Abstract
Green plants seem to form two sister lineages: Chlorophyta, comprising the green algal classes Prasinophyceae, Ulvophyceae, Trebouxiophyceae, and Chlorophyceae, and Streptophyta, comprising the Charophyceae and land plants. We have determined the complete chloroplast DNA (cpDNA) sequence (200,799 bp) of Nephroselmis olivacea, a member of the class (Prasinophyceae) thought to include descendants of the earliest-diverging green algae. The 127 genes identified in this genome represent the largest gene repertoire among the green algal and land plant cpDNAs completely sequenced to date. Of the Nephroselmis genes, 2 (ycf81 and ftsI, a gene involved in peptidoglycan synthesis) have not been identified in any previously investigated cpDNA; 5 genes [ftsW, rnE, ycf62, rnpB, and trnS(cga)] have been found only in cpDNAs of nongreen algae; and 10 others (ndh genes) have been described only in land plant cpDNAs. Nephroselmis and land plant cpDNAs share the same quadripartite structure—which is characterized by the presence of a large rRNA-encoding inverted repeat and two unequal single-copy regions—and very similar sets of genes in corresponding genomic regions. Given that our phylogenetic analyses place Nephroselmis within the Chlorophyta, these structural characteristics were most likely present in the cpDNA of the common ancestor of chlorophytes and streptophytes. Comparative analyses of chloroplast genomes indicate that the typical quadripartite architecture and gene-partitioning pattern of land plant cpDNAs are ancient features that may have been derived from the genome of the cyanobacterial progenitor of chloroplasts. Our phylogenetic data also offer insight into the chlorophyte ancestor of euglenophyte chloroplasts.
Complete chloroplast DNA (cpDNA) sequences have been determined for five land plants (Marchantia polymorpha, ref. 1; Pinus thunbergii, ref. 2; Nicotiana tabacum, ref. 3; Oryza sativa, ref. 4; and Zea mays, ref. 5) and for six algae from different lineages (the glaucocystophyte Cyanophora paradoxa, ref. 6; the red alga Porphyra purpurea, ref. 7; the heterokont Odontella sinensis, ref. 8; the cryptophyte Guillardia theta, ref. 9; the euglenophyte Euglena gracilis, ref. 10; and the green alga Chlorella vulgaris, ref. 11). These sequence data support the view that all chloroplasts were derived from a single primary endosymbiotic event involving the capture of a cyanobacterium (12, 13). The chloroplasts of glaucocystophytes, red algae, and green algae are thought to be direct products of this primary endosymbiotic event, and secondary endosymbioses involving the capture of photosynthetic algae have been postulated for the origin of heterokont, cryptophyte, and euglenophyte chloroplasts (12, 13). A phylogenetic analysis of 45 protein-coding genes common to all cpDNAs sequenced thus far, except those of Chlorella and Guillardia, indicates that the primary chloroplasts of glaucocystophytes evolved before the divergence of the red and green algae and that the secondary chloroplasts of heterokonts were derived from a red alga (14). Given the important differences in structure, gene content and gene organization between the cpDNAs examined thus far, it is difficult to predict the cpDNA architecture of the common ancestor of glaucocystophytes, red algae, and green algae. With 251 genes, Porphyra cpDNA is the most gene-rich cpDNA; in contrast, Euglena, Chlorella, and land plant cp-DNAs contain many fewer genes (97–110). Gene loss has been an ongoing process in all algal lineages, with independent parallel losses occurring in multiple lineages (14).
Considering that the unicellular progenitors of land plants lie within the green algae, deciphering the evolutionary history of cpDNA within this algal group is of particular interest. Phycologists generally recognize five green algal classes: Charophyceae, which include the closest relatives of land plants, Chlorophyceae, Trebouxiophyceae, Ulvophyceae, and a nonmonophyletic group known as Prasinophyceae (13, 15, 16). All extant green algae are thought to belong to two major evolutionary lineages: Streptophyta, containing the charophytes, land plants, and possibly some prasinophytes, and Chlorophyta, containing the rest of the algae. Prasinophytes are considered to represent the earliest offshoots of the chlorophyte lineage.
The cpDNA sequence of the trebouxiophyte Chlorella (11) and the fragmentary cpDNA data available from a few members of the Charophyceae, Ulvophyceae, and Chlorophyceae indicate that green algal cpDNAs evolve in a much less conservative fashion compared with their land plant counterparts (17). These data preclude any prediction regarding the cpDNA architecture of the most ancestral green algae. Whether one of the structural landmarks of the ancestral green algal genome was an rRNA operon-encoding inverted repeat (IR) homologous to that dominating the architecture of most land plant cpDNAs is unknown (12, 17).
