 (100 μg) were analyzed as described (10).
Respiratory Enzyme Assays and Detection. Activities for succinate–cytochrome c oxidoreductase (SDH–Cyt bc1) and NADH–cytochrome c oxidoreductase (NADH–Cyt bc1) were measured in purified mitochondria (11) according to procedures described in ref. 19, except 2,6-dichloroindophenol (DCIP) (extinction coefficient of 21 mM−1⋅cm−1) was used in place of Cyt c. Cytochrome oxidase (Cyt ox) activity was determined as described in ref. 19. except reduced Cyt c (extinction coefficient of 19.2 mM−1⋅cm−1) was prepared as described (20). Cyt bc1 activity was measured according to described procedures (21). Aconitase activity was determined essentially as described (22). Cells were grown to an OD600 of 2–3 in YPGal at 34°C. The equivalent of 20 OD units of cells were harvested, washed, resuspended in 0.3 ml of 20 mM potassium phosphate (pH 7.2), and broken by glass bead homogenization. After a 10-min spin at 16,000 × g, 20 μl of extract was added to 0.28 ml of aconitase reaction buffer [20 mM Tris⋅HCl, pH 7.2/100 mM NaCl/1 mM cis-aconitate (extinction coefficient of 4.88 mM⋅cm−1)], and the decrease in absorbance at 240 nm was measured in a cuvette with a pathlength of 1 mm over time.
Antisera were kindly provided by B. Lemire (Univ. of Alberta) (Sdh1, Sdh2) (23) and B. Trumpower (Dartmouth Medical School) (Cyt c1; Rip1) (24). CoxII-specific monoclonal antiserum was obtained from Molecular Probes.
Measurement of Mitochondrial Iron.
Mitochondria were purified from strains grown at 34°C in YP media containing galactose (11). Iron levels in a suspension of purified mitochondria (25 μl) containing 0.2–0.4 mg of mitochondrial protein in buffer A (600 mM sorbitol/10 mM Tris buffer, pH 7.5) were determined colorimetrically with ferene after wet ashing with ultrapure H2SO4 and H2O2 and neutralization following described procedures (25, 26). Data were normalized to the protein content of the samples. Protein determinations were performed by using the Bio-Rad protein assay with ovalbumin as a standard.
RESULTS
nfu1 Is Synthetic Lethal with ssq1.
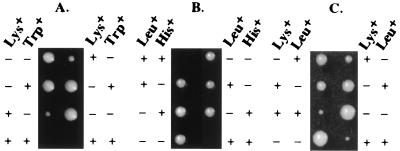
To identify gene products, which either functionally interact or are redundant with Ssq1 in mitochondria, a synthetic-lethal screen was conducted. From ≈12,000 independent insertional mutants, 1 candidate was shown to depend on the presence of SSQ1 for viability. To identify the mutation, the DNA adjacent to the transposon was cloned and sequenced. The insertion is located at nucleotide +246 with respect to the start of translation in the yeast ORF YKL040c, which encodes a 256-aa protein. To verify that inactivation of YKL040c is synthetically lethal with a deletion of SSQ1, a defined YKL040c∷TRP1 deletion was constructed in a diploid strain heterozygous for ssq1∷LYS2. Tetrad analysis showed that all Trp+ Lys+ spores were able to germinate, but stopped growing after a few rounds of replication, reproducing the phenotype of the original insertional mutation (Fig. 1A).
Figure 1.
Tetrad analysis of mutant strains. Growth of spores dissected onto YPD is shown. Plates were replica plated to synthetic media with the indicated amino acids omitted; growth (+) or lack of growth (−). Asci of heterzygous strains Δssq1Δnfu1 at 34°C, (A) Δisu1Δisu2 at 30°C (B), and Δssq1Δisu1 at 34°C (C).
Relationship to Domains of NifU of Nitrogen-Fixing Bacteria. The YKL040c protein is related to the C-terminal domain of NifU from nitrogen-fixing bacteria. Amino acids 155–227 of YKL040c are 32 and 40% identical with the NifU proteins from Azotobacter vinelandii and Anabaena azollae, respectively (Fig. 2A, data not shown). Thus, we have named this ORF NFU1. Caenorhabditis elegans and Rickettsia prowazekii each possess an ORF that shows at least 30% identity to yeast Nfu1 over their entire length, however, the ORFs from S. cerevisiae and C. elegans appear to have N-terminal extensions (Fig. 2A, data not shown).
