Abstract
During myogenesis, reductions in trophic factor availability signal most myoblasts to fuse, up-regulate the expression of muscle-specific genes, and form myotubes. Those cells failing to differentiate into myotubes initiate apoptosis and rapidly die. At present, the signal-transduction molecules that determine whether myoblasts should differentiate or die are largely unknown. In this report, we describe the cloning and characterization of DALP, a small LIM-only type zinc-finger protein that is induced when the intersegmental muscles (ISMs) of the moth Manduca sexta become committed to die at the end of metamorphosis. Forced expression of death-associated LIM-only protein (DALP) in Drosophila results in skeletal muscle atrophy. Ectopic expression of DALP, or its mammalian ortholog Hic-5, blocks differentiation and induces apoptosis in mouse C2C12 myoblasts. Both of these effects can be overcome by contact with normal myoblasts or by ectopic expression of the muscle-specific transcription factor MyoD. Hic-5 expression is specifically and dramatically induced in normal myoblasts that die after removal of trophic support. Taken together, these data suggest that DALP and Hic-5 act upstream of MyoD and function as phylogenetically conserved “switches” to block muscle differentiation and induce death.
One of the fundamental problems faced by developing organisms is that many more cells are produced than are ultimately needed (1–3). Although this provides the organism with considerable developmental plasticity to match the size of interacting populations of cells, it does have a deleterious down side. Deregulation of the cell death machinery can result in the retention of defective, mitotically competent cells, which in turn can facilitate the generation of neoplasms.
During muscle development, myoblasts function as muscle stem cells and initially provide a pool of renewable cells that can participate in muscle development and repair (4). During normal development in vertebrates, these cells are produced in the somites and then migrate to more peripheral locations in the trunk and limbs. In response to the loss of trophic support, myoblasts exit the cell cycle, fuse, and up-regulate a variety of genes that commit them to form striated skeletal muscle. Those cells that fail to activate appropriate survival and differentiation programs instead rapidly die by apoptosis (5). The ability to use cell–cell interactions to select a subset of valuable cells in this and other lineages facilitates normal organogenesis.
Once a myoblast has made the decision to survive, it exits from the cell cycle and dramatically induces the expression of the cyclin-dependent kinase inhibitor p21 (6), which confers an apoptosis-resistant phenotype (5). The retinoblastoma protein (Rb), which acts downstream of p21 in this pathway, also plays an essential role in myoblast resistance to apoptosis (7, 8). In addition, phosphatidylinositol 3-kinase, which acts as a survival factor in several systems, is induced in forming myotubes and is essential for the induction of muscle-specific gene expression (9). However, despite these insights, almost nothing is known about the molecular machinery that allows a myoblast to decide whether it should survive and activate survival mechanisms or to instead commit suicide.
To gain insight into the molecular mechanisms that mediate the death of skeletal muscle, we have examined the intersegmental muscles (ISMs) from the tobacco hawkmoth Manduca sexta (10). The ISMs are composed of giant fibers, each one of which is 5 mm long and up to 1 mm in diameter. These muscles are used for the eclosion (emergence) behavior of the adult moth at the end of metamorphosis and then die during the subsequent 30 h. The death of the ISMs is induced by a decline in the circulating titer of the insect molting hormone 20-hydroxyecdysone (11). The death of the ISMs depends on de novo gene expression, and several death-associated transcripts have been identified (12). A number of these proteins have been shown to be phylogenetically conserved and capable of functioning in cells from a wide variety of taxa (13). In this report, we describe the cloning and analysis of DALP (death-associated LIM-only protein) and demonstrate that it and its mammalian ortholog Hic-5 (14) act as negative regulators of muscle differentiation in insects and mammals.
MATERIALS AND METHODS
Cloning and Structural Analysis of DALP.
A cDNA library generated from day 18 ISM RNA was constructed in the λ ZapII (Stratagene) vector (12). The library was differentially screened by using plus/minus hybridization with 32P-labeled cDNA probes generated from day 15 (precommitment) and day 18 (postcommitment) ISM RNA (15). The initial DALP cDNA was truncated at the 5′ end, and inverse reverse transcription–PCR was used to recover the missing region. The presumptive full-length recombinant was constructed and sequenced.