Here, we report the complete cpDNA sequence of the prasinophyte Nephroselmis olivacea. This green algal cpDNA and its land plant homologs share the same quadripartite structure—which is defined by the presence of two copies of an IR sequence separated by unequal single-copy regions—and very similar sets of genes in corresponding genomic regions. As Nephroselmis is a member of the Chlorophyta, our results provide insight into the cpDNA organization of the ancestral green algae that appeared before the divergence of the Chlorophyta and Streptophyta. Our results also suggest that the IR and pattern of gene partitioning in the ancestral green algal genome trace their origin to the chloroplasts of the common ancestor of glaucocystophytes, red algae, and green algae.
MATERIALS AND METHODS
As the nuclear and organelle DNAs of N. olivacea (NIES-484) could not be resolved in CsCl-bisbenzimide gradients, a library of total cellular DNA was prepared by ligating partially digested and partially filled-in MboI fragments to LambdaGEM-11 (Promega; ref. 18). Clones carrying cpDNA inserts were first identified by hybridization with Chlamydomonas cpDNA fragments specific to various genes. These clones served as starting points for genome walks undertaken to assemble a collection of clones encompassing Nephroselmis cpDNA—by using, as probes, PCR-amplified fragments complementary to the termini of selected inserts or long PCR-amplified fragments covering gaps between contigs. Numerous clones from each round of hybridization were selected for DNA isolation (18).
All sequences were determined with the PRISM dye terminator cycle sequencing kit (Applied Biosystems) on the ABI model 373 DNA sequencer (Applied Biosystems). Both ends of each insert were sequenced with T7 and SP6 primers, whereas internal regions were sequenced by the primer-walking method with specific 22-mer oligonucleotides. Genomic regions not represented in the clones analyzed were sequenced from PCR-amplified fragments. Sequences were assembled with autoassembler 1.4.0 (Applied Biosystems) and analyzed with the Genetics Computer Group software (Version 9.1, Madison, WI). Statistical analyses of codon usage and base composition at first and third codon positions were performed as described (19).
Multiple protein alignments were carried out with clustalw 1.74 (20). Protein alignments were concatenated; ambiguously aligned regions were deleted; and phylogenetic analyses were performed with protml in molphy 2.3b3 (J. Adachi and M. Hasegawa, Institute of Statistical Mathematics, Tokyo) and the JTT-F model of amino acid replacement. The bootstrap method of resampling of the estimated log-likelihood was used to assess the statistical significance of tree topologies.
RESULTS AND DISCUSSION
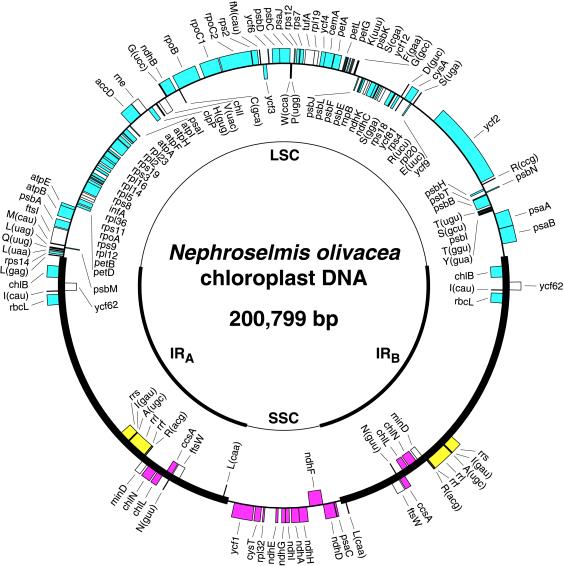
Structure and Gene Content of Nephroselmis cpDNA. The NephroselmiscpDNA sequence assembles as a circle of 200,799 bp, with an overall A + T content of 57.9% (Fig. 1). Two identical copies of a large sequence (46,137 bp) containing the rRNA operon are present in inverted orientation and are separated from one another by LSC (92,126 bp) and SSC (16,399 bp) regions. Such a quadripartite structure is found in the completely sequenced cpDNAs of Cyanophora, Odontella, Guillardia, and land plants, as well as in numerous algal and land plant cpDNAs that have been characterized by physical and gene mapping (17). In NephroselmiscpDNA, the orientation of the IR relative to the single-copy regions is the same as that found in Odontella, Guillardia, and land plant cpDNAs, i.e., the 5S rRNA genes (rrf) are proximal to the SSC region and distal to the LSC region. In CyanophoracpDNA, the IR is in a reverse orientation.
Figure 1.