Figure 2.
Sequence comparisons of Nfu1, Isu1, and Isu2. (A) Alignment of the amino acid sequence (155–245) of Nfu1 (gp:x69584) from S. cerevisiae (Sc) with that of NifU (pid:g762779) of A. azollae (Aa) (230–311), pid:g2635719 of B. subtilis (Bs) (41–111), pid:g3879150 of C. elegans (Ce), and pid:e1342949 of R. prowazekii (Rp). (B) Alignment of IscU homologs from S. cerevisiae, E. coli (Ec) (gp:AE000339), and Homo sapiens (Hs) (gp:U47101). ∗ indicates conserved cysteine residues. Identical residues between at least two of the sequences are indicated by the black boxes with white letters. Gaps that were inserted during the alignment are denoted by dashes. Alignments were performed by using megalign DNAstar (Madison, WI).
The A. vinelandii genome contains not only nifU, which is involved specifically in the maturation of nitrogenase but also genes that are only related to one of the three domains of NifU. These smaller gene products are thought to be involved in the maturation or repair of other Fe–S cluster-containing proteins (8). There are also ORFs in Escherichia coli and Bacillus subtilis that are related only to the C-terminal domain of NifU (Fig. 2A, data not shown), suggesting that there is a conserved function for the C-terminal domain of NifU, which is required for functions other than nitrogen fixation.
The Azotobacter genome also contains a separate ORF, iscU, that is only related to the N terminus of NifU. When we searched the S. cerevisiae database for related sequences, two nifU homologs, YPL135w and YOR226c, were found. For consistency, we have named YPL135w as ISU1 and YOR226c as ISU2. Isu1 and Isu2 are 75% identical to each other and at least 70% identical to proteins from E. coli and human, which suggests an evolutionarily conserved role for Isu proteins (Fig. 2B). There were no yeast ORFs identified as homologs to the middle domain of NifU, which has been shown to bind a 2Fe–2S cluster (6).
Nfu1, Isu1, and Isu2 Are Mitochondrial Proteins Located in the Matrix.
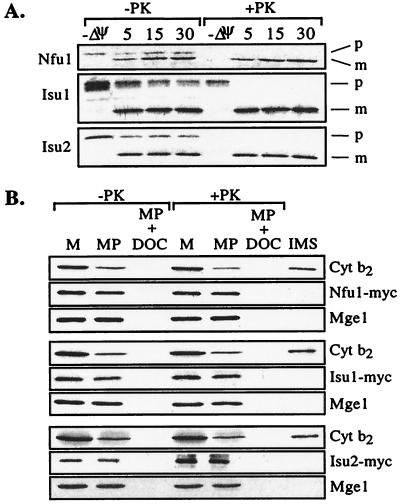
Because NFU1 was identified through its genetic interaction with SSQ1, which encodes a mitochondrial protein, we tested whether the products of these genes were also located in the mitochondria. Mitochondrial localization seemed likely because the Nfu1, Isu1, and Isu2 proteins have N-terminal extensions relative to their bacterial homologs, which had features characteristic of mitochondrial presequences (Fig. 2B, data not shown). To determine whether Nfu1, Isu1, and Isu2 can be imported into mitochondria in vitro, radiolabeled proteins were synthesized in rabbit reticulocyte lysate and added to energized mitochondria. Nfu1, Isu1, and Isu2 were all efficiently translocated into the mitochondria in a reaction dependent on a membrane potential, and they underwent processing to lower-molecular-weight products (Fig. 3A). Therefore, we conclude that Nfu1, Isu1, and Isu2 can be imported into mitochondria. To directly determine their normal cellular location, we constructed versions of these proteins tagged with a myc epitope. Mitochondria purified from strains containing the myc-tagged versions of the proteins were subjected to hypotonic treatment to disrupt the outer membrane. The myc-tagged proteins remained within the mitoplasts, becoming susceptible to proteinase K only after disruption of the inner membrane with detergent (Fig. 3B). Together, these results indicate that Nfu1, Isu1, and Isu2 are proteins of the mitochondrial matrix.
