Abstract
The genomes of modern humans are riddled with thousands of endogenous retroviruses (HERVs), the proviral remnants of ancient viral infections of the primate lineage. Most HERVs are nonfunctional, selectively neutral loci. This fact, coupled with their sheer abundance in primate genomes, makes HERVs ideal for exploitation as phylogenetic markers. Endogenous retroviruses (ERVs) provide phylogenetic information in two ways: (i) by comparison of integration site polymorphism and (ii) by orthologous comparison of evolving, proviral, nucleotide sequence. In this study, trees are constructed with the noncoding long terminal repeats (LTRs) of several ERV loci. Because the two LTRs of an ERV are identical at the time of integration but evolve independently, each ERV locus can provide two estimates of species phylogeny based on molecular evolution of the same ancestral sequence. Moreover, tree topology is highly sensitive to conversion events, allowing for easy detection of sequences involved in recombination as well as correction for such events. Although other animal species are rich in ERV sequences, the specific use of HERVs in this study allows comparison of trees to a well established phylogenetic standard, that of the Old World primates. HERVs, and by extension the ERVs of other species, constitute a unique and plentiful resource for studying the evolutionary history of the Retroviridae and their animal hosts.
Retroviruses are unique among RNA viruses in their ability to integrate DNA copies of their genomes into the genome of the infected cell. On occasion, integration takes place in a germ-line cell, giving rise to an endogenous retrovirus (ERV), which can be inherited by the offspring of the infected host, and may eventually become fixed in the gene pool of the host population (1). The genomes of vertebrate species contain dozens to thousands of ERV sequences (2), some of which were acquired in evolutionarily recent times, whereas others derive from “ancient” times, as indicated by their identical site of integration in more than one species (1, 3, 4). Typically, ancient proviruses have sustained numerous point mutations, deletions, and insertions, rendering them incapable of expressing virus. No biologically active viruses have been associated with the ancient proviruses.
Despite their abundance in vertebrate genomes, and some other especially useful features described below, ERVs have rarely been exploited as phylogenetic markers (5–10). In a few instances integration site polymorphisms have served as a source of phylogenetic signal (6), or as markers for linkage analysis (11), but the usefulness of orthologous ERV nucleotide sequences has never been fully explored. Here we report the application of ancient human endogenous retrovirus (HERV) sequences to phylogenetic analysis on a time scale spanning recent primate evolution.
HERVs can be organized into at least a dozen distinct groups, which vary in size from one to thousands of members (1, 12). Cross-hybridization and PCR studies consistently reveal that most HERV families are also found in other primates, including apes and Old World monkeys (OWMs) (12–19). Many HERVs, including the ones used in this study, are the result of integration events that took place between 5 and 50 million years ago, as indicated by the distribution of specific proviruses at the same integration sites (or “loci”) among related species. The evolution of primates has been the subject of intense study for well over a century, providing a well established phylogenetic consensus with which to compare and evaluate the performance of ERVs as phylogenetic markers.
METHODS
Preparation of Genomic DNA Samples.
Human DNA samples came from the National Institute of General Medical Sciences Human Genetic Mutant Cell Repository. Great-ape genomic DNA was extracted from whole blood (Yerkes Regional Primate Research Center) or immortalized cell lines, including WES (chimpanzee), ROK (gorilla), PUTI (orangutan), MLA144 (gibbon), and COS-7 (African green monkey) cells (American Type Culture Collection; Manassas, VA). Additional blood samples were purchased from the New England Regional Primate Research Center (Southborough, MA) and included rhesus macaque, cynomolgus macaque, baboon, marmoset, and spider monkey samples. Genomic DNA was extracted from cells by digestion with SDS/Pronase and extraction with phenol or from whole blood by using the QIAamp Blood Kit (Qiagen, Santa Clarita, CA.).
PCR and Sequencing. All PCR mixtures contained 200–400 ng of genomic DNA, 1.5–2.0 mM MgCl2, 200 μM each dNTP, 0.2 μM each primer, and 4 units of Taq DNA polymerase (Perkin–Elmer).