Gene map of Nephroselmis cpDNA and compared patterns of gene partitioning in Marchantia and Nephroselmis cpDNAs. Thick lines represent the two copies of the IR (IRA and IRB). The relative orientation of the small single-copy (SSC) and large single-copy (LSC) regions is arbitrary in the representation shown. Genes on the outside of the map are transcribed in a clockwise direction; those inside the map are transcribed counterclockwise. The color-code denotes the genomic regions containing the homologous genes in Marchantia: cyan, LSC region; magenta, SSC region; and yellow, IR. Transfer RNA genes are indicated by the one-letter amino acid code followed by the anticodon in parentheses. The ORFs unique to Nephroselmis cpDNA are not indicated (see GenBank accession no. AF137379).
A total of 127 genes are encoded by Nephroselmis cpDNA (Table 1). This gene repertoire is the largest among the green algal and land plant cpDNAs sequenced to date, representing 18 and 7 additional genes compared with the gene complements of Chlorella and Marchantia cpDNAs, respectively (see Fig. 3). In Nephroselmis cpDNA, seven genes are absent from all previously investigated green algal and land plant cpDNAs (Table 1). These include the gene encoding the RNA component of RNase P (rnpB), trnS(cga), and five protein-coding genes. The latter five genes consist of ftsI and ftsW, each encoding a component of the chloroplast division machinery; rne, encoding RNase E, an enzyme probably participating in processing of rRNA precursors and mRNAs; and finally ycf62 and ycf81, two genes encoding proteins of unknown function. All seven genes, except ftsI and ycf81, have been found in nongreen algal cpDNAs (14). Before our study, ycf81 and ftsI had only been identified in bacterial genomes, with ycf81 having been detected in Synechocystis sp. PCC6803, Bacillus subtilis, and Thermus aquaticus and with ftsI having been found in many bacteria.
Table 1.
Genes encoded by Nephroselmis cpDNA
| Gene products | Genes |
|---|---|
| Photosystem I | psaA, B, C, I, J |
| Photosystem II | psbA, B, C, D, E, F, H, I, J, K, L, M, N, T |
| Cytochrome b6/f | petA, B, D, G, L |
| ATP synthase | atpA, B, E, F, H, I |
| Chlorophyll biosynthesis | chlB, I, L, N |
| Rubisco | rbcL |
| NADH oxidoreductase | ndhA, B, C, D, E, F, G, H, I, K |
| Large subunit ribosomal proteins | rpl2, 5, 12, 14, 16, 19, 20, 23, 32, 36 |
| Small subunit ribosomal proteins | rps2, 3, 4, 7, 8, 9, 11, 12, 14, 18, 19 |
| RNA polymerase | rpoA, B, C1, C2 |
| Translation factors | infA, tufA |
| Chloroplast division | ftsI, ftsW, minD |
| Other proteins | accD, ccsA, cemA, clpP, cysA, cysT, rne |
| Proteins of unknown function | ycf1, 2, 3, 4, 6, 9, 12, 62, 81 |
| Ribosomal RNAs | rrf, rrl, rrs |
| Transfer RNAs | trnA(ugc), C(gca), D(guc), E(uuc), F(gaa), G(gcc), G(ucc), H(gug), I(cau), I(gau), K(uuu), L(caa), L(gag), L(uaa), L(uag), fM(cau), M(cau), N(guu), P(ugg), Q(uug), R(acg), R(ccg), R(ucu), S(cga), S(gcu), S(gga), S(uga), T(ggu), T(ugu), V(uac), W(cca), Y(gua) |
| Other small RNAs | rnpB |
Underlined genes have not been found in previously investigated green algal and land plant cpDNAs. The ORFs unique to Nephroselmis cpDNA are not listed.
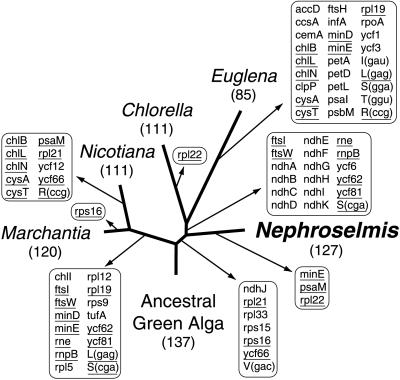
Figure 3.
Phylogenetic distribution of gene loss from cpDNA in Chlorophyta and Streptophyta. Loss events were mapped on the tree shown in Fig. 2. Numbers below species names indicate numbers of genes in corresponding cpDNAs. Note that intron-encoded genes were not considered here. Genes that were lost independently in different lineages are underlined.