Figure 3.
Location of Nfu1, Isu1, or Isu2 in mitochondria. (A) Translocation of preproteins into isolated mitochondria was carried out in the presence of a membrane potential, Δψ, for 5, 15, and 30 min and in the absence of a membrane potential, −Δψ. PK, proteinase K; p, precursor; m, mature. (B) Mitochondria (M) from cells expressing Myc-tagged Nfu1 (Top), Isu1 (Middle), or Isu2 (Bottom) were separated into mitoplast (MP) and intermembrane space (IMS) fractions. An equivalent portion of mitoplasts was disrupted by addition of deoxycholate detergent (MP + DOC). Equivalent amounts of the fractions were either treated (+PK) or not treated (−PK) with proteinase K. Immunoblot analysis was carried out on the fractions with antibodies specific for the MYC epitope, cytochrome b2 (Cyt b2) as a marker for the intermembrane space, or the matrix protein Mge1 after separation by electrophoresis.
ISU1 and ISU2 Are an Essential Gene Pair.
To determine the consequence of the lack of Isu protein, strains containing defined ISU1 and ISU2 deletions were constructed. Deletion of either ISU1 or ISU2 resulted in cells without any obvious growth defect on rich glucose-based media (data not shown). To determine whether cells required at least one copy of an iscU homolog, a Δisu1∷LEU2 strain was crossed with a Δisu2∷HIS3 strain. No Leu+His+ progeny were recovered after dissection of 20 asci (Fig. 1B), indicating that the expression of at least one of these closely related genes is required for cell viability. Thus, Isu proteins carry out an essential function in mitochondria.
Genetic Interactions Among NFU1, ISU, and SSQ1 Genes.
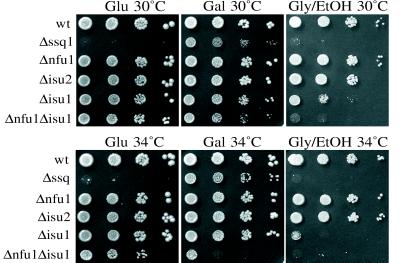
Because we found a synthetic-lethal interaction between SSQ1 and NFU1, we tested whether SSQ1 and/or NFU1 had a genetic interaction with the ISU genes by crossing the Δssq1∷LYS2 and Δnfu∷TRP1 strains with Δisu1∷LEU2 and Δisu2∷HIS3 strains. No difference in growth of the Δssq1Δisu2 double-deletion strain was observed compared with Δssq1. However, tetrad analysis revealed a genetic interaction between SSQ1 and ISU1 because the Lys+Leu+ (Δssq1Δisu1) haploids grew more slowly than Δssq1 cells under all conditions tested (Fig. 1C; data not shown). Genetic interactions were also observed between NFU1 and ISU1 but not between NFU1 and ISU2. The Δnfu1Δisu1 strain is temperature-sensitive on synthetic media, showing reduced growth at 34°C and no growth at 37°C (Fig. 4; data not shown).
Figure 4.
Growth of mutant strains. Each of the strains was serially diluted 1:10 and spotted onto complete synthetic media with either glucose (Glu), galactose (Gal), or glycerol/ethanol (Gly/EtOH) as the carbon source. Plates were grown at the indicated temperatures for 4 days.