HERV-K(HML6.17) (GenBank accession nos. U60268 and U60269). Primers for amplifying the 5′ long terminal repeat (LTR) from human, chimpanzee, bonobo, gorilla, and gibbon were HML1 (5′-TTGCCTTCTCAAGACAATATGGGC-3′) and HML2 (5′-AGGCGCTGACCTCATGTGCGC-3′). The 5′ LTR from orangutan, African green monkey, baboon, cynomolgus macaque, and rhesus macaque was amplified with HML7 (5′-GGGCAAAGTGAGTACATTTAGTGG-3′) and HML8 (5′-GTTCAGACTGTCGCCTGTTTGAG-3′). The 3′ LTR from human, chimpanzee, bonobo, gorilla, orangutan, and African green monkey was amplified with HML5 (5′-GTCTCTGTCTTCTGTTTATAGTCTG-3′) and HML6 (5′-GTTTTGTGCCTTTACAACATAGGTAC-3′); from baboon, cynomolgus macaque, and rhesus macaque, with HML5 and HML4 (5′-ATAGGTACTAACTTAACTTCAACAAC-3′). Screening for cellular target sequence in marmoset and squirrel monkey used primers HML1 or HML7 (forward) and HML4 or HML6 (reverse).
RTVL-Ia (accession nos. M92067 and M34038). The 5′ LTR was amplified from chimpanzee, bonobo, gorilla, orangutan, and gibbon with primers RTVLIa-1 (5′-CCTAAAGGTGAATTATCACAAAATACT-3′) and RTVLIa-5 (5′-GGACAATGTTTTCCCTGTAT-3′). The 5′ LTR of African green monkey was amplified with RTVLIa-8 (5′-TACTAGAAATAACCACATAAGTGTTT-3′) and RTVLIa-5. The 3′ LTR was amplified from chimpanzee, bonobo, gorilla, and orangutan by using RTVLIa-3 (5′-AAGACCCAAGTAGAATAACAGAGCC-3′) and RTVLIa-7 (5′-CCTCTACTTCTTGAAATTTTCC-3′); the 3′ LTR was amplified from gibbon and African green monkey by using RTVLIa-3 and RTVLIa-2 (5′-TTTCCATAATCAAAGATTCCTTAAAT-3′).
Published RTVL-H sequences were used to query databases for RTVL-H loci with flanking sequences suitable for designing PCR primers. RTVL-Ha is an RTVL-H element spanning nucleotides 14602–20350 of cosmid clone N28H9 (accession no. Z71183). 5′ LTRs were amplified from human, chimpanzee, bonobo, and gorilla with RTVLH-1 (5′-CTGACCGATGCTGACAATGGC-3′) and RTVLH-2 (5′-CTCACGGAGCAAAGAACAGGAGG-3′). The 3′ LTRs of the same species were amplified with primers RTVLH-3 (5′-GAAACATCGCCCATTCTCTCTCC-3′) and RTVLH-4 (5′-GATCAATGGCAGGTTTTCAACCTC-3′). The uninterrupted cellular sequence was amplified from orangutan and African green monkey by using primers RTVLH-1 and RTVLH-4. RTVL-Hb is between bases 75–5817 of human bacterial artificial chromosome (BAC) clone RG341D10 (accession no. AC002530). The 5′ LTRs were amplified by using RTVLHb-1 (5′-GCTCTAAGTTAATTTATAGGTCAC-3′) and RTVLHb-2 (5′-CGATCCGAGTCACGGCACC-3′); the 3′ LTRs were amplified with RTVLHb-3 (5′-CTAATCCCGCTTGAAGCAGCC-3′) and RTVLHb-4 (5′-AACAGCCCTGCATGAAGTCAC-3′).