Forced Expression of DALP in Drosophila.
DALP cDNA was cloned into the BglII–XhoI sites 3′ to the GAL4 upstream activating sequence of the pUAST P element vector (16) and used to generate P{UAS-DALP} transgenic flies according to standard protocols. A second UAS strain was generated that expressed mutant DALP specifically lacking the WHPEHF motif (described below). P{UAS-DALP} females were mated to P{GawBG487}IS-1, which expresses in an anterior–posterior gradient in body wall musculature (17). The highest levels of expression are seen in muscle 31 (ventral internal 1), which is located in and is specific for the first abdominal segment. The organization of actin filaments was visualized with FITC-labeled phalloidin (Sigma).
Mammalian Myoblast Differentiation.
C2C12 myoblasts (18) were maintained at 37°C in DMEM (GIBCO/BRL) supplemented with 10% FBS (Atlanta Biologicals, Norcross, GA) and 100 units/ml penicillin-streptomycin (GIBCO/BRL). Myogenesis was induced by incubating the cells in low serum differentiation medium consisting of DMEM, 0.1% FBS, 5 μg/ml insulin (Sigma), 5 μg/ml transferrin (Sigma), and antibiotics. In some experiments, trypan blue exclusion was used to determine the percentage of dead cells 3 days after transfer to differentiation medium. Three separate fields were counted from each plate with a minimum of 100 cells per field.
Transfection of Constructs into Myoblasts.
DALP was subcloned into both the 1704 vector, which uses Moloney murine leukemia virus long terminal repeat to drive the expression of the target gene as well as bacterial lacZ gene from an IRES sequence (19) and pFlag-CMV-2 (Kodak), which creates an N-terminal Flag epitope-tagged protein. Hic-5 subcloned into both pRc/CMV (14) and pFlag-CMV-2. Muscle LIM protein (MLP) cDNA was subcloned into the pcDNA3 plasmid. Transfections into C2C12 myoblasts were performed by using linearized plasmids and Lipofectamine (GIBCO/BRL) at a ratio of 5 μl/ug of DNA for 4 h. Antibiotic-resistant stable transformants were selected with either G418 (500 μg/ml) or puromycin (3 μg/ml). Tranformation efficiency was ≈10% under these conditions. In separate experiments, MyoD cDNA was subcloned into the pBabe-puromycin retroviral vector which, after packaging, was used to infect C2C12 cells. For each construct transfected in this study, six individual monoclonal cell lines were generated and analyzed. Data are presented for individual cell lines.
Cell Mixing Experiments.
In some experiments, transfected cells were labeled for 20 h with BrdUrd (Sigma) at a final concentration of 50 μM. After labeling, transfected cells were incubated in growth medium for an additional 24 h. Both wild-type and labeled transfected cells then were trypsinized, mixed at a ratio of 1:1, and replated in growth medium. Four hours later, cells were switched to differentiation medium and examined after 3 days by BrdUrd immunohistochemistry. Rat-1 cells were maintained at 37°C in DMEM supplemented with 10% FBS and antibiotics as described above.
Immunocytochemistry.
Cells were grown on coverslips, fixed with 2% paraformaldehyde for 20 min at room temperature, and reacted overnight at 4°C with monoclonal antisera against myosin heavy chain (1:100; MF20; Development Studies Hybridoma Bank, Iowa City) and desmin (D76; Development Studies Hybridoma Bank), or BrdUrd (1:50; G3G4; Development Studies Hybridoma Bank). A rabbit polyclonal antiserum generated against Hic-5 prepared as described (14) also was used for immunohistochemistry at a 1:2,000 dilution. Biotin-labeled secondary antibodies and horseradish peroxidase-conjugated avidin (Vector Laboratories) were used for detection.
Western Blots.
