Abstract
cDNA fragments of the homologues of the Drosophila head homeotic genes labial (lab), proboscipedia (pb), and Deformed (Dfd) have been isolated from the crustacean Porcellio scaber. Because the accumulation domains of the head homeotic complex (Hox) genes had not been previously reported for crustaceans, we studied the expression patterns of these genes in P. scaber embryos by using in situ hybridization. The P. scaber lab homologue is expressed in the developing second antennal segment and its appendages. This expression domain in crustaceans and in the homologous intercalary segment of insects suggests that the lab gene specified this metamere in the last common ancestor of these two groups. The expression domain of the P. scaber pb gene is in the posterior part of the second antennal segment. This domain, in contrast to that in insects, is colinear with the domains of other head genes in P. scaber, and it differs from the insect pb gene expression domain in the posterior mouthparts, suggesting that the insect and crustacean patterns evolved independently from a broader ancestral domain similar to that found in modern chelicerates. P. scaber Dfd is expressed in the mandibular segment and paragnaths (a pair of ventral mouthpart structures associated with the stomodeum) and differs from insects, where expression is in the mandibular and maxillary segments. Thus, like pb, Dfd shows a divergent Hox gene deployment. We conclude that homologous structures of the mandibulate head display striking differences in their underlying developmental programs related to Hox gene expression.
Homologues of the Drosophila homeotic complex (HOM-C) genes from various arthropods have been the subject of intense research and comparative analysis (1–10). A goal of these studies has been to understand the role of these genes in the morphological evolution of this group. The function of Hox gene products as selective transcription factors is apparently highly conserved in animals (1, 2), and the expression patterns of these genes have been used as stable molecular markers for judging evolutionary relationships, such as homologies, of structures along the anteroposterior axis (5, 7, 8). Nevertheless, for the Hox genes to serve as useful tools for phylogenetic comparisons, a better understanding of their particular evolutionary history is needed. Moreover, observations on their expression patterns should ideally be coupled with tests for developmental function (6, 10, 11). Thus far, such comparative analyses have been done primarily on Hox genes in insects, on “trunk” Hox genes in the crustacean Artemia franciscana, and, most recently, in the chelicerates (3–7). Some models for the roles of Hox genes in the evolution of arthropods have already been proposed (2, 4, 5). However, the vast phylogenetic distances between and among the groups studied and the small assortment of genes analyzed make it difficult to create a complete picture of the evolution of Hox genes in arthropods.
In Drosophila, the Hox genes specify structures along the anteroposterior body axis; three are necessary for the correct development of head segments and appendages: the labial (lab), proboscipedia (pb), and Deformed (Dfd) genes. The expression patterns of these genes in insects show small, distinct, and usually nonoverlapping expression domains covering one to two segments. This pattern is in sharp contrast to the expression patterns of their homologues in vertebrate (lab, pb, and Dfd are homologous to the genes of vertebrate classes 1, 2, and 4 respectively) and chelicerate embryos, where expression domains are broadly overlapping and many segments long (7, 8, 12). The fact that the trunk genes Ultrabithorax, abdominal-A, and Abdominal-B are also expressed in extended overlapping domains in insects, chelicerates, and vertebrates suggests that the insect head genes have undergone evolution to limit their expression domains. Unfortunately, in the Mandibulata, which includes the Insecta, Crustacea, and Myriapoda, no data exist for Hox head gene expression patterns for any group other than Insecta.
We have chosen the crustacean Porcellio scaber, order Isopoda, as a noninsect model organism to study the expression patterns of the head Hox genes. It is important to note that our model organism belongs to the subclass Malacostraca (higher crustaceans) and is as derived as insects are in its body plan and tagmatization, relative to phylogenetically more basal groups. Moreover, the interpretations of the expression patterns reported here are based on the assumptions that: (i) the Insecta is monophyletic, with the order Thysanura a basal group; (ii) the Mandibulata are monophyletic with the Crustacea, a sister group of the Insecta; and (iii) the Chelicerata is an outgroup to the Insectan–Crustacean clade (based on refs. 13–17; reviewed in refs. 18 and 19).