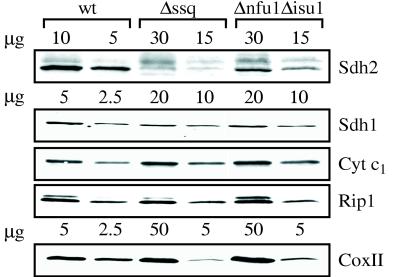
Respiratory Enzyme Activities Are Reduced in Δssq1 and Δnfu1Δisu1. The impaired growth of Δnfu1Δisu1 cells provided a viable genetic background in which to test the possible role of the NFU1 and ISU genes in iron metabolism in yeast. If the NFU1 and ISU genes are involved in Fe–S biogenesis, the absence of one or more should have an affect on the synthesis of those proteins that contain this metal center. When we measured the specific activity of the Fe–S containing aconitase (component of the tricarboxylic acid cycle) and SDH and Cyt bc1 (components of the electron transport chain), we found reduced activity. Aconitase activity was reduced to 36% in Δnfu1Δisu1 cells compared with wt cells (Table 1). In Δnfu1 and Δisu1 cells, activity was 48% and 63% of wt, respectively. Consistent with a previous report (6), aconitase levels were 11% of wt levels in Δssq1 cells. Loss of these proteins also reduced SDH–Cyt bc1 and NADH–Cyt bc1 activities. The Δnfu1Δisu1 and Δssq1 mitochondria possessed only 4–6% of wt SDH–Cyt bc1 activity and maintained wt levels of activity for NADH–Cyt bc1, suggesting a strong defect in SDH activity (Table 1). A direct assay of Cyt bc1 (ubiquinone–Cyt bc1) activity showed that Δnfu1Δisu1 and Δssq1 mitochondria maintained only 55 and 28% of wt activity, respectively (Table 1). Therefore, the Δnfu1Δisu1 and Δssq1 mutations have a more dramatic effect on SDH activity than Cyt bc1 activity. The other single mutants showed less than a 50% decrease in SDH–Cyt bc1 activity and no decrease in Cyt bc1 activity (Table 1). To determine whether the reduced activities observed in the Δssq1 and Δnfu1Δisu1 strains were caused by inactive proteins or reduced protein levels, immunoblot analyses were performed by using antibodies specific to Sdh1 and Sdh2 of the SDH complex and Cyt1 and Rip1 of the Cyt bc1 complex (Fig. 5). Δnfu1Δisu1 and Δssq1 mitochondria showed reduced levels of all proteins examined. Compared with wt mitochondria, mutant mitochondria had only 10% of Sdh2 (the Fe–S containing subunit), 25% of Sdh1, and 50% of Rip1 (the Fe–S-containing subunit) and Cyt1. Protein levels were normal in the single-deletion strains (data not shown). Therefore, much of the reduction in activity can be attributed to the absence of the proteins.
Table 1.
Respiratory enzyme activities and mitochondrial iron content in wild-type and mutant strains
| Strain | Aconitase* | SDH–Cytbc1* | NADH–Cytbc1* | Ubi–Cytbc1* | Cyt ox* | Iron content† |
|---|---|---|---|---|---|---|
| wt | 473 ± 67 | 96 ± 18 | 1011 ± 192 | 1351 ± 158 | 962 ± 332 | 1 |
| Δssq1 | 50 ± 10 | 6 ± 1 | 1050 ± 78 | 376 ± 56 | 152 ± 48 | 10.6 ± 3.9 |
| Δnfu1 | 225 ± 21 | 46 ± 14 | 999 ± 28 | 1568 ± 66 | 568 ± 56 | 1.3 ± 0.2 |
| Δisu2 | 433 ± 48 | 56 ± 28 | 1154 ± 163 | 1647 ± 25 | 682 ± 228 | 1.3 ± 0.2 |
| Δisu1 | 296 ± 10 | 42 ± 20 | 1173 ± 62 | 1177 ± 24 | 526 ± 106 | 1.2 ± 0.1 |
| Δnfu1Δisu1 | 170 ± 4 | 4 ± 2 | 1082 ± 22 | 725 ± 105 | 180 ± 42 | 4.4 ± 0.9 |
Values obtained are an average of at least three independent mitochondrial preparations; ± indicates range observed.
Activity: aconitate, converted⋅min−1⋅mg−1 cell lysate; SDH–Cytbc1, nmol of DCIP reduced⋅min−1⋅mg−1 of mitochondrial protein; Ubi–Cytbc1, nmol of Cyt c reduced⋅min−1⋅mg−1 of mitochondrial protein; Cyt ox, nmol of Cyt c oxidized⋅min−1⋅mg−1 of mitochondrial protein.
Values are relative to wt.
Figure 5.