C2C12 cells were harvested at various times after transfer to differentiation medium. Aliquots of protein (30 μg) were fractionated by SDS/10% PAGE, transferred to Immobilon P membrane (Millipore), and reacted with the anti-Hic-5 rabbit polyclonal antiserum. Horseradish peroxidase-labeled secondary antisera were detected with the enhanced chemiluminescence (ECL) Western blotting detection system (Amersham Pharmacia) and x-ray film (Eastman Kodak).
RESULTS AND DISCUSSION
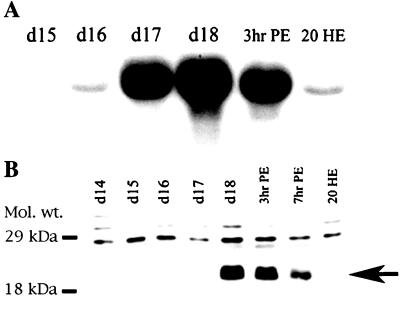
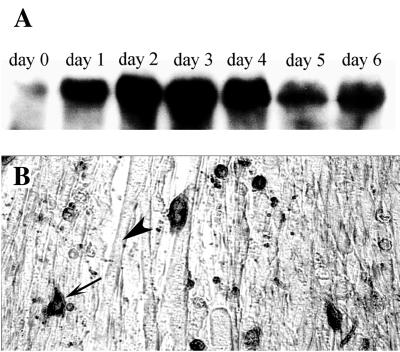
DALP was isolated in a screen designed to identify death-associated transcripts from the ISMs of the moth M. sexta (15). Northern blot analysis revealed that DALP mRNA (≈1.7 kilobase) accumulated dramatically late on day 17 of pupal/adult development, when the ISMs become committed to die, and continued to be present when the cells initiated death late on day 18 (Fig. 1A). DALP mRNA appeared in the muscles before any other known death-associated transcripts. The DALP protein is ≈20 kDa in size and was observed only in ISMs that were either committed to die or actively dying on day 18 (Fig. 1B). DALP expression was restricted to the ISMs and was not detected in flight muscle, fat body, Malpighian tubules, ovary/eggs, or male sexual accessory gland (data not shown). In addition, the expression of DALP was correlated with the commitment of the ISMs to die, because injecting animals with the steroid hormone 20-hydroxyecdysone on the day before emergence (day 17 of pupal/adult development), delays both cell death (10) and the expression of DALP (Fig. 1A).
Figure 1.
Expression of Manduca DALP. (A) A developmental Northern blot of ISM RNA was hybridized with 32P-labeled DALP cDNA. ISMs become committed to die late on day 17 and begin to die during the hours after adult emergence late on day 18. Treatment of day-17 animals with 25 μg of the steroid 20-hydroxyecdysone (20 HE) delays both ISM death and the expression of death-associated transcripts (11). d, day of pupal-adult development; hr, hours post-adult emergence (PE). (B) Western blot of ISM proteins during the period before and after the onset of cell death. Arrow indicates the migration of DALP that was developmentally regulated. The identity of the higher molecular weight, constitutively expressed proteins is unknown. Protein markers are in kDa.
DNA sequence analysis revealed that the DALP cDNA contains a 612-bp ORF encoding a 204-aa protein product (Fig. 2). The most significant structural feature of the DALP protein is the presence of one perfect and two imperfect LIM domains (Fig. 3), structural motifs that consist of paired zinc fingers that participate in protein–protein interactions (20). Whereas many LIM proteins also possess a homeodomain, DALP belongs to the subfamily of LIM-only proteins (LMO) that lack this DNA binding motif (21). LMOs have been shown to function in signal transduction, focal adhesion, and differentiation (21).
Figure 2.
The complete DNA sequence and presumptive ORF of DALP. The first methionine indicates the start of translation,∗ marks the stop codon, and the presumptive polyadenylation site is underlined. Cysteine and histidine residues that define the zinc-finger motifs are in bold, and the conserved WHPEHF motif is double underlined.
Figure 3.
Comparison of Manduca DALP with mouse and human Hic-5. Cysteine and histidine residues that define the LIM domains are underlined, and the individual LIM motifs are numbered. The conserved WHPEHF motifs are boxed. Note that the spacings within and between LIM domains are highly conserved. Zinc-finger domains 5 and 6 define a perfect LIM motif, whereas the others have minor substitutions (21).