HERV-K18 (M12853 and M12852). Primers were derived from the HERV-K18 published flanking sequence (HERVK-1 and HERVK-4) conserved HERV-K family gag and env sequences (HERVK-2 and HERVK-3) (internal sequences of HERV-K18 have not been published). The 5′ LTR was amplified from human, chimpanzee, bonobo, and gorilla with primers HERVK-1 (5′-TGGCATGTACAACATAAGCGGAATC-3′) and HERVK-2 (5′-CTCCACGTTGGGCACCAGATG-3′). The 3′ LTR was amplified from the same species by using primers HERVK-3 (5′-TGTCTGTTGTTAGTCTGCAGGTGTACC-3′) and HERVK-4 (5′-CTGTGATTACCGCCTTACAGGATTTC-3′).
PCR products were gel purified and sequenced directly on both strands by using the ABI Prism Dye-Terminator Cycle Sequencing Ready Reaction Kit and analyzed with an ABI 373 Stretch automated sequencer (Perkin–Elmer).
Each of the proviruses used in this study maps to a different human chromosome, according to information available in the sequence databases, or by PCR screening of monochromosomal human × rodent somatic cell hybrids (data not shown).
Phylogenetic Analysis.
Sequences were aligned with clustal w (ref. 20; version 1.7) and adjusted by hand. Maximum parsimony, neighbor-joining, and maximum likelihood trees were generated by using PAUP* (21).
RESULTS AND DISCUSSION
Building Phylogenetic Trees from ERV LTR Sequences
Endogenous retrovirus loci provide no less than three sources of phylogenetic signal, which can be used in complementary fashion to obtain much more information than simple distance estimates of homologous sequences. First, the distribution of provirus-containing loci among taxa dates the insertion. Given the size of vertebrate genomes (>1 × 109 bp) and the random nature of retroviral integration (22, 23), multiple integrations (and subsequent fixation) of ERV loci at precisely the same location are highly unlikely (24). Therefore, an ERV locus shared by two or more species is descended from a single integration event and is proof that the species share a common ancestor into whose germ line the original integration took place (14). Furthermore, integrated proviruses are extremely stable: there is no mechanism for removing proviruses precisely from the genome, without leaving behind a solo LTR or deleting chromosomal DNA. The distribution of an ERV among related species also reflects the age of the provirus: older loci are found among widely divergent species, whereas younger proviruses are limited to more closely related species. In theory, the species distribution of a set of known integration sites can be used to construct phylogenetic trees in a manner similar to restriction fragment length polymorphism (RFLP) analysis.
Second, as with other sequence-based phylogenetic analyses, mutations in a provirus that have accumulated since the divergence of the species provide an estimate of the genetic distance between the species. Because, for any given provirus, it is highly unlikely that there will be selection for or against any specific sequence, it is safe to assume that the rate of accumulation of mutations approximates the rate of their occurrence, with appropriate corrections for reversion. Analysis of closely related proviruses integrated at different sites should also reveal regional differences in mutation rates.
Third, sequence divergence between the LTRs at the ends of a given provirus provides an important and unique source of phylogenetic information. The LTRs are created during reverse transcription to regenerate cis-acting elements required for integration and transcription. Because of the mechanism of reverse transcription, the two LTRs must be identical at the time of integration, even if they differed in the precursor provirus (Fig. 1A). Over time, they will diverge in sequence because of substitutions, insertions, and deletions acquired during cellular DNA replication. Although it has been noted that the divergence between the two LTRs of an ERV can serve as a molecular clock (8, 15, 18, 25), there are no reported prior attempts to utilize the LTRs of individual ERV loci as a source of phylogenetic signal.
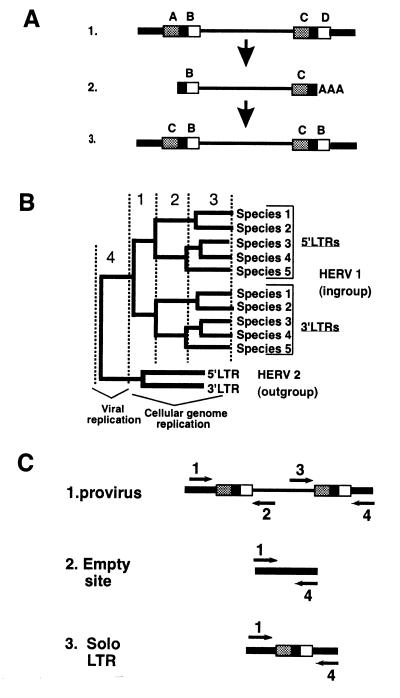
Figure 1.