Given the biochemical evidence that RNase P from spinach chloroplasts lacks an RNA component (21) and the phylogenetic evidence that Nephroselmis represents an early offshoot of the Chlorophyta (see below), the finding of rnpB in this chlorophyte cpDNA implies that the RNase P RNA was lost specifically in Streptophyta. The presence of ftsI in Nephroselmis cpDNA was most unexpected, considering that in bacteria the ftsI product catalyzes growth of the peptidoglycan layer during septum formation (22) and that chloroplast envelopes of green algae have not been reported to contain any peptidoglycan layer. Only the chloroplasts of Cyanophora have been shown to feature a peptidoglycan cell wall (23) and to contain a second gene required for septum formation (ftsW). The finding of ftsI in Nephroselmis suggests that a peptidoglycan layer or vestige of this layer may be more widespread than previously documented in algal chloroplasts. Moreover, the presence of ftsI and ftsW as well as minD (in both Nephroselmis and Chlorella) and minE (in Chlorella) in green algae provides molecular evidence for the conservation of division mechanisms during the evolution of chloroplasts from their endosymbiotic progenitors.
Another surprising feature of the Nephroselmis chloroplast gene repertoire is the presence of 10 homologs of mitochondrial genes encoding subunits of the NADH:ubiquinone oxidoreductase (ndh; Table 1). These genes, as well as ndhJ, have been found in four of the five completely sequenced land plant cpDNAs; however, no ndh genes have been reported previously in an algal cpDNA (12, 17). The independent losses of all chloroplast ndh genes in algal lineages together with the complete loss of the same genes in Pinus raise questions about the functional significance of these genes. It has recently been shown that Ndh proteins from tobacco chloroplasts are part of a functional complex, which is dispensable for plant growth under optimal conditions (24, 25). This complex may participate in cyclic electron flow around the photosystem I complex in the light and in chlororespiration in the dark.
In Nephroselmis cpDNA, 33 ORFs of more than 80 codons have also been identified. Of these ORFs, 20 reside in a 20.9-kilobase (kb) segment of the IR that lacks any standard chloroplast genes. The density of coding sequences in this region (55%) is significantly lower than that in the rest of the genome (79%). Even though the 20.9-kb segment shows no significant difference with respect to A + T content (59% versus 57% for the rest of the genome), codon usage as well as base composition at the first and third codon positions were found to be atypical in statistical analyses. This result suggests that the 20.9-kb segment was acquired by lateral transfer as proposed for the long extra sequence in the IR of the chlorophycean green alga Chlamydomonas moewusii (26). A separate (6.9-kb) segment of the Nephroselmis IR that lacks standard chloroplast genes and contains only two ORFs might have also been acquired by lateral transfer, as it is significantly richer in A + T (67%) compared with typical regions of the chloroplast genome.
Like its Porphyra, Odontella, and Guillardia homologs, Nephroselmis cpDNA contains no introns. This fact is consistent with the idea that there was no more than one intron in the cpDNA of the common ancestor of chlorophytes and streptophytes and that chloroplast introns proliferated independently in Chlorophyta and Streptophyta (12, 17). About 20 introns, most of which belong to the group II intron family, are present in land plant cpDNAs (17). Although introns are also abundant in Chlamydomonas cpDNAs, they are located in entirely different positions and mostly fall within the group I family (12). Only three group I introns are present in Chlorella cpDNA (11); the one inserted in trnL(caa) seems to have been inherited vertically from the cyanobacterial ancestor of chloroplasts, because a similar intron at an identical position in the same tRNA gene is found in cyanobacteria and in the cpDNAs of Cyanophora, land plants, and some charophytes (27, 28).
Phylogenetic Position of Nephroselmis Chloroplasts.
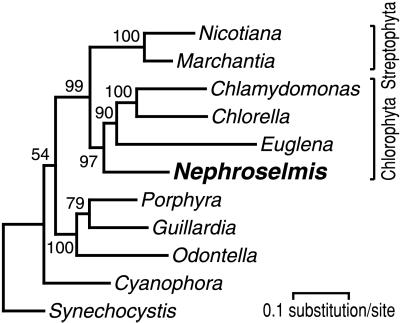
To determine this position, we analyzed a data set containing the concatenated sequences of 37 cpDNA-encoded proteins (Fig. 2). These sequences represent all of the protein-coding genes shared by the 10 selected cpDNAs and the Synechocystis genome (29), with the exception of rbcL, which was excluded because it has undergone a number of duplications and transfer events during chloroplast evolution (12). Although the cpDNA of Chlamydomonas reinhardtii (Chlorophyceae) has not been sequenced completely, it was used to generate a phylogeny comprising at least two chlorophytes (the other being Chlorella). As shown in Fig. 2, Nephroselmis clusters robustly with Chlorella, Chlamydomonas, and Euglena, and, in agreement with phylogenies inferred from nuclear-encoded 18S rRNA gene sequences (15, 30), Nephroselmis represents a basal branch of the Chlorophyta. The inclusion of Euglena in Chlorophyta confirms other evidence that euglenophyte chloroplasts were acquired secondarily from a green alga (12, 13). It is likely that this green algal ancestor was a prasinophyte more advanced than Nephroselmis or a member of the class (Ulvophyceae) believed to have emerged before the divergence of the Trebouxiophyceae and Chlorophyceae (15, 16). Fig. 2 also shows that the chloroplasts of Odontella, Guillardia, and Porphyra form a strongly supported clade, thus supporting the hypothesis that the secondary chloroplasts of heterokonts and cryptophytes are of red algal origin (12, 13).