The sequence of DALP shares 36% identity and 52% similarity with mouse Hic-5, a hydrogen peroxide- and transforming growth factor β-inducible LMO (Fig. 3; ref. 14). In addition to conserved placement of cysteine and histidine residues that define the LIM domains, both of these proteins contain the sequence WHPEHF in the first LIM domain (see Fig. 3). Hic-5 differs from DALP in that it contains one more LIM domain than DALP, a second WHPEHF motif and a proline-rich N-terminal domain. The only other protein found by database analysis that contains the WHPEHF motif is the cytoskeletal LIM protein paxillin (data not shown).
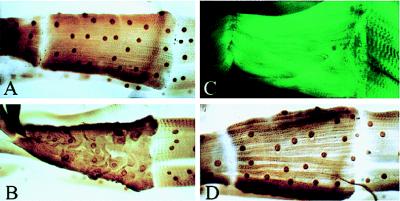
Although Manduca is an ideal system for biochemical analysis, it lacks the tools required for genetic studies. Consequently, we analyzed the function of DALP by using the Gal4 upstream activating sequence (UAS) P element system (16) that allows gene expression to be targeted to specific tissues in the fruit fly Drosophila. We generated a transformant fly strain P{UAS–DALP} and mated these flies to the P{GawBG487}IS-1 line, which targets ectopic gene expression in an anterior–posterior gradient in the abdominal musculature of third-instar larvae (Fig. 4; ref. 17). Ectopic expression of moth DALP resulted in the disorganization of the contractile apparatus and subsequent atrophy in muscle 31 in the anterior abdominal segment (Fig. 4 B and C). It is not clear why these cells were preferentially affected by DALP expression, although these cells are the ones that display the highest levels of expression with this Gal4 line (V. Budnik, personal communication). The effects of ectopic DALP expression depended on expression of the intact protein, because selective deletion of the WHPEHF motif blocked muscle atrophy (Fig. 4D).
Figure 4.
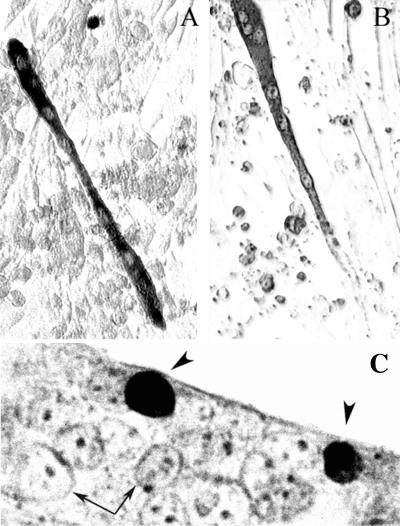
The effects of ectopic DALP expression in Drosophila skeletal muscle. (A) Bacterial β-galactosidase was expressed in Drosophila abdominal muscles by using the Gal4/UAS expression system. Expression was monitored with anti-β-galactosidase immunocytochemistry. (B) The same Gal4 enhancer trap line was used to coexpress β-galactosidase and moth DALP in the ISMs. Expression was monitored by β-galactosidase immunohistochemistry. (C) The organization of the contractile apparatus was visualized by using FITC-labeled phalloidin in separate animals expressing ectopic DALP. (D) The WHPEHF motif was deleted from DALP and coexpressed with β-galactosidase in the abdominal muscles. Muscles retained normal appearance and were contractile. The triangular muscle in these assays is muscle 31 and is about 260 μm.