The six-segmented mandibulate head is believed to be a primitive character shared by all three extant mandibulate groups (20–22). This feature allows reliable identification of homologous segments and appendages, and it allows predictions regarding expression patterns and possible developmental functions of the Hox genes. Based on the available insect and chelicerate expression pattern data, we proposed two conflicting predictions: (i) because malacostracan crustaceans and insects are related mandibulate groups with morphologically similar head structures, their Hox expression patterns should be similar, if not identical; or (ii) the crustacean patterns could be intermediate between those seen in chelicerates and in insects (6–8). Whereas much is known about the expression patterns for the trunk genes of the crustacean A. franciscana (4), no expression patterns of the head genes are known for any crustacean group, although partial sequences have been published (5, 23). Here, we describe the expression patterns of lab, pb, and Dfd in the crustacean P. scaber. Our observations show that neither of the two predictions presented above is borne out. Rather, a combination of these results and our previously published data on the expression pattern of the head Hox gene Sex combs reduced (Scr) (24) demonstrates that the expression patterns of these genes in this crustacean are divergent from those seen in insects. Moreover, the observed crustacean pattern does not appear to represent a chelicerate–insect intermediate. We conclude that the divergent crustacean/insectan expression patterns reveal that unexpectedly dissimilar developmental processes likely underlie the specification of homologous and morphologically similar mandibulate mouthparts.
MATERIALS AND METHODS
Cloning of the cDNA Fragments and Sequence Analysis.
Total mRNA was isolated from two broods (about 50 embryos) of P. scaber embryos by using the TRIzol reagent (Life Technologies) and following the manufacturer’s instructions. The cDNA was screened by PCR with degenerate primers designed against conserved amino acid regions of the head Hox protein (see Fig. 2). Multiple independent clones produced only repeat copies of the same sequence for the lab, pb, and Dfd genes. All primer pairs and PCR protocols have been previously described (6, 10, 11). A related PCR screen for homeoboxes that used degenerate ELEKEF- and WFQNRR-encoding primers produced single unique copies of the lab, pb, and Dfd homeoboxes matching those cloned with the more specific primers. A total of 39 and 27 homeobox fragments have been cloned from the cDNA pool and from genomic DNA, respectively. For example, the Hom-C fragments cloned from cDNA include 12 Ubx, 9 pb, 3 Scr, 3 Abd-A, and 5 lab homeobox segments with nucleotide sequences identical to the previously cloned orthologous copies of these genes. A short cDNA fragment of the Hox3 class homologue was also cloned and will be described elsewhere. The sequences were compared with our homeobox sequences from miscellaneous arthropods (unpublished data), cloned in the laboratory, and with those available in the National Center for Biotechnology Information database.
Figure 2.
Alignments of the deduced head Hox protein sequences. Hexapeptide + variable region and homeodomain sequences are aligned for four insect species and the crustaceans A. franciscana and P. scaber (some sequences were not available). All sequences are compared with their Drosophila counterparts. Dashes indicate sequence identity, breaches in the sequences indicate introduced gaps. Species are indicated in the first column: Dm, Drosophila melanogaster; Of, Oncopeltus fasciatus; Ad, Acheta domestica; Td, Thermobia domestica; Ps, P. scaber; Af, A. franciscana. Gene names are indicated in the second column: LAB, labial; PB, proboscipedia; DFD, Deformed.
Whole-Mount in Situ Hybridization, Microscopy, and Photography.
In situ hybridization was performed as previously described by Panganiban et al. (26), but with modifications (10), and with the exception that protease K treatment was 10–15 min instead of 1 hr. The size of the in situ probes ranged from 230 bp (pb) to 260 bp (lab). Probes of similar size have been used successfully to reveal specific expression domains of a number of arthropod genes (4, 6, 8, 10, 11, 24). All procedures for mounting and photographing embryos have been described (10, 27).
RESULTS
Development of the P. scaber Head as Revealed by Antibody to Engrailed (EN) Protein.
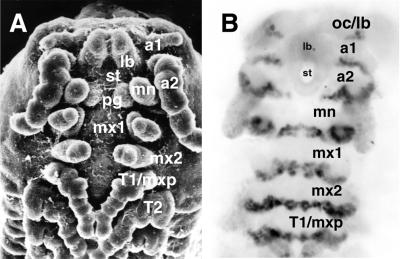
To assign the P. scaber head appendages to specific segments (Fig. 1A), we used the monoclonal antibody Mab4D9, which recognizes EN, to indicate the posterior of the segmental borders (28, 29). The antibody revealed six segments in the embryonic head of P. scaber: ocular, first antennal, second antennal, mandibular, first maxillary, and second maxillary (Fig. 1B). The most anterior region of the head, the labrum, develops as a pair of small appendage-like structures that fuse medially at about the 65–70% stage of development. In early embryos, the EN stripe of the ocular segment is not complete and is interrupted on the ventral side by the labrum. The labrum itself does not express EN and, in this respect, appears to be continuous with the stomodeum. The first antennal segment bears a pair of small uniramose antennae, which are reduced in the adult. The second antennae are the largest pair of the appendages on the head. The stomodeal opening protrudes at the level of the posterior first antennal (a1) segment and extends to the posterior of the second antennal (a2) segment, which results in a ventral interruption of the EN stripes of the a1 and a2 segments. The broad mandibular EN band is seen in the developing posterior mandibular appendages. EN is expressed similarly in the first and second maxillary segments.