Levels of mitochondrial proteins. Mitochondrial proteins from each strain were separated by electrophoresis in the amounts indicated, transferred to nitrocellulose, and subjected to immunoblot analyses by using antibodies specific to Sdh1, Sdh2, CoxII, Cyt c1, and Rip1. Densitometry of the autoradiographs was performed by using Ofoto (Light Source Computer Images) and Scan Analysis (Biosoft) software packages.
Because the activity of the electron-transport chain is thought to be coordinately regulated, we tested the activity of complex IV (Cyt ox), which lacks a Fe–S cluster. Δssq1 and Δnfu1Δisu1 mitochondria possessed only 15–20% of wt Cyt ox activity (Table 1) and about 15% of wt CoxII protein levels (Fig. 5). The single deletion strains, Δnfu1, Δisu1, and Δisu2 had between 55–70% of the wt Cyt ox activity and a slight decrease in CoxII protein levels (Table 1, data not shown). Possible reasons for this decrease are considered in the Discussion.
The observed reduction in activity of components of the electron-transport chain would be expected to compromise growth under conditions requiring respiration. As expected, growth of Δnfu1Δisu1 and Δssq1 cells on nonfermentable carbon sources was severely compromised (Fig. 4). We were surprised to find that growth of Δnfu1 and Δisu1 cells on media containing only glycerol or lactate as the nonfermentable carbon source was compromised, because they both had nearly wt respiratory enzyme activities (data not shown). Δisu1 cells also showed poor growth on ethanol as the nonfermentable carbon source, whereas Δnfu1 cells grow as well as wt cells. Either the combination of slightly reduced activities of Sdh and Cyt ox reduces growth by respiration for the Δnfu1 and Δisu1 cells or some other physiological defect is present in these strains.
Mitochondria from Δnfu1 Δisu1 Strains Accumulate Iron.
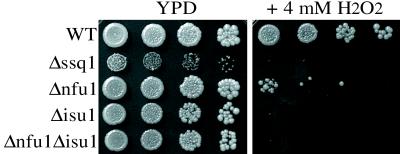
Because there appeared to be a functional interaction between Nfu1 and Isu with Ssq1, a protein that is involved in mitochondrial iron homeostasis, we tested whether the absence of these proteins also altered mitochondrial iron metabolism. Mitochondria isolated from Δnfu1, Δisu1, or Δisu2 cells had near-wt levels of iron (5.0 pmol of Fe/ug of mitochondrial protein) (Table 1). In contrast, the Δssq1 strain resulted in an ≈11-fold accumulation of iron in mitochondria (Table 1; ref. 5). Moreover, mitochondria from the Δnfu1Δisu1 strain contained ≈4.5-fold more iron than wt mitochondria. The accumulation of iron in Δnfu1Δisu1 mitochondria suggests that Nfu1 and Isu1 may function together with Ssq1 in iron metabolism in mitochondria. Because accumulation of mitochondrial iron causes strains to be hypersensitive to oxidative stress (27), we also tested the sensitivity of the deletion strains to H2O2. As expected, the Δssq1 and Δnfu1Δisu1 strains were extremely sensitive to H2O2 compared with wt (Fig. 6). In this assay, Δnfu1 showed moderate sensitivity. Surprisingly, Δisu1 cells were also extremely sensitive to H2O2, even though their iron levels were very similar to those of wt, indicating that the sensitivity is caused by something other than the accumulation of iron.
Figure 6.
Sensitivity to H2O2. Cells were serially diluted 1:10 and spotted onto YPD +/− 4 mM H2O2 and incubated at 34°C for 3 days.
DISCUSSION
Our results lead us to propose that the S. cerevisiae genes NFU1, ISU1, and ISU2, which are the focus of this report, play fundamental roles in iron metabolism in mitochondria. Their sequence relationship to nifU from nitrogen-fixing bacteria, which has previously been shown to be necessary for the maturation or repair of the Fe–S-containing nitrogenases, in addition to the phenotypes of strains containing deletion of these genes, supports this hypothesis.