Evolution of ERV LTR sequences. (A) The LTRs of an ERV are identical at the time of integration. Diagram D1, hypothetical provirus with spontaneous LTR mutations at points A, B, C, and D. Diagram D2, a newly transcribed viral RNA genome will contain only the mutations at points B and C. Diagram D3, after reverse transcription and integration, the new provirus has identical LTRs with mutations B and C found in both LTRs. (B) Hypothetical phylogeny based on ERV LTR sequences. The tree contains four distinct types of substitutions. 5′ and 3′ LTRs from the same provirus are expected to cluster separately, because of substitutions that predate all speciation events (“1”). The node joining the two clusters represents the time of integration, when the two LTRs were identical. Substitutions that occur between speciation events (synapomorphies) fall exclusively within one or the other cluster and define the topology of the cluster according to the evolutionary history of the different species (“2”). Assuming that both LTRs have evolved independently and at the same rate, the two clusters will have the same branching pattern, revealing the phylogeny of the input species. Substitutions unique to one species (autoapomorphies) are phylogenetically uninformative (“3”). Branches separating distinct ERV loci, e.g., between the ingroup provirus HERV 1 and the outgroup provirus HERV 2, represent errors accumulated during viral replication (“4”). Thus trees containing sequences from more than one HERV can be rooted at the node joining the two proviruses, because the two loci share a common viral ancestor. Rooting the tree with another ERV is therefore independent of any assumptions about host species phylogeny. (C) PCR amplification of ERV LTRs from genomic DNA. Arrows indicate 5′ → 3′ orientation of PCR primers; thick lines, cellular flanking sequences; thin lines, ERV sequences; boxes, LTR sequences. LTRs and adjacent cellular sequences are amplified by using one flanking sequence-specific primer and one provirus-specific primer (depicted as primers 1 and 2 for the 5′ LTR and primers 3 and 4 for the 3′ LTR). If neither LTR can be detected the presence of the uninterrupted cellular sequence or solo LTR can be determined by using primers 1 and 4. Proviral integration is essentially random, so flanking sequences amplified with each LTR confirm that homologous loci are being compared. Integration also results in a duplication of target sequences (4–6 bp) flanking each provirus, which can be used to confirm that the amplified 5′ LTR and 3′ LTR are from the same provirus.
Assuming that the LTRs of an ERV are evolving independently, at approximately the same rate, and in the absence of rearrangement events, a phylogenetic tree containing 5′ and 3′ LTRs derived from the same ERV locus is predicted to have a topology similar to that depicted in Fig. 1B. The most useful feature of the predicted tree is the separate clustering of the 5′ and the 3′ LTRs. The node joining the 5′ and 3′ LTR clusters must be the deepest within the ingroup, since it represents the time of integration, when the two LTRs were identical. Furthermore, both clusters are predicted to have similar branching patterns as determined by the phylogenetic history of the host species, with similar branch lengths. Thus, each tree displays two estimates of host phylogeny, both of which are derived from the evolution of an initially identical sequence (compare the 5′ LTR and 3′ LTR clusters in Fig. 1B). As we shall see, deviation of actual trees from this prediction provides a powerful means of testing the assumptions and detecting events other than neutral accumulation of mutations in the evolutionary history of a species.