Figure 2.
Phylogenetic position of Nephroselmis as inferred from 37 chloroplast proteins. The figure shows the best tree that was computed with protml from a data set of 7,449 amino acid positions. Bootstrap values are indicated at the nodes. Following are the names of the genes analyzed, with the lengths of alignment used indicated in parentheses: atpA (497), atpB (456), atpE (114), atpH (80), petB (213), petG (33), psaA (740), psaB (708), psaC (78), psaJ (31), psbA (344), psbB (506), psbC (448), psbD (342), psbE (67), psbF (33), psbH (52), psbI (32), psbJ (35), psbK (39), psbL (35), psbN (41), psbT (29), rpl14 (111), rpl16 (132), rpl20 (99), rpoB (734), rpoC2 (459), rps3 (113), rps4 (134), rps7 (139), rps11 (117), rps12 (116), rps18 (47), rps19 (91), ycf4 (150), and ycf9 (54).
Pattern of Gene Losses from cpDNA in Chlorophyta and Streptophyta.
Given that Nephroselmis represents an early branch of the Chlorophyta, any similarity between its cpDNA and those of its land plant counterparts can be interpreted as reflecting a feature of the cpDNA architecture from the common ancestor of chlorophytes and streptophytes. Based on the tree topology shown in Fig. 2, it can be predicted that the ancestral green algal cpDNA carried a minimum of 137 genes (100 protein-coding genes, 33 tRNA genes, 3 rRNA genes, and rnpB) and that only 10 events of gene loss led to the chloroplast gene content of Nephroselmis (Fig. 3). With 17 events of gene loss accounting for its gene content, Marchantia cpDNA is the second closest genome to that of the common ancestor of chlorophytes and streptophytes. Multiple genes were independently lost in Streptophyta and Chlorophyta, including the previously mentioned ndh genes and the seven genes unique to Nephroselmis cpDNA (Fig. 3).
Gene Organization.NephroselmiscpDNA has gene clusters that are conserved in most or all other cpDNAs examined to date and often in cyanobacteria. The largest of these clusters (rpl23 to rps9) includes 13 genes and corresponds to the str-S10-spc-α operons of Escherichia coli. Particularly noteworthy is the finding that Nephroselmis ndh genes are arrayed in a fashion mirroring the order of its homologs in land plant cpDNAs. Of the 10 Nephroselmis ndh genes, 9 form three linkage groups (ndhC–K, ndhH–A–I–G–E, and psaC-ndhD) that are identical to those found in land plants, with the exception that the ndhC cluster lacks ndhJ. The ndhC–K and ndhH–A–I–G–E clusters are also present in Synechocystis.
Apart from the presence of ancestral clusters, the overall gene order in Nephroselmis cpDNA differs markedly from the gene arrangements in Chlorella and land plant cpDNAs (Table 2). Nephroselmis and Chlorella cpDNAs share only seven sets of genes that are not conserved in other completely sequenced cpDNAs; seven gene clusters also are shared specifically between Nephroselmis and land plant cpDNAs. A single chloroplast gene cluster of noncyanobacterial origin is shared specifically between Nephroselmis and a nongreen alga. This cluster, ftsW/trnN(guu), is found in Cyanophora.
Table 2.
Nephroselmis chloroplast gene clusters shared specifically with Chlorella and land plant (as represented by Marchantia) cpDNAs
| Compared group | Shared chloroplast gene clusters |
|---|---|
| Chlorella | rpl23-rpl2-rps19-rps3-rpl16-rpl14-rpl5-rps8- infA-rpl36-rps11-rpoA-rps9-rpl12 C(gca)/rpoB-rpoC1-rpoC2 W(cca)-P(ugg)/psaJ-rps12-rps7-tufA-rpl19-ycf4petA-petL-petGE(uuc)-rpl20ycf5/chlL-chlNrpl32-cysT-ycf1 |
| Land plants | ycf4-cemA-petA |
| ndhC-ndhK | |
| cysA/E(uuc) | |
| ycf9/S(uga) | |
| rrs-I(agu)-A(ugc)-rrl-rrf-R(acg) | |
| psaC-ndhD | |
| ndhH-ndhA-ndhI-ndhG-ndhE |
Contiguous genes encoded on the same DNA strand are separated by hyphens, whereas contiguous genes encoded on different strands are separated by slashes.