Figure 1.
Head appendages and segments of the crustacean P. scaber, ventral view, with anterior at the top. (A) A scanning electron micrograph of a developing (45–50%) embryo (×50.) The segments indicated are as follows: a1, first antennae; a2, second antennae; lb, labrum; mn, mandibles; pg, paragnaths; mx1, first maxillae; mx2, second maxillae; T1/mxp, first thoracic appendages/maxillipeds (B) The head of the embryo stained with Mab4D9 anti-EN antibody. The segments indicated are as follows: oc/lb, ocular/labral; a1, first antennal; a2, second antennal; mn, mandibular; mx1, first maxillary; mx2, second maxillary; T1, first thoracic segments.
The Orthologues of the lab, pb, and Dfd Genes.
Fig. 2 shows an alignment of the predicted partial amino acid sequences for the recovered P. scaber genes lab, pb, and Dfd and their homologues from several insect orders and from A. franciscana. Because of high levels of Porcellio Hox sequence conservation, each cDNA can be unambiguously assigned to its specific HOM-C gene class. Analysis of the alignment in Fig. 2 reveals that the lab homologues from Drosophila melanogaster, Oncopeltus fasciatus, Thermobia domestica, and P. scaber are highly similar immediately downstream of the YKWM motif and in most of the homeobox; however, the N-terminal arm of the homeodomain and the “variable” region just upstream are not conserved among the orthologues. All pb homologues share essentially identical homeobox and N-terminal arm regions (see Fig. 2). Porcellio has the largest variable region of all the sequences shown. Porcellio Dfd is very similar to its homologues in insects and A. franciscana. Its variable region appears to be more similar to that of insects than to that of Artemia.
Hox Gene Expression Patterns. The expression patterns of lab, pb, and Dfd were obtained by using whole mount in situ hybridization to reveal transcript distribution. Expression can be detected from the 30% to the 80% stage but is best seen at the 50–60% stage. All embryos shown in Fig. 3 are at the 50–60% developmental stage. At this stage the embryo begins dorsal closure and its appendages develop unique morphologies. Additionally, the first pair of the thoracic appendages begins to transform into the mouthpart maxillipeds (24).
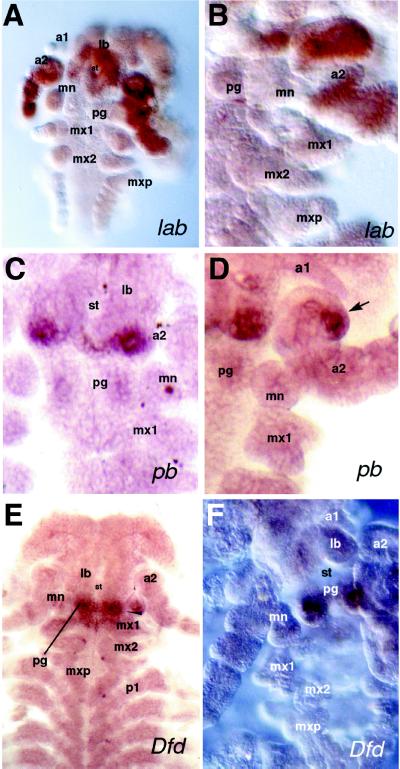
Figure 3.
Embryonic expression patterns of the P. scaber homologues of the Drosophila genes lab, pb, and Dfd, as revealed by in situ hybridization. Abbreviations as in Fig. 1. Ventral view of embryos with anterior at the top in all panels. (A and B) 50–60% Stage embryos showing the lab expression pattern; whole embryo (A) and close-up of embryonic head (B). (C and D) 50–60% Stage embryos showing pb expression in close-up views of the embryonic head revealing details of the expression domain. Arrow points to the pb expression in the posterior-lateral second antennae (D). (E and F) 50–60% Stage embryos showing Dfd mRNA distribution with dissected embryonic head (F) showing strong Dfd expression in the paragnaths. (A and E, ×50; B, C, D, and F, ×100.)
Expression of P. scaber lab is clear in the developing a2 segment and its appendages (Fig. 3A). The embryo in Fig. 3A shows no expression in the first antennae, mandibles, or any other mouthparts. Closer examination reveals that P. scaber lab expression is continuous within the a2 segment and circles the stomodeum (Fig. 3A). Very strong expression just anterior to the stomodeum and in the posterior part of the labrum either suggests that the anterior boundary is ventrally parasegmental or that parts of the aforementioned organs originate from the a2 segment. Fig. 4A shows the P. scaber lab expression domain as a colored overlay on a scanning electron micrograph.