Species Distribution of HERV Loci
Fig. 1C depicts the PCR strategy used to determine the distribution of 6 unlinked HERV proviruses among the genomes of 12 primate species. The presence of each HERV in a given species was determined by PCR amplification of both the 5′ and 3′ LTRs of the HERV from genomic DNA. Two genomic DNA samples from each species were screened, except for humans (12 individuals) and bonobo (1 individual). In some cases, the absence of a HERV from a species was confirmed by PCR amplification of the uninterrupted cellular target sequence (Fig. 1C). Three of the loci, HERV-KC4, HERV-KHML6.17, and RTVL-Ia, were detectable in the genomes of OWMs and hominoids, but not New World monkeys, and therefore integrated into the germ line of a common ancestor of the Old World lineages. HERV-K18, RTVL-Ha, and RTVL-Hb were found exclusively in humans, gorillas, chimpanzees, and bonobos, and thus are consistent with a gorilla/chimpanzee/human clade. None of the loci was detected in New World monkeys.
Evolution of HERV LTR Sequences
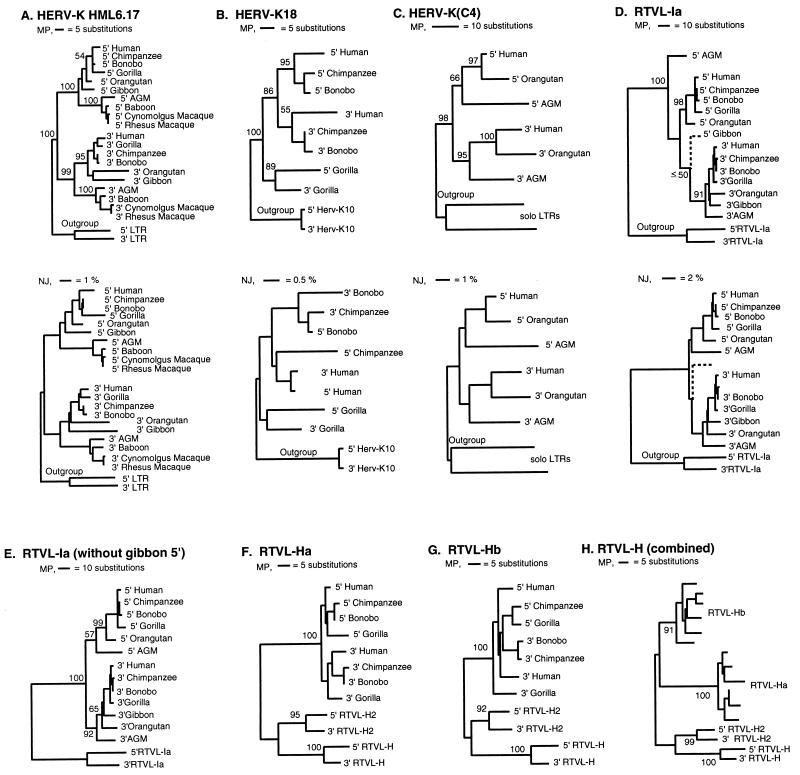
For each HERV locus, the amplified LTRs from each species were directly sequenced, and the aligned sequences were used to generate phylogenetic trees (Fig. 2). The 5′ and 3′ LTRs of HERV-KHML6.17 fell into two distinct clusters, in accord with prediction (Fig. 2A). Moreover, both LTR cluster topologies are consistent with established versions of primate species phylogeny (26–29). As has been the case with numerous nuclear DNA markers, there was no consensus among the HERV trees for the relationship among humans, chimpanzees, and gorillas (30). The remaining trees displayed interesting deviations from the predicted separation of the 5′ and 3′ LTR sequences.
Figure 2.