The data we report are consistent with the idea that Isu and Nfu1 are directly involved in the mobilization of iron for insertion into Fe–S-containing proteins. If so, the role of Isu genes must be critical because cell viability depends on the presence of either Isu1 or Isu2. The N-terminal domain of bacterial NifU, which is related to these proteins, has been shown to bind iron, which supports a role for Isu proteins in iron mobilization (8). In addition, yeast NFS1, related to iscS and nifS, is an essential gene (28). NifS and IscS from nitrogen-fixing bacteria have been shown to catalyze the removal of sulfur from cysteine as biologically active elemental sulfur (8, 29). Thus, Isu and Nfs1 proteins could be involved in mobilization of Fe and S for the assembly of Fe–S clusters in the mitochondria. Nfu1, on the other hand, is not essential; no other proteins with obvious similarity are encoded by the yeast genome. Therefore, unless a functional homolog exists, Nfu1 may play an auxiliary role in this process.
The viability, but compromised growth, of the Δnfu1Δisu1 strain allowed us to test the idea that the product of these genes are involved in Fe–S protein maturation/repair. The 10-fold- and 3-fold-lower activity of SDH and aconitase, respectively, in Δnfu1Δisu1 mitochondria is consistent with a defect in Fe–S cluster assembly. The decrease in steady-state protein levels of Sdh2 may be because of decreased stability without the insertion of a Fe–S. The fact that activity of the Cyt bc1 complex is only reduced to 55% of wt levels may reflect varying requirements among different proteins for assistance in Fe–S cluster formation or the existence of more specialized factors.
The increased mitochondrial iron levels in Δnfu1Δisu1 mitochondria directly support a role for ISU and NFU1 genes in iron metabolism. The genetic interactions between Δssq1 and Δnfu1 and between Δssq1 and Δisu1 may be caused by their combined defects in iron metabolism, because increased mitochondrial iron levels are found in Δssq1 mitochondria, which can be seen as electron-dense deposits within the matrix (5). The increased iron levels in Δssq1 mitochondria are presumably the result of the role of Ssq1 in the import and/or maturation of Yfh1, the yeast homolog of the human frataxin gene. Yfh1 appears to modulate mitochondrial iron levels by regulating export of iron from the organelle (3). Therefore, lowering of Yfh1 levels in mitochondria would be expected to increase intramitochondrial iron levels.
The reason for the increased iron levels in Δnfu1Δisu1 mitochondria is less obvious. If these proteins are involved in maintaining iron in a biologically active form that can be assembled into Fe–S clusters, failure to assemble iron efficiently may result in feedback regulation and thus iron accumulation in the matrix. However, it is also possible that Nfu1 and Isu1/2 are more directly involved in regulation of iron metabolism and loss of activity directly disturbs iron homeostasis. According to this scenario, the decrease in enzyme activities found in the mutants could be indirect, because Fe–S centers are particularly sensitive to damage by oxygen radicals whose generation is enhanced by high levels of iron (30). In support of this hypothesis, it has been shown that yeast cells lacking Yfh, as well as human cells having a defective frataxin protein, have greatly reduced activities for aconitase and components of the electron-transport chain (4). This reduced activity was attributed to iron overload causing increased oxygen radical formation and thus, inactivation of the labile Fe–S clusters.
The decrease in activity of Cyt ox cannot be simply explained, as it contains no Fe–S cluster. It is possible that coordinate regulation of components of the electron transport chain is responsible, as it is known that reduction of Cyt bc1 levels causes a proportional decrease in Cyt ox activity. In a Cyt bc1 deletion mutant, Cyt ox has a basal level of 15% (31). However, the coordinate regulation of Cyt ox and Cyt bc1 cannot completely explain the reduction of Cyt ox levels to 15% of wt in the Δssq1 and Δnfu1Δisu1 strains, because Cyt bc1 levels are only reduced to 55%. There are several other possible explanations. A role for Isu, Nfu1, and Ssq1 proteins in the maturation of other iron-containing proteins of the mitochondria, such as the heme-containing cytochromes, has not been excluded. Possibly, other Fe–S containing proteins of the mitochondria, which have yet to be identified, may play some role in maturation of protein complexes of the respiratory chain. A more detailed analysis is required to separate the direct effects of the deletion of NFU1 and ISU genes, from indirect effects caused by the primary physiological defect.