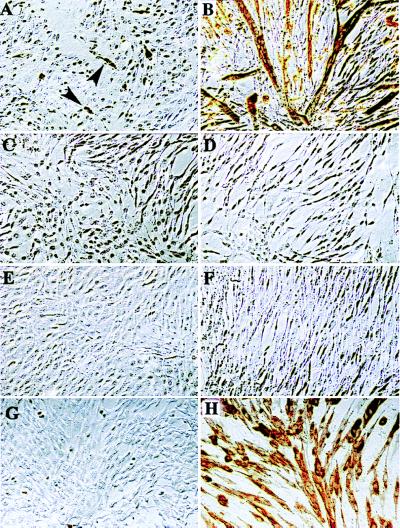
To gain further insight into the function of DALP, we turned to an in vitro cell culture system. The mouse myoblast cell line C2C12 has been extensively used as a model for examining skeletal muscle differentiation in mammals (18). C2C12 cells can be maintained as a stable, nontransformed line that ceases cycling and differentiates into multinucleated myotubes after incubation in a low serum differentiation medium (5, 18) (Fig. 5 A and B). Viable, stable C2C12 lines forced to constitutively express DALP from a retroviral promoter were generated for analysis. Western blot analysis verified that these cells expressed DALP (data not shown). Control C2C12 cells in growth medium displayed low levels of muscle differentiation markers such as myosin heavy chain (Fig. 5A) and desmin (data not shown). However, the DALP-expressing cells had no detectable basal levels of these two marker proteins (Fig. 5C and data not shown). When these cells were switched to differentiation medium, they failed to up-regulate muscle markers or form a single multinucleated myotube (Fig. 5D). Instead, when induced to differentiate, these cells underwent apoptosis and rapidly died (Fig. 6).
Figure 5.
Effects of DALP and Hic-5 expression on C2C12 myoblasts. C2C12 myoblasts were grown in growth (A, C, E, and G) or differentiation (B, D, F, and H) medium and then stained with a mAb against myosin heavy chain (brown). (A and B) Wild-type cells. (C and D) C2C12 line constitutively expressing DALP. (E and F) C2C12 line constitutively expressing exogenous Hic-5. No myotubes were formed from DALP- or Hic-5-expressing cells. (F and G) Hic-5-expressing cells infected with a MyoD-expressing retroviral construct. Ectopic MyoD expression can abrogate the effects of Hic-5 on myotube formation. Arrowheads point to myosin heavy chain-positive mononucleated myoblasts in growth medium.
Figure 6.
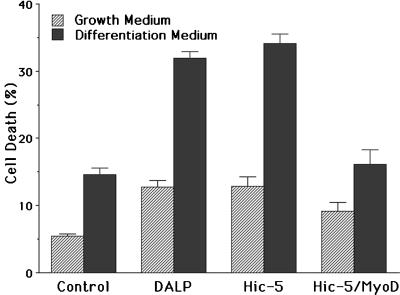
Blockade of myotube formation is correlated with enhanced rates of cell death. Numbers of dying cells were determined for control C2C12 cells and those expressing DALP, Hic-5, or Hic-5 plus MyoD in growth (hatched bars) and differentiation (filled bars) media.
To better evaluate the developmental role assumed by this class of LMO in myogenesis, we generated C2C12 cells that stably expressed ectopic Hic-5 protein, which was verified by Western blot analysis (data not shown). As with the DALP-expressing cells, normal basal levels of muscle differentiation markers were also absent (Fig. 5E). In addition, these cells were also incapable of forming myotubes (Fig. 5F) and instead died when exposed to differentiation medium (Fig. 6). Therefore, both DALP and Hic-5 were able to fully block the differentiation of stably transformed mouse myoblasts.
When wild-type C2C12 cells are placed in differentiation medium, the myogenic transcription factor MyoD is induced dramatically, which in turn drives muscle differentiation (22). However, C2C12 cells that express either ectopic DALP or Hic-5 failed to express detectable levels of MyoD protein, even after several days in differentiation medium (data not shown). To determine whether Hic-5 functions upstream of MyoD in the myogenic pathway, Hic-5-expressing C2C12 cells were stably transfected with a MyoD expression construct. Forced expression of ectopic MyoD did not induce muscle differentiation in C2C12 cells cultured in growth medium, nor did it alter endogenous levels of Hic-5. However, when these cells were transferred to differentiation medium, muscle differentiation was reinstated as demonstrated by myotube formation, myosin heavy chain expression (Fig. 5H), and reduced rates of apoptosis (Fig. 6).