Phylogenies of seven HERV loci. Maximum parsimony (MP) trees are shown for each locus. Neighbor-joining (NJ) and maximum likelihood (ML) analyses yielded essentially identical results (data not shown). Insertions and deletions were weighted equal to 1 substitution. Tree searches were performed by using the branch-and-bound option [HERV-K(HML6.17), RTVL-Ia, and RTVL-H] or the exhaustive search option [HERV-K18 and HERV-K(C4)]. Final trees were rooted by designating outgroup sequences. Numbers indicate bootstrap values for major nodes (n = 100). AGM, African green monkey. (A) HERV-K(HML6.17) One of three minimum MP trees of length (L) = 258 aligned HERV-K(HML6.17) sequences. Outgroup LTRs are from a provirus related to HERV-K(HML6.17) found in human BAC110P12 (bases 115, 102 to 122, and 217; accession no. U95626) by blast homology search. (B) Single most-parsimonious tree for the HERV-K18 locus (L = 128). The published HERV-K10 LTR sequences were used as an outgroup (31). (C) Single most-parsimonious tree (L = 199) for the HERV-K(C4) sequences. Two HERV-K(C4)-related solo LTRs were included as outgroups. (D) One of eight equally parsimonious trees (L = 393) for the RTVL-Ia locus. Dashed lines indicate the unexpected placement of the gibbon 5′ LTR branch. The outgroup contains RTVL-Ib LTR sequences (14). (E) One of four equally parsimonious trees (L = 372) for the RTVL-Ia locus after excluding the gibbon 5′ LTR. (F) One of seven equally parsimonious trees (L = 145) for RTVL-Ha. (G) One of two equally parsimonious trees (L = 126) for RTVL-Hb. The 5′ LTR from bonobo was not amplified. (H) One of seven equally parsimonious trees containing both the RTVL-Ha and RTVL-Hb loci (L = 200). Published RTVL-H and RTVL-H2 LTRs were designated as outgroups for rooting the final trees in F, G, and H (36, 37).
Fig. 2B shows the trees for the HERV-K18 LTR sequences. Contrary to expectation, the 5′ and 3′ LTRs of the gorilla provirus cluster together instead of with their counterparts from the other three species (compare Fig. 1B and Fig. 2B). The gorilla LTRs are separated from the other HERV-K18 LTRs by substitutions at 11 sites (Fig. 3A). Assuming that the two LTRs of an HERV locus are evolving independently, every substitution within the ingroup should be manifest as a difference between the 5′ and 3′ LTRs (compare the 5′ and 3′ LTR patterns above the white arrows in Fig. 3A). Substitution patterns at the 11 sites in question, however, do not differ between the 5′ and 3′ LTRs within a species (black arrows in Fig. 3A). For example, a substitution at site 242 appears in both the 5′ and 3′ gorilla LTRs. Although it is possible that any one position may suffer an identical substitution in both LTRs by chance, the probability of 11 positions undergoing identical substitutions in both LTRs is exceedingly low. It is far more likely that most of the 11 substitutions occurred only once, and that the two LTRs were homogenized by gene conversion. The tree in Fig. 2B is consistent with gene conversion between both LTRs of either the gorilla provirus or the human/chimpanzee provirus.
Figure 3.
Identifying conversion of LTR sequences. Open circles represent sites that have retained the ancestral sequence, and filled circles indicate substitutions. Black and white arrows indicate changes discussed in the text. Uninformative sites are not shown. (A) HERV-K18. For clarity, chimpanzee species and humans have been collapsed to a single branch. The boxes highlight 11 sites that cluster the gorilla LTRs separately from their human and chimpanzee counterparts (see Fig. 2B). Although the figure depicts the substitutions as occurring in gorilla, the ancestral sequence (and thus the direction of change) is ambiguous, as indicated by question marks (?) and half-filled circles (◑). (B) RTVL-Ia. Apes are depicted as a single branch. AGM, African green monkey. The pattern of substitutions indicates that a portion of the RTVL-Ia LTR was replaced by conversion with sequences from the 3′ LTR. The pattern between bases 417 and 469 of the gibbon 5′ LTR is identical to the 3′ LTRs (boxed areas).
Alternatively, the topology of the tree in Fig. 2B may indicate that the HERV-K18 provirus of gorillas and the HERV-K18 provirus of humans/chimpanzees are not true orthologues. There are at least two mechanisms to explain this possibility:
(i) The proviruses are derived from two independent integration events (xenology). This possibility would require two nearly identical viruses (differing by no more than 11 substitutions within the LTRs) to integrate into precisely the same nucleotide position in two different lineages—a highly unlikely possibility. A similarly unlikely variation on this possibility is independent integrations into very similar cellular target sequences.