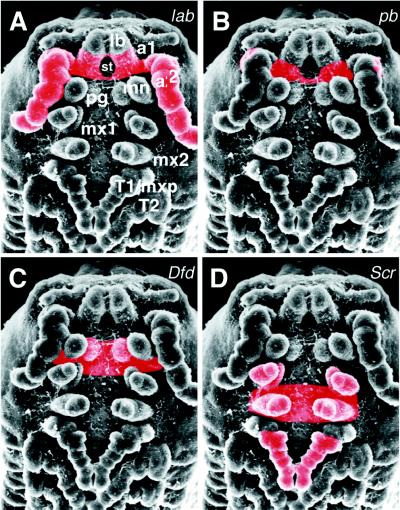
Figure 4.
Summary of Hox expression domains in P. scaber. The main domains of expression in the ectoderm are shown in red: (A) lab is expressed in the second antennal segment and its appendages. (B) pb is expressed in the posterior second antennal segment. (C) Expression of Dfd is limited to the ventral mandibular segment and paragnaths. (D) Scr mRNA distribution in the first maxillary appendages, second maxillary segment, and appendages and distal half of T1 leg (24).
The expression domain of P. scaber pb appears as a narrow stripe in the posterior a2 segment; it is reminiscent of the ventral portion of the EN stripe in that segment (Fig. 1 and Fig. 3C). Like the EN stripe, the P. scaber pb stripe is interrupted medially by the stomodeum. Careful examination of the embryonic head reveals additional strong expression in a small group of cells in the posterior-lateral portion of the a2 appendages (Fig. 3D). These antennal cells form a small bud-like structure, possibly representing a rudimentary antennal exopod (Fig. 3D). The P. scaber pb stripe does not extend into the remainder of the a2 appendage and is restricted to the ventrolateral portion of the a2 segment. Based on the segment morphology and direct comparison to P. scaber lab stained embryos, we conclude that P. scaber pb expression in the a2 segment overlaps that of P. scaber lab. The exact extent of the overlap between the domains of the two genes is difficult to ascertain because of the lack of good morphological markers in the posterior a2 segment. Embryos that were stained for several more hours showed additional weak expression in the paragnaths, ventral mouthpart structures associated with the stomodeum, but this expression appears to be mesodermal (Fig. 3C). No other expression domains can be seen, even in overstained embryos. A summary of the pb expression domains is shown in Fig. 4B.
In embryos of all stages that were examined, the P. scaber Dfd homologue is expressed in the developing paragnaths (Fig. 1; and Fig. 3 E and F; see Discussion). Weak expression in the mandibles is mesodermal. There is also P. scaber Dfd expression in the ventral portion of the mandibular segment, which may be parasegmental; it extends from the mid-mandibular segment into the anterior half of the first maxillary (mx1) (Fig. 3E). A much weaker expression domain in the mesoderm can be recognized in the mandibular appendages (Fig. 3E). There is not the detectable accumulation in the mx1 appendages or posterior half of the mx1 segment that one might have predicted based on the expression of Dfd in the homologous maxillary segment of insects (6). These data are summarized in Fig. 4C.
The pattern of P. scaber Scr expression has been described (24). We note here only that colinearity of expression is exhibited by this gene. The anterior border of Scr accumulation is in the posterior of the mx1 segment, whereas the posterior border of Dfd resides in the anterior of this same segment. It is possible that there is some overlap of expression in the middle of mx1 but a demonstration of this point awaits further experimentation.
DISCUSSION
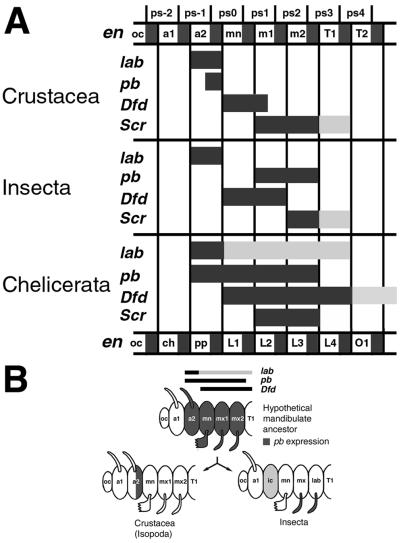
Two comparative analyses of insect and chelicerate Hox expression patterns have been published recently (7, 8). These analyses are intriguing, but they point to the necessity for studies on crustacean and myriapod Hox head genes to gain a better understanding of their evolution in arthropods (7, 8). To partially fill this need, we have determined the embryonic expression patterns for the genes lab, pb, and Dfd in P. scaber, a malacostracan crustacean, and compared our results with data from insects and chelicerates. In general, we found the P. scaber expression patterns to be well-defined and discrete, but not identical to those of insects or chelicerates (Fig. 5).
Figure 5.
(A) Diagram of the Hox gene expression pattern boundaries in the Crustacea (P. scaber), Insecta, and Chelicerata (refs. 6–8; unpublished data). Detailed expression pattern of the Scr homologue of P. scaber has been described elsewhere (24). Columns represent homologous segments. Parasegments and mandibulate segments (as in Crustacea) are labeled at the top of the diagram and chelicerate segments are labeled at the bottom. Bars indicate Hox gene expression patterns. Black bars indicate primary domains (strong expression) and gray bars indicate secondary domains (weak, transient, late, or variable expression). (B) Model for evolution of the homeotic gene pb expression pattern. Schematic head segments and structures are shown for insects, P. scaber, and a hypothetical mandibulate ancestor. Segmental expression of pb homologues is shown in dark gray. The segments are labeled according to the accepted conventions in the respective taxa. Abbreviations as in Figs. 1–4, except that in the schematic Insecta head, ic marks the intercalary segment and lab indicates the labial segment.
Comparison of Insect and Crustacean Hox Expression Domains.lab expression in the Drosophila head is in the intercalary segment, a small metamere that is devoid of appendages, located posterior to the antennal segment and anterior to the mandibular segment. The exact role of lab in this segment is unclear, because mutants do not show an obvious homeotic transformation. Nevertheless, the gene is important for the formation of the embryonic and adult head in Drosophila, because mutants do show defects in the development of cephalic structures (for review, see ref. 30). lab expression in the intercalary segment is conserved in all of the insect species examined thus far (for review, see ref. 6).
Comparison of the P. scaber lab expression domain to that of insects reveals both conservation and change. P. scaber expression is restricted to the first postantennular metamere, i.e., the a2 segment, which, based on morphological and molecular (EN expression) data, is thought to be homologous to the intercalary (22, 29–32). Moreover, innervation of this first postantennular segment is associated with the tritocerebral ganglion of the brain in all mandibulates (31). The fact that lab is expressed in a homologous segment in insects and crustaceans suggests that it was recruited for and might be involved in conserved and homologous developmental processes. However, in Porcellio, lab is expressed in the a2 appendages, whereas the adult insect intercalary segment is limbless. Interestingly, the embryos of some insects develop small transitory appendages on this segment, which might be atavisms to a more primitive state (34). It is not clear, however, whether lab was directly involved in appendage loss in the insects; mutations in this gene do not cause limb growth from the intercalary (30). Nevertheless, it is possible that in crustaceans, lab contributes to the unique morphology of the second as compared with the first pair of antennae. For example, in P. scaber, the first antennae are greatly reduced in size, whereas the second antennae are large and more leg-like.
The pb gene is located upstream of lab in the HOM-C and is expressed in insects in the appendages of the maxillary and labial segments, where it has been shown to specify the posterior mouthparts in Drosophila and Tribolium castaneum (3, 35). Oddly, the main expression domain of pb in Drosophila is not colinear with that of lab and Dfd. In all of the insects surveyed, the anterior boundary of Dfd, the next upstream HOM-C gene relative to pb, is in the mandibular segment (10), and, based on the position of pb in the complex, one might expect its anterior border of expression to be at or anterior to this point. To rationalize this fact, it has been suggested that an ancestral insect pb gene lost its colinear expression pattern and gained a new, appendage-specific role in the maxillae and labium (35). Studies of the expression patterns of pb in several insect orders revealed another, albeit weaker, expression domain in the ventral portion of the intercalary (6). The significance of this domain is not clear, and in some groups, including Drosophila, it was found to be mesodermal. In the apterygote insect T. domestica, there is epidermal pb expression in the intercalary, but it is weak, appears late, and is transient (6, 35). These latter, more anterior patches of expression may be remnants of the posited ancestral, more extensive expression domain.
The pb expression domain of P. scaber was found to be quite dissimilar from the insect pattern. In this crustacean, pb accumulation is restricted to the posterior part of the a2 segment and includes neither the mx1 nor the mx2 segments or appendages (Figs. 3A and 4B). The first and second maxillae of crustaceans are homologous to the maxillae and labium of insects, respectively (30). As pb is required for maxillary and labial appendage development in insects, it clearly cannot be performing a similar function in the P. scaber embryo. At this point, it is difficult to discern a possible developmental function for pb in P. scaber embryos.
In D. melanogaster, the expression domain of Dfd includes the mandibular and maxillary segments and appendages. It has been shown genetically to be required for the normal development of both mandibles and maxillae in Drosophila (30). As noted above, Dfd expression has been found to be very similar in all insect groups studied, including the basal insect T. domestica (10, 11), and the mandibles and maxillae of insects are homologous to the mandibles and first maxillae (maxillulae) of crustaceans (31).
Comparison of the Dfd expression domains in P. scaber and insects reveals that the crustacean domain is smaller (Figs. 3E and 4C). P. scaber Dfd is expressed strongly in the paragnaths and the mandibular segment, but not the mandibular appendages. The paragnaths are associated with the stomodeum, but their exact embryonic origin is obscure. Some authors have concluded that these structures are sternal protrusions of the mandibular segment associated with the mouth, reduced appendages associated with the mandibles, or even structures homologous to the insect hypopharynx (31, 37, 38). Whatever their allegiance, paragnaths are found in a diversity of crustaceans (37). Thus, P. scaber Dfd is not expressed in the mandibles or maxillae (only mesodermal expression is detected in mandibles), where Dfd function is required in insects, suggesting that Dfd has a different developmental function in the crustacean head. It is not known which, if any, selector gene is expressed in the ectoderm of the crustacean mandibles proper.
Comparison with Chelicerate Hox Gene Expression Domains. A comparison of the crustacean/insect and chelicerate patterns of Hox gene expression is made difficult by the uncertainty of segmental homologies between the two groups. The more traditional view, based on anatomy and patterns of innervation, concludes that the segment associated with the deuterocerebral ganglion of the central nervous system is greatly reduced or absent in modern chelicerates (31). If this conclusion is correct, the more anterior ocular/protocerebral segment would be homologous to the same segment in insects and crustaceans, whereas the next posterior segment in the spider would correspond to the intercalary/tritocerebral segment of insects and to the second antennal segment of crustaceans. Thus, the homologue of the insect antennal and the crustacean first antennal segment would be absent in chelicerates (31). More recently, this question of head–segment homology has been revisited by using the patterns of Hox gene expression as a basis for determining the presence or absence of the deuterocerebral segment in chelicerates (7, 8). These authors, using the spider Cupiennius salei, the mite Archegozetes longisetosus, and several head Hox genes as probes, have concluded that this segment is present (7, 8). For the purposes of this discussion we will take this point of view. However, we must stress that the use of only the expression patterns of the Hox genes as a primary reference for anatomical homology is inappropriate. Indeed, we show in this work that boundaries of expression patterns can and do change during evolution, and only some general tendencies, such as colinearity, persist. Nonetheless, in chelicerates, lab is expressed anteriorly in the developing pedipalps (homologous to the appendages of the intercalary segment). This expression extends posteriorly to the fourth pair of walking limbs, which would correspond to the first pair of the thoracic appendages in insects and crustaceans (maxillipeds in P. scaber) (7, 8). Therefore, the anterior boundary of lab expression appears to be conserved among chelicerates, crustaceans, and insects. It is notable, however, that the posterior limit of expression is not conserved. The integumentary expression of lab is limited to the intercalary or second antennal segment in insects and crustaceans, respectively, whereas in chelicerates, expression extends posteriorly a further four full segments (Fig. 5A).
In A. longisetosus, pb is expressed in a broad domain from the pedipalps to the third pair of walking legs, where it is accumulated in the appendages (7). This boundary is colinear with both lab and Dfd, and is thus similar to the relative expression domains of these genes in vertebrates, annelids, and Porcellio (39–41); but it is dissimilar to that seen in insects (6). Thus, the anterior boundary of pb expression in the a2 segment of P. scaber appears to resemble that seen in chelicerates rather than that in insects. However, it should also be noted that, as for lab, the expression domain of chelicerate pb extends further posteriorly than that in insects and crustaceans (Fig. 5A).
Dfd expression has been examined in both C. salei and A. longisetosus. In C. salei, Dfd is expressed in all walking legs (7). The mite shows a similar expression domain in the L1–L4 legs, with additional accumulation covering all opistosomal (abdominal) segments except the most terminal ones (8). The anterior boundary of Dfd accumulation appears to be conserved and located in the mandibular (insects) and homologous L1 (chelicerates) segments (Fig. 5A). If one assigns the paragnaths to the mandibular segment, the anterior boundary of Dfd expression then appears to be similar, albeit not identical, in P. scaber vis-a-vis insects and chelicerates. However, both the chelicerate and insect Dfd domains are clearly broader than those seen in the crustacean.
Hox Genes and the Evolution of Mandibulate Head Structures. As noted above, the extended and broadly overlapping expression domains in chelicerates are reminiscent of those in vertebrates and are probably closer to an ancestral state. In contrast, the expression domains in insects and crustaceans are more resolved and segment-specific (Fig. 4). Based on morphological and recent molecular evidence, the Crustacea belongs to the monophyletic group Mandibulata, which is a close sister group to the Insecta (13–19). Thus, crustaceans represent an ideal case for study of the evolution of the homologous head Hox gene expression patterns and possible functions in the homologous structures of insects.
Comparison of the expression patterns of the crustacean and insect Hox genes demonstrates that there is conservation of segment affinity (e.g., lab) and spatial colinearity (e.g., lab, pb, Dfd, and Scr) of expression (Figs. 4 and 5A; and ref. 24). In addition, the anterior boundaries of the lab and Dfd genes appear to be conserved in insects, crustaceans, and chelicerates. However, there is also divergence of the observed expression domains (e.g., pb and Dfd) (Figs. 4 and 5A). Consequently, substantial variation in the deployment of the Hox genes, and presumably in the developmental processes regulated by them, can be seen in homologous and morphologically similar crustacean and insect head structures. Genes involved in the development of mandibles and posterior mouthparts in insects are expressed in novel, though still colinear domains. For example, in insects, the maxillary and labial mouthparts express pb, whereas in P. scaber, the homologous appendages both express and probably depend on Scr, a different head homeotic gene (refs. 6, 11, 24; also see Fig. 4).
We hypothesize that the mandibulate head evolved prior to the establishment of the defined head Hox gene expression domains, which have been recruited to their current regions and developmental functions independently in crustaceans and insects (Fig. 5B). This model involves an intermediate hypothetical mandibulate ancestor that did not have segment-specific expression domains and probably resembled the pattern of expression seen in modern chelicerates. The specification of individual segments and mouthparts in such an animal would depend on the redundant and/or fractional functions of multiple Hox genes, and would be facilitated by the subsequent evolution of more distinct expression domains (Figs. 4 and 5). That is, the head Hox genes functioned in a manner analogous to the genes of the D. melanogaster Bithorax complex. To test this model and to better understand the evolution of the Hox genes and head structures, further studies across different crustacean and myriapod groups will be required.
Acknowledgments
We thank R. Turner for the scanning electron microscopy; M. Peterson and A. Popadic for providing primers and protocols; Thierry Rigaud for identifying the woodlouse species and culture suggestions; N. Patel for the EN/INV antibody; Dee Verostko for administrative assistance; and G. Scholtz, Kevin Cook, S. Glueck, and T. Powers for valuable comments on the manuscript. This work was supported by the Howard Hughes Medical Institute. T.C.K. is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- a1
first antennal (segment)
- a2
second antennal (segment)
- EN
engrailed
- Hox
homeotic complex (genes)
- mx1
first maxillary (segment)
- Ps
P. scaber
Footnotes
References
- 1.Carroll S B. Nature (London) 1995;376:479–485. doi: 10.1038/376479a0. [DOI] [PubMed] [Google Scholar]
- 2.Warren R, Carroll S B. Curr Opin Genet Dev. 1995;5:459–465. doi: 10.1016/0959-437x(95)90049-m. [DOI] [PubMed] [Google Scholar]
- 3.Beeman R W, Stuart J J, Brown S J, Denell R E. BioEssays. 1993;15:439–444. doi: 10.1002/bies.950150702. [DOI] [PubMed] [Google Scholar]
- 4.Averof M, Akam M. Nature (London) 1995;376:420–423. doi: 10.1038/376420a0. [DOI] [PubMed] [Google Scholar]
- 5.Akam M. Philos Trans R Soc Lond B. 1995;349:313–319. doi: 10.1098/rstb.1995.0119. [DOI] [PubMed] [Google Scholar]
- 6.Rogers B T, Kaufman T C. Int Rev Cytol. 1997;174:1–84. doi: 10.1016/s0074-7696(08)62115-4. [DOI] [PubMed] [Google Scholar]
- 7.Damen W G M, Hausdorf M, Seyfarth E-A, Tautz D. Proc Natl Acad Sci USA. 1998;95:10665–10670. doi: 10.1073/pnas.95.18.10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Telford M J, Thomas R H. Proc Natl Acad Sci USA. 1998;95:10671–10675. doi: 10.1073/pnas.95.18.10671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelsh R, Weinzierl R O J, White R A H, Akam M. Dev Genet. 1994;15:19–31. doi: 10.1002/dvg.1020150104. [DOI] [PubMed] [Google Scholar]
- 10.Rogers B T, Peterson M D, Kaufman T C. Development (Cambridge, U.K.) 1997;124:149–157. doi: 10.1242/dev.124.1.149. [DOI] [PubMed] [Google Scholar]
- 11.Peterson M D, Rogers B T, Popadic A, Kaufman T C. Dev Genes Evol. 1999;209:77–90. doi: 10.1007/s004270050230. [DOI] [PubMed] [Google Scholar]
- 12.Manak J R, Scott M P. Development. U.K.: Cambridge; 1994. Suppl. 1994, 61–71. [Google Scholar]
- 13.Friedrich M, Tautz D. Nature (London) 1995;376:165–167. doi: 10.1038/376165a0. [DOI] [PubMed] [Google Scholar]
- 14.Osorio D, Averof M, Bacon J P. TREE. 1995;10:449–454. doi: 10.1016/s0169-5347(00)89178-8. [DOI] [PubMed] [Google Scholar]
- 15.Ballard J W, Olsen G J, Faith D P, Odgers W A, Rowell D M, Atkinson P W. Science. 1992;258:1345–1348. doi: 10.1126/science.1455227. [DOI] [PubMed] [Google Scholar]
- 16.Boore J L, Collins T M, Stanton D, Daehler L L, Brown W M. Nature (London) 1995;376:163–165. doi: 10.1038/376163a0. [DOI] [PubMed] [Google Scholar]
- 17.Boore J L, Lavrov D V, Brown W M. Nature (London) 1998;392:667–668. doi: 10.1038/33577. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert S F, Raunio A M. Embryology: Constructing the Organism. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 19.Zrzavy J, Hypsa V, Vlaskova M. In: Arthropod Relationships. Fortey R A, Thomas R H, editors. London: Chapman & Hall; 1997. pp. 97–108. [Google Scholar]
- 20.Nielsen C. Animal Evolution: Interrelationships of the Living Phyla. Oxford: Oxford Univ. Press; 1989. [Google Scholar]
- 21.Telford M J, Thomas R H. Nature (London) 1995;376:123–124. [Google Scholar]
- 22.Scholtz G. In: Arthropod Relationships. Fortey R A, Thomas R H, editors. London: Chapman & Hall; 1997. pp. 317–332. [Google Scholar]
- 23.Mouchel-Vielh E, Rigolot C, Gibert J-M, Deutsch J S. Mol Phylogenet Evol. 1998;9:382–389. doi: 10.1006/mpev.1998.0498. [DOI] [PubMed] [Google Scholar]
- 24.Abzhanov A, Kaufman T C. Development (Cambridge, U.K.) 1999;126:1121–1128. doi: 10.1242/dev.126.6.1121. [DOI] [PubMed] [Google Scholar]
- 25.Whitington P M, Leach D, Sandeman R. Development (Cambridge, U.K.) 1993;118:449–461. doi: 10.1242/dev.118.2.449. [DOI] [PubMed] [Google Scholar]
- 26.Panganiban G, Nagy L, Carroll S. Curr Biol. 1994;4:671–675. doi: 10.1016/s0960-9822(00)00151-2. [DOI] [PubMed] [Google Scholar]
- 27.Gorman M J, Kaufman T C. Genetics. 1995;140:557–572. doi: 10.1093/genetics/140.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel N H, Martin-Blanco E, Coleman K G, Pole S J, Ellis M C, Kornberg T B, Goodman C S. Cell. 1989;58:955–968. doi: 10.1016/0092-8674(89)90947-1. [DOI] [PubMed] [Google Scholar]
- 29.Scholtz G. Zoology. 1995;98:104–114. [Google Scholar]
- 30.Kaufman T C, Seeger M A, Olsen G. Adv Genet. 1990;27:309–362. doi: 10.1016/s0065-2660(08)60029-2. [DOI] [PubMed] [Google Scholar]
- 31.Beklemishev W N. Principles of the Comparative Anatomy of Invertebrates. Chicago: Univ. of Chicago Press; 1964. pp. 21–25. , 311–315. [Google Scholar]
- 32.Schmidt-Ott U, Technau G M. Development (Cambridge, U.K.) 1992;116:111–125. doi: 10.1242/dev.116.1.111. [DOI] [PubMed] [Google Scholar]
- 33.Rogers B T, Kaufman T C. Development (Cambridge, U.K.) 1996;122:3419–3432. doi: 10.1242/dev.122.11.3419. [DOI] [PubMed] [Google Scholar]
- 34.Tamarelle M. Int J Insect Morphol Embryol. 1984;13:331–336. [Google Scholar]
- 35.Denell R E, Brown S J, Beeman R W. Semin Cell Dev Biol. 1996;7:527–538. [Google Scholar]
- 36.Peterson M D. Ph.D. thesis. Bloomington: Indiana Univ.; 1998. [Google Scholar]
- 37.Kaestner A. Invertebrate Zoology. Vol. 3. New York: Interscience; 1970. [Google Scholar]
- 38.Schram F R. Crustacea. Oxford: Oxford Univ. Press; 1986. [Google Scholar]
- 39.Hanken J. Am Zool. 1993;33:448–456. [Google Scholar]
- 40.Keynes R. Annu Rev Neurosci. 1994;17:109–132. doi: 10.1146/annurev.ne.17.030194.000545. [DOI] [PubMed] [Google Scholar]
- 41.Matthew J K, Master V A, Lokhorst D K, Nardelli-Haefliger D, Wedeen C J, Martindale M Q, Shankland M. Dev Biology. 1997;190:284–300. doi: 10.1006/dbio.1997.8689. [DOI] [PubMed] [Google Scholar]