It is known that MyoD can bind to MLP, another LMO, which in turn acts as a positive regulator of MyoD-dependent gene expression (23). This raised the possibility that Hic-5 might compete with MLP for binding to MyoD, thereby providing a mechanism for how Hic-5 acts as a negative regulator of differentiation. To test this hypothesis, Hic-5-expressing C2C12 cells were stably transfected with an MLP expression construct. In contrast to the results obtained with MyoD, no myotubes were formed, and muscle marker expression was not detected (data not shown). Consequently, it appears that Hic-5 acts upstream of MyoD expression in muscle differentiation and does not appear to compete with MLP.
Several experiments demonstrated that the blockade of myotube formation induced by forced expression of DALP or Hic-5 could be overcome by contact with normal myoblasts. C2C12 cells were transiently transfected with constructs that coexpressed DALP and β-galactosidase (Fig. 7A) or Flag epitope-tagged Hic-5 (Fig. 7B) and were then subsequently placed in differentiation medium. In both cases, labeled myotubes were formed in the presence of nontransfected C2C12 cells, despite strong expression of these LMO proteins.
Figure 7.
Effects of DALP and Hic-5 expression on myotube formation in C2C12 cells. Myoblasts were transiently transfected with expression vectors that coexpressed DALP and β-galactosidase (A) or Flag-tagged Hic-5 (B). After 48 hr, cultures were transferred to differentiation medium. After an additional 3 days, cells were processed for β-galactosidase (A) or Flag (B) immunohistochemistry. In separate experiments, BrdUrd-labeled Hic-5-expressing C2C12 cells were mixed with wild-type cells and then placed in differentiation medium. Arrows, nuclei from wild-type cells; arrowheads, BrdUrd-positive nuclei. The micrograph is overexposed to enhance the visualization of the control nuclei.
In a separate series of experiments, Hic-5-expressing C2C12 cells were cultured in the presence of BrdUrd to label their nuclei. These cells then were cocultured with wild-type C2C12 cells. After incubation in differentiation medium, myotubes were observed in the cultures. BrdUrd immunohistochemistry demonstrated the presence of labeled nuclei, suggesting that Hic-5-expressing cells could participate in myogenesis if they were allowed to contact wild-type cells (Fig. 7C). Coculture experiments with Rat-1 fibroblasts and Hic-5-expressing C2C12 myoblasts failed to support the formation of any myotubes. These data suggest that there is a requirement for cell type-specific, contact-dependent signals to overcome Hic-5- or DALP-mediated inhibition of muscle differentiation.
These data strongly suggest that Hic-5 can dramatically influence the patterns of myoblast survival and differentiation. However, for this observation to be meaningful in a physiological context, it is essential that myoblasts actually express Hic-5 during differentiation. Western blot analysis demonstrated that low levels of endogenous Hic-5 protein were present in cycling wild-type C2C12 cells (Fig. 8A). However, Hic-5 levels increased dramatically when cells were placed in differentiation medium. Immunocytochemical analysis of C2C12 cultures in differentiation medium demonstrated that these high levels of Hic-5 protein were restricted to mononucleated and apoptotic myoblasts relative to multinucleated myotubes (Fig. 8B).
Figure 8.
Expression of Hic-5 in wild-type C2C12 cells. (A) Western blot analysis of endogenous Hic-5 protein levels in C2C12 myoblasts transferred to differentiation medium. Time is expressed as days in differentiation medium. (B) Immunohistochemical analysis of endogenous Hic-5 expression in C2C12 cells incubated in differentiation medium for 3 days. Expression is seen predominantly in mononucleated cells (arrow) as opposed to myotubes (arrowhead).
Taken togther, these data suggest that moth DALP and its mammalian ortholog Hic-5 encode phylogenetically conserved proteins that act as negative regulators of muscle differentiation. In Drosophila, and perhaps in Manduca, expression of DALP is associated with muscle atrophy, which may play an important role in the dissolution of the ISMs at the end of metamorphosis. In mammalian myoblasts, Hic-5 expression blocks differentiation and facilitates the initiation of apoptosis. Hic-5 has a variety of effects in other cell types in vitro, including enhancing senescence in human diploid fibroblasts and facilitating differentiation of MC3T3-E1 osteoblastic cells (24, 25). Other LMOs also have been shown to influence differentiation within specific lineages in mammals, usually as negative regulators (26–28).
In C2C12 cells, Hic-5 acts to block the expression of MyoD, although where it acts in the upstream signal-transduction cascade that leads to myotube formation is still unknown. Data presented here suggest that cell–cell contact may influence the ability of Hic-5 to regulate differentiation. In this regard, it is interesting to note that Hic-5 shares 62% identity with the LIM domains of the focal-adhesion protein paxillin (29). Paxillin can target focal-adhesion kinase (pp125FAK) to adhesion complexes and undergo pp125FAK-dependent phosphorylation (30). Paxillin phosphorylation is blocked by Hic-5 (31), which may block essential integrin-dependent signals required for activating myoblast survival and differentiation programs, including MyoD expression.
It has been suggested that all cells are on the verge of committing suicide unless signaled to activate survival programs after contact with the appropriate neighbors (1). In fact, a general feature of organogenesis is that many more cells are produced in most lineages than are ultimately required to form a given tissue. Some mechanism(s) must exist to ensure that valuable cells are retained while potentially dangerous mitotic cells are selectively lost when they are either present in surplus numbers or reside in ectopic locations. Expression of DALP/Hic-5 may serve as a control point to ensure the removal of myoblasts that end up in inappropriate locations during development. Our data suggest the following model. Myoblasts express low levels of Hic-5 and continue to cycle as long as trophic support is provided. Once trophic factors become rate-limiting, Hic-5 is induced. If myoblasts receive appropriate cell–cell contact from neighbors that are destined to differentiate into myotubes, they can overcome Hic-5-mediated inhibition. If cells are in more peripheral locations, Hic-5 acts to inhibit both survival and subsequent myotube formation. Faced with the “choice” of differentiation or death, these cells undergo apoptosis. In this way, cell–cell signaling during development can be used to ensure that only those myoblasts in the appropriate location are retained, whereas potentially dangerous, mitotically competent cells are lost.
Acknowledgments
We thank K. Murphy for generating the final figures for the manuscript, C. Valavanis and Michaela Heeb for helpful discussions and a critical reading of the manuscript, S. Tapscott for helpful discussions, M. Shibanuma for the gift of the Hic-5 expression plasmids, and P. Caroni and G. Engelmann for provision of MLP cDNA. This work was supported by National Institutes of Health grants to J.R.N. and L.M.S.
ABBREVIATIONS
- DALP
death-associated LIM-only protein
- ISMs
intersegmental muscles
- LMO
LIM-only proteins
- MLP
muscle LIM protein
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF172845).
References
- 1.Raff M C. Nature (London) 1992;356:397–400. doi: 10.1038/356397a0. [DOI] [PubMed] [Google Scholar]
- 2.Oppenheim R W. Annu Rev Neurosci. 1991;14:453–501. doi: 10.1146/annurev.ne.14.030191.002321. [DOI] [PubMed] [Google Scholar]
- 3.Milligan C M, Schwartz L M. Brit Med Bull. 1997;53:570–590. doi: 10.1093/oxfordjournals.bmb.a011631. [DOI] [PubMed] [Google Scholar]
- 4.Walsh K. Prog Cell Cycle Res. 1997;3:53–58. doi: 10.1007/978-1-4615-5371-7_5. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Walsh K. Science. 1996;273:359–361. doi: 10.1126/science.273.5273.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo K, Wang J, Andres V, Smith R C, Walsh K. Mol Cell Biol. 1995;15:3823–3829. doi: 10.1128/mcb.15.7.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zacksenhaus E, Jiang Z, Chung D, Marth J D, Phillips R A, Gallie B L. Genes Dev. 1996;10:3051–3064. doi: 10.1101/gad.10.23.3051. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Guo K, Wills K N, Walsh K. Cancer Res. 1997;57:351–354. [PubMed] [Google Scholar]