(ii) In one of the two lineages, HERV-K18 was largely replaced by recombination with a separate (but nearly identical) provirus. Such recombination would have been restricted to sequences within the provirus, as the flanking cellular sequences are identical in both lineages. It should be noted that there are hundreds of HERV-K LTR sequences within the primate genome (31–34).
The HERV-K(C4) LTR sequences (Fig. 2C) give the predicted topology; however, as noted previously (15, 16), the provirus was missing altogether from gorilla and chimpanzee DNA, in which only an unoccupied integration site was detectable. HERV-K(C4) is found in some ape and OWM species, proving that integration occurred in a common ancestor of apes and OWMs (15, 16). The provirus is located within the human C4B gene, which arose by duplication before the separation of the apes and OWMs. The absence of HERV-K(C4) from some species is most likely caused by frequent homogenization of the C4-CYP21 locus (35), resulting in conversion back to the unoccupied integration site. Both alleles of the C4 locus (with and without the HERV-K(C4) provirus) have been identified within more than one species, suggesting that such conversions have occurred multiple times during primate evolution (35).
The RTVL-Ia tree (Fig. 2D) deviates from expectation by the joining of the outgroup to the ingroup at a node that separates the 5′ African green monkey sequences from all the other ingroup sequences. Inspection of the alignment reveals 10 substitutions on the RTVL-Ia tree that contribute to this unexpected branching pattern (dashed line in Fig. 2D). Seven of these sites fall within a 52-bp stretch (arrows in Fig. 3B). Within this segment, the gibbon 5′ LTR is identical to the 3′ LTRs (including the gibbon 3′ LTR). After the gibbon lineage branched off from the other primate lineages, a portion of the 3′ LTR must have been transferred to the 5′ LTR by gene conversion. Because of this conversion, the most parsimonious tree identifies the gibbon 5′ LTR as the progenitor of all the 3′ LTRs, and incorrectly invokes parallel evolution (homoplasy) to explain the appearance of identical substitutions in the African green monkey 5′ LTR and the 5′ LTRs of orangutan and apes. After eliminating the hybrid gibbon 5′ LTR from the analysis, the most parsimonious explanation for the sequence at these sites is shared, derived evolution (Fig. 2E). Moreover, all the new trees have the predicted topology (including those derived by neighbor-joining and maximum likelihood methods), with the root of the tree separating the 5′ and 3′ LTRs of the ingroup into two distinct lineages.
The trees in Fig. 2 F, G, and H contain proviruses of the very large RTVL-H family (36, 37). The two loci were identified by searching the genome databases for RTVL-H-related sequences and are referred to here as RTVL-Ha and RTVL-Hb. The RHTVL-Ha provirus tree conforms well to the expected topology (Fig. 2F); however, the RTVL-Hb cluster (Fig. 2G) bears no resemblance to primate species phylogeny. Most of the substitutions fall on terminal branches and provide no phylogenetic signal. One interpretation is that the RTVL-Hb sequences are recombining with other RTVL-H loci, which would have the effect of homogenizing the sequences in a type of concerted evolution. RTVL-H is the largest known HERV family, containing over 1,000 members (18), which may serve as a source of sequences for recombination.
The tree in Fig. 2H is a particularly effective illustration of the principle that LTRs derived from the same provirus cluster together. This tree contains LTRs from four related proviruses, RTVL-Ha, RTVL-Hb, and the proviruses designated as outgroups, RTVL-H and RTVL-H2. All LTRs cluster exclusively with sequences from the same provirus, with a high level of bootstrap support for the nodes separating the four loci. The RTVL-Ha and RTVL-Hb clades do not differ significantly from the trees generated for the two proviruses separately in Fig. 2 F and G, respectively.
Nucleotide Substitution Patterns
Some authors have suggested that methyl-CG deamination has evolved as a specific defense against colonization of the genome by ERVs (38). Existence of such a mechanism should be manifest as a bias toward C⋅G → T