The relationship between the yeast genes discussed here and genes from other organisms is complex. The yeast Nfu1 and Isu proteins are related to the C-terminal and N-terminal domains, respectively, of NifU protein of nitrogen-fixing bacteria. NifU is proposed to mobilize iron for its insertion into the nitrogenases (7, 32). In these bacteria, nifU is a member of a cluster of genes required for fixing nitrogen. However, these bacteria, as well as non-nitrogen-fixing bacteria such as E. coli, contain related genes proposed to carry out less specialized roles in the formation and maintenance of other Fe–S proteins. These genes, called iscU in prokaryotes, have homology to the N terminus of NifU (8). Proteins encoded by iscU are extraordinarily well conserved; the E. coli and the two yeast homologs are 70% identical in sequence. iscU resides in a gene cluster with homologs of NifS (iscS) DnaJ (hscB), an Hsp70 DnaK (hscA), and ferredoxin in A. vinelandii, E. coli, and Haemophilus influenzae (8, 33, 34). The coexpression of the isc genes with chaperone genes has been suggested to indicate the presence of a macromolecular system for the formation and insertion of Fe–S clusters into Fe–S-containing proteins and their subsequent folding (8). The genetic interactions reported here among the SSQ1 Hsp70, ISU, and NFU1 genes, all of which encode mitochondrial proteins, strengthens this idea and suggests that this system has been conserved through evolution, being maintained in the mitochondria of eukaryotes. The identification of this system in mitochondria raises the question of the formation of Fe–S clusters in the cytosol. There are no obvious homologs of Isu and Nfu1 encoded in the yeast genome. Perhaps distantly related proteins carry out this function.
Whether defects in related genes in humans are the cause of any mitochondrial myopathies of unknown etiology having decreased activity of mitochondrial Fe–S proteins remains an open question (35). Because decreased levels of frataxin leads to increased mitochondrial iron levels, clinically manifesting itself as Friedreich’s ataxia (4), this possibility should be explored. The yeast system should be a useful model in which to better understand the role of these conserved genes in iron homeostasis and assembly of macromolecular complexes containing essential Fe–S moieties.
Acknowledgments
We thank T. Donohue, P. Kiley, and R. Eisenstein for thoughtful comments on the manuscript, B. Trumpower and B. Lemiere for gifts of antibodies, and M. Ohlson for expert technical help. Work was supported by GM27870 from National Institutes of Health to E.A.C.
ABBREVIATIONS
- wt
wild type
- SDH
succinate dehydrogenase
- Cyt bc1
cytochrome bc1 complex
- Cyt ox
cytochrome oxidase
- hsp
heat shock protein
Note Added in Proof
An article by Lill and co-workers (36) was published after this manuscript had been submitted. Evidence is presented suggesting that Nfs1 of the mitochondrial matrix is important for cytosolic Fe-S cluster formation, raising the question of the role of Isu1/2 and Nfu1 in the assembly of Fe-S clusters for cytosolic proteins.
References
- 1.Tzagoloff A. Mitochondria. New York: Plenum; 1982. [Google Scholar]
- 2.Radisky D C, Kaplan J. J Biol Chem. 1999;274:4481–4484. doi: 10.1074/jbc.274.8.4481. [DOI] [PubMed] [Google Scholar]
- 3.Radisky D C, Babcock M C, Kaplan J. J Biol Chem. 1999;274:4497–4499. doi: 10.1074/jbc.274.8.4497. [DOI] [PubMed] [Google Scholar]
- 4.Rötig A, de Lonlay P, Chretien D, Foury F, Koenig M, Sidi D, Munnich A, Rustin P. Nat Genet. 1997;17:215–217. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- 5.Knight S A B, Sepuri N B V, Pain D, Dancis A. J Biol Chem. 1998;273:18389–18393. doi: 10.1074/jbc.273.29.18389. [DOI] [PubMed] [Google Scholar]
- 6.Strain J, Lorenz C R, Bode J, Garland S, Smolen G A, Ta D T, Vickery L E, Culotta V C. J Biol Chem. 1998;273:31138–31144. doi: 10.1074/jbc.273.47.31138. [DOI] [PubMed] [Google Scholar]