- 9.Jiang B-H, Zheng J Z, Vogt P K. Proc Natl Acad Sci USA. 1998;95:14179–14183. doi: 10.1073/pnas.95.24.14179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz L M. J Neurobiol. 1992;23:1312–1326. doi: 10.1002/neu.480230918. [DOI] [PubMed] [Google Scholar]
- 11.Schwartz L M, Kosz L, Kay B K. Proc Natl Acad Sci USA. 1990;87:6594–6598. doi: 10.1073/pnas.87.17.6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwartz L M, Myer A, Kosz L, Engelstein M, Maier C. Neuron. 1990;5:411–419. doi: 10.1016/0896-6273(90)90080-y. [DOI] [PubMed] [Google Scholar]
- 13.Sun D, Sathyanarayana U G, Johnston S A, Schwartz L M. Dev Biol. 1996;173:499–509. doi: 10.1006/dbio.1996.0043. [DOI] [PubMed] [Google Scholar]
- 14.Shibanuma M, Mashimo J, Kuroki T, Nose K. J Biol Chem. 1994;269:26767–26774. [PubMed] [Google Scholar]
- 15.Schwartz L M, Milligan C E, Bielke W, Robinson S J. Methods Cell Biol. 1995;46:107–138. doi: 10.1016/s0091-679x(08)61927-5. [DOI] [PubMed] [Google Scholar]
- 16.Brand A, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 17.Budnik V, Koh Y H, Guan B, Hartmann B, Hough C, Woods D, Gorczyca M. Neuron. 1996;17:627–640. doi: 10.1016/s0896-6273(00)80196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yaffe D, Saxel O. Nature (London) 1977;270:725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- 19.Ghattas I R, Sanes J R, Majors J E. Mol Cell Biol. 1991;11:5848–5859. doi: 10.1128/mcb.11.12.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmeichel K L, Beckerle M C. Cell. 1994;79:211–219. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 21.Dawid I B, Breen J J, Toyama R. Trends Genet. 1998;14:156–162. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- 22.Tapscott S J, Davis R L, Thayer M J, Cheng P-F, Weintraub H, Lassar A B. Science. 1988;242:405–411. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- 23.Kong Y, Flick M J, Kudla A J, Konieczny S F. Mol Cell Biol. 1997;17:4750–4760. doi: 10.1128/mcb.17.8.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shibanuma M, Mochizuki E, Maniwa R, Mashimo J, Nishiya N, Imai S, Takano T, Oshimura M, Nose K. Mol Cell Biol. 1997;17:1224–1235. doi: 10.1128/mcb.17.3.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shibanuma M, Nose K. Int J Biochem Cell Biol. 1998;30:39–45. doi: 10.1016/s1357-2725(97)00155-6. [DOI] [PubMed] [Google Scholar]
- 26.Visvader J E, Mao X, Fujiwara Y, Hahm K, Orkin S H. Proc Natl Acad Sci USA. 1997;94:13707–13712. doi: 10.1073/pnas.94.25.13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kenny D A, Jurata L W, Saga Y, Gill G N. Proc Natl Acad Sci USA. 1998;95:11257–11262. doi: 10.1073/pnas.95.19.11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taniguchi Y, Furukawa T, Tun T, Han H, Honjo T. Mol Cell Biol. 1998;18:644–654. doi: 10.1128/mcb.18.1.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuya M, Sasaki H, Aoto H, Mitaka T, Nagura K, Ohba T, Ishino M, Takahashi S, Suzuki R, Sasaki T. J Biol Chem. 1998;273:1003–1014. doi: 10.1074/jbc.273.2.1003. [DOI] [PubMed] [Google Scholar]
- 30.Tachibana K, Sato T, D’Avirro N, Morimoto C. J Exp Med. 1995;182:1089–1099. doi: 10.1084/jem.182.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujita H, Kamiguchi K, Cho D, Shibanuma M, Morimoto C, Tachibana K. J Biol Chem. 1998;273:26516–26521. doi: 10.1074/jbc.273.41.26516. [DOI] [PubMed] [Google Scholar]