Gene Partitioning. The pattern of gene partitioning in NephroselmiscpDNA is remarkably similar to that found in most land plant cpDNAs containing an IR structure. In MarchantiacpDNA, homologs of all the Nephroselmis genes mapping to the SSC or LSC region, with the exception of trnL(uag), are located in the corresponding single-copy region (Fig. 1 and Table 3). Although several differences in gene content are observed at the level of the IR, most of these differences can be attributed to the well known property of the IR to expand and contract by gene conversion or recombinational repair of double-strand breaks (31). Consistent with the idea that the NephroselmisIR has been enlarged compared with that of Marchantia, each of its terminal regions features a number of genes that reside in the immediately adjacent single-copy region in MarchantiacpDNA (Fig. 1). One exception is trnL(caa), which is found near the SSC region in Nephroselmis but is present in the LSC region in land plants.
Table 3.
Comparison of gene contents in the smallest segment delimited by the two copies of the rRNA operon in IR-containing cpDNAs
| Gene* | Cyanophora | Guillardia | Odontella | Nephroselmis | Marchantia | Nicotiana |
|---|---|---|---|---|---|---|
| ccsA | ● | ● | ● | ● | ● | ● |
| chlL | ● | ● | ● | |||
| chlN | ● | ● | ● | |||
| cysT | ● | ● | ||||
| ftsW | ● | ● | ||||
| minD | ● | ● | ||||
| ndhA | ● | ● | ● | |||
| ndhD | ● | ● | ● | |||
| ndhE | ● | ● | ● | |||
| ndhF | ● | ● | ● | |||
| ndhG | ● | ● | ● | |||
| ndhH | ● | ● | ● | |||
| ndhI | ● | ● | ● | |||
| psaC | ● | ● | ● | ● | ● | ● |
| psbA | ○ | ● | ● | ○ | ○ | ○ |
| rbcR | ● | ● | ● | |||
| rpl21 | ● | ● | ● | ● | ||
| rpl27 | ● | ● | ||||
| rpl32 | ● | ● | ● | ● | ● | |
| rpl34 | ● | ● | ● | |||
| rps6 | ● | ● | ● | |||
| rps15 | ● | ● | ||||
| secA | ● | ● | ||||
| ycf1 | ● | ● | ● | |||
| ycf27 | ● | ○ | ||||
| ycf32 | ● | ● | ● | |||
| ycf35 | ● | ● | ● | |||
| ycf44 | ● | ● | ||||
| ycf46 | ● | ● | ||||
| L(uag) | ● | ● | ● | ○ | ● | ● |
| L(caa) | ○ | ○ | ● | ○ | ○ | |
| N(guu) | ● | ● | ● | ● | ● | ● |
| R(acg) | ○ | ○ | ○ | ● | ● | ● |
Presence of a gene in the smallest segment delimited by the rRNA operons is denoted by ●, whereas its presence in the largest segment delimited by these operons is denoted by ○. Absence of a gene from a genome is indicated by the absence of symbol.
Only the genes encoded by more than one of the compared cpDNAs are indicated. The 41 genes forming the cluster extending from rps16 to clpC in Odontella cpDNA are not indicated, because their homologs are found in the largest segment delimited by the rRNA operons.
From the striking similarities reported above, we infer that the cpDNA of the common ancestor of chlorophytes and streptophytes contained an rRNA-encoding IR with the same orientation as the IR in Nephroselmis and land plant cpDNAs and that the ancestor’s pattern of gene partitioning closely matched the pattern found in the later cpDNAs. The IR structure is thus a more stable element than previously thought. In Streptophyta, it has survived the long evolutionary period separating the common ancestor of chlorophytes and streptophytes from extant land plants, although it has been lost on a number of separate occasions (17). The similar gene content observed in the corresponding genomic regions of Nephroselmis and land plant cpDNAs also implies that gene relocations from one single-copy region to the other via inversions of sequences encompassing one copy of the IR sequence have occurred rarely. Such events are rare, probably because the direct repeats produced after inversion of the IR sequence are prone to homologous recombination, a process resulting in loss of essential chloroplast genes (17). In the cpDNA of the common ancestor of Chlamydomonads (Chlorophyceae), inversion of one copy of the IR has presumably occurred without the fatal loss of the chloroplast genome, thus yielding, after a second inversion event, single-copy regions of about equal sizes containing radically different gene contents compared with the equivalent regions in Nephroselmis and land plant cpDNAs (32).
To our surprise, close inspection of the patterns of gene partitioning in the IR-containing cpDNAs of Cyanophora and Guillardia revealed striking similarities with the pattern observed in Nephroselmis and land plant cpDNAs. As shown in Table 3, all genes in the smallest segment delimited by the rRNA operons in Cyanophora and Guillardia cpDNAs, with the exception of psbA, trnL(uag), trnL(caa), and trnR(acg), have their homologs in this same genomic region in Nephroselmis, Marchantia, and Nicotiana cpDNAs. Odontella cpDNA has a similar pattern of gene partitioning, although this pattern is less evident because of gene scrambling. Homologs of all Odontella genes in the smallest segment delimited by the rRNA operons, except psbA, trnL(uag), trnR(acg), and a cluster of 41 contiguous genes extending from rps16 to clpC, are located in this same region in other completely sequenced cpDNAs (Table 3). Transfer of the latter gene cluster from the LSC region during chloroplast genome evolution may account for the larger size of the Odontella SSC region compared with the same region in other cpDNAs.
The structural similarities reported here strongly suggest that the IRs of all chloroplast genomes have a single origin and that gene transfers between different single-copy regions were rare events during the very long evolutionary period separating Cyanophora from the common ancestor of chlorophytes and streptophytes. These similarities thus reinforce the conclusion, based primarily on phylogenetic evidence (12–14), that all primary plastids trace back to a single, primary endosymbiotic event. Our view on the origin of the IR is not consistent with the idea that the large direct repeat found in Porphyra cpDNA represents an ancestral feature (9, 33) and that the IR of Guillardia cpDNA resulted from an inversion of one copy of this structure after recombination (9). The patterns of gene partitioning in these two cpDNAs as well as the presence of an IR in at least two red algal cpDNAs [those of Porphyra yezoensis (ref. 34) and Cyanidium caldarium (ref. 35)] are more consistent with the hypothesis that the direct repeats of Porphyra appeared after the inversion of a cpDNA segment encompassing one copy of an ancestral IR and adjacent single-copy sequences. Interestingly, the Cyanidium IR has the same orientation as its homologs in Guillardia, Odontella, Nephroselmis, and land plants (35).
The presence of a rRNA-encoding IR in the genome of Synechocystis (29) makes credible the hypothesis that the IR of all chloroplast genomes originated from a similar structure in the cyanobacterial ancestor of chloroplasts. Although the orientation of the Synechocystis IR is the same as that of Cyanophora, the pattern of gene partitioning in the cyanobacterial genome differs in many ways from that of the glaucocystophyte cpDNA. These differences may be attributed to gene rearrangements that took place during the independent evolution of cyanobacteria, because the presence of a large number of transposase genes in Synechocystis suggests that cyanobacterial genomes are highly plastic in their gene organization (29).
Acknowledgments
We thank Michel Guertin for methodological advice, Stéphane Lévesque for preparing DNA templates, and Jeffrey D. Palmer for constructive criticism. When this research was performed, M.T. and C.L. were Scholars in the Program in Evolutionary Biology of the Canadian Institute for Advanced Research, for which we are grateful for salary support. This work was supported by Natural Sciences and Engineering Research Council of Canada Grants GP0003293 (to M.T.) and GP0002830 (to C.L.).
ABBREVIATIONS
- cpDNA
chloroplast DNA
- IR
inverted repeat
- LSC
large single-copy
- SSC
small single-copy
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF137379).
References
- 1.Ohyama K, Fukuzawa H, Kohchi T, Shirai H, Sano T, Sano S, Umesono K, Shiki Y, Takeuchi M, Chang Z, et al. Nature (London) 1986;322:572–574. [Google Scholar]
- 2.Wakasugi T, Tsudzuki J, Ito S, Nakashima K, Tsudzuki T, Sugiura M. Proc Natl Acad Sci USA. 1994;91:9794–9798. doi: 10.1073/pnas.91.21.9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwonge J, Obokata J, Yamaguchi-Shinozaki K, et al. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiratsuka J, Shimada H, Whittier R, Ishibashi T, Sakamoto M, Mori M, Kondo C, Honji Y, Sun C-R, Meng B-Y, et al. Mol Gen Genet. 1989;217:185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- 5.Maier R M, Neckermann K, Igloi G L, Kossel H. J Mol Biol. 1995;251:614–628. doi: 10.1006/jmbi.1995.0460. [DOI] [PubMed] [Google Scholar]
- 6.Stirewalt V, Michalowski C, Löffelhardt W, Bohnert H, Bryant D. Plant Mol Biol Rep. 1995;13:327–332. [Google Scholar]
- 7.Reith M, Munholland J. Plant Mol Biol Rep. 1995;13:333–335. [Google Scholar]
- 8.Kowallik K V, Stoebe B, Schaffran I, Kroth-Panic P, Freier U. Plant Mol Biol Rep. 1995;13:336–342. [Google Scholar]
- 9.Douglas S E, Penny S L. J Mol Evol. 1999;48:236–244. doi: 10.1007/pl00006462. [DOI] [PubMed] [Google Scholar]
- 10.Hallick R B, Hong L, Drager R G, Favreau M R, Monfort A, Orsat B, Spielmann A, Stutz E. Nucleic Acids Res. 1993;21:3537–3544. doi: 10.1093/nar/21.15.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wakasugi T, Nagai T, Kapoor M, Sugita M, Ito M, Ito S, Tsudzuki J, Nakashima K, Tsudzuki T, Suzuki Y, et al. Proc Natl Acad Sci USA. 1997;94:5967–5972. doi: 10.1073/pnas.94.11.5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmer J D, Delwiche C F. In: Molecular Systematics of Plants II DNA Sequencing. Soltis D E, Soltis P S, Doyle J J, editors. Norwell, MA: Kluwer; 1998. pp. 375–409. [Google Scholar]
- 13.Bhattacharya D, Medlin L. Plant Physiol. 1998;116:9–15. [Google Scholar]
- 14.Martin W, Stoebe B, Goremykin V, Hansmann S, Hasegawa M, Kowallik K V. Nature (London) 1998;393:162–165. doi: 10.1038/30234. [DOI] [PubMed] [Google Scholar]
- 15.Friedl T. Plant Syst Evol, Suppl. . 1997;11:87–101. [Google Scholar]
- 16.Chapman R L, Buchheim M A, Delwiche C F, Friedl T, Huss V A, Karol K G, Lewis L A, Manhart J, McCourt R M, Olsen J L, et al. In: Molecular Systematics of Plant II DNA Sequencing. Soltis D E, Soltis P S, Doyle J J, editors. Norwell, MA: Kluwer; 1998. pp. 508–540. [Google Scholar]
- 17.Palmer J D. In: The Molecular Biology of Plastids. Bogorad L, Vasil K, editors. San Diego: Academic; 1991. pp. 5–53. [Google Scholar]
- 18.Turmel, M., Lemieux, C., Burger, G., Lang, B. F., Otis, C., Plante, I. & Gray, M. W. (1999) Plant Cell, in press. [DOI] [PMC free article] [PubMed]
- 19.Lawrence J G, Ochman H. J Mol Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- 20.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas B C, Gao L, Stomp D, Li X, Gegenheimer P A. Nucleic Acids Symp Ser. 1995;33:95–98. [PubMed] [Google Scholar]
- 22.Donachie W D. Annu Rev Microbiol. 1993;47:199–230. doi: 10.1146/annurev.mi.47.100193.001215. [DOI] [PubMed] [Google Scholar]
- 23.Aitken A, Stanier R Y. J Gen Microbiol. 1979;212:218–223. [Google Scholar]
- 24.Burrows P A, Sazanov L A, Svab Z, Maliga P, Nixon P J. EMBO J. 1998;17:868–876. doi: 10.1093/emboj/17.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shikanai T, Endo T, Hashimoto T, Yamada Y, Asada K, Yokota A. Proc Natl Acad Sci USA. 1998;95:9705–9709. doi: 10.1073/pnas.95.16.9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemieux C, Turmel M, Lee R W, Bellemare G. Plant Mol Biol. 1985;5:77–84. doi: 10.1007/BF00020089. [DOI] [PubMed] [Google Scholar]
- 27.Kushel M G, Strickland R, Palmer J D. Science. 1990;250:1570–1573. doi: 10.1126/science.2125748. [DOI] [PubMed] [Google Scholar]
- 28.Xu M Q, Kathe S D, Goodrich-Blair H, Nierzwicki-Bauer S A, Shub D A. Science. 1990;250:1566–1570. doi: 10.1126/science.2125747. [DOI] [PubMed] [Google Scholar]
- 29.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 30.Steinkötter J, Bhattacharya D, Semmelroth I, Bibeau C, Melkonian M. J Phycol. 1994;30:340–345. [Google Scholar]
- 31.Goulding S E, Olmstead R G, Morden C W, Wolfe K H. Mol Gen Genet. 1996;252:195–206. doi: 10.1007/BF02173220. [DOI] [PubMed] [Google Scholar]
- 32.Boudreau E, Otis C, Turmel C. Plant Mol Biol. 1994;24:585–602. doi: 10.1007/BF00023556. [DOI] [PubMed] [Google Scholar]
- 33.Reith M, Munholland J. Curr Genet. 1993;24:443–450. doi: 10.1007/BF00351855. [DOI] [PubMed] [Google Scholar]
- 34.Shivji M S. Curr Genet. 1991;19:49–54. [Google Scholar]
- 35.Maid U, Zetsche K. Plant Mol Biol. 1992;19:1001–1010. doi: 10.1007/BF00040531. [DOI] [PubMed] [Google Scholar]