- 7.Fu W, Jack R F, Morgan T V, Dean D R, Johnson M K. Biochemistry. 1994;33:13455–13463. doi: 10.1021/bi00249a034. [DOI] [PubMed] [Google Scholar]
- 8.Zheng L, Cash V L, Flint D H, Dean D R. J Biol Chem. 1998;273:13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- 9.James P, Pfund C, Craig E. Science. 1997;275:387–389. doi: 10.1126/science.275.5298.387. [DOI] [PubMed] [Google Scholar]
- 10.Schilke B, Forster J, Davis J, James P, Walter W, Laloraya S, Johnson J, Miao B, Craig E. J Cell Biol. 1996;134:603–614. doi: 10.1083/jcb.134.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gambill B D, Voos W, Kang P J, Miao B, Langer T, Craig E A, Pfanner N. J Cell Biol. 1993;123:109–117. doi: 10.1083/jcb.123.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherman F, Fink G R, Hicks J B. Laboratory Course Manual for Methods in Yeast Genetics. 1986. [Google Scholar]
- 13.Bender A, Pringle J. Mol Cell Biol. 1991;11:1295–1305. doi: 10.1128/mcb.11.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burns N, Grimwade B, Ross-Macdonald P B, Choi E-Y, Finberg K, Roeder G S, Snyder M. Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- 16.Burns N, Ross-Macdonald P, Roeder G S, Snyder M. In: Microbial Genome Methods. Adolph K W, editor. Boca Raton, FL: CRC; 1996. pp. 61–79. [Google Scholar]
- 17.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider B L, Seufert W, Steiner B, Yang Q H, Futcher A B. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- 19.Watson K, Bertoli E, Griffiths D E. Biochem J. 1975;146:401–407. doi: 10.1042/bj1460401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poyton R O, Goehring B, Droste M, Sevarino K A, Allen L A, Zhao X-J. Methods Enzymol. 1995;260:97–116. doi: 10.1016/0076-6879(95)60133-3. [DOI] [PubMed] [Google Scholar]
- 21.Graham L A, Trumpower B L. J Biol Chem. 1991;266:22485–22492. [PubMed] [Google Scholar]
- 22.Fansler B, Lowenstein J M. Methods Enzymol. 1969;13:26–30. [Google Scholar]
- 23.Robinson K M, Kieckebusch-Guck A v, Lemire B D. J Biol Chem. 1991;266:21347–21350. [PubMed] [Google Scholar]
- 24.Nett J H, Denke E, Trumpower B L. J Biol Chem. 1997;272:2212–2217. doi: 10.1074/jbc.272.4.2212. [DOI] [PubMed] [Google Scholar]
- 25.Beinert H. Methods Enzymol. 1978;24:435–445. doi: 10.1016/s0076-6879(78)54027-5. [DOI] [PubMed] [Google Scholar]
- 26.Kennedy M C, Kent T A, Emptage M, Merkle H, Beinert H, Münck E. J Biol Chem. 1984;259:14463–14471. [PubMed] [Google Scholar]
- 27.Babcock M, de Silva D, Oaks R, Davis-Kaplan S, Jiralerspong S, Montermini L, Pandolfo M, Kaplan J. Science. 1997;276:1709–1712. doi: 10.1126/science.276.5319.1709. [DOI] [PubMed] [Google Scholar]
- 28.Kolman C, Söll D. J Bacteriol. 1993;175:1433–1442. doi: 10.1128/jb.175.5.1433-1442.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng L, White R H, Cash V L, Jack R F, Dean D R. Proc Natl Acad Sci USA. 1993;90:2754–2758. doi: 10.1073/pnas.90.7.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fridovitch I. Annu Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 31.Boumans H, Berden J A, Grivell L A, van Dam K. Biochem J. 1998;331:877–883. doi: 10.1042/bj3310877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dean D R, Bolin J T, Zheng L. J Bacteriol. 1993;175:6737–6744. doi: 10.1128/jb.175.21.6737-6744.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blattner F R, Plunkett G, III, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 34.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 35.Hall R E, Henriksson K G, Lewis S F, Haller R G, Kennaway N G. J Clin Invest. 1993;92:2660–2666. doi: 10.1172/JCI116882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kispal G, Csere P, Prohl C, Lill R. EMBO J. 1999;18:3981–3989. doi: 10.1093/emboj/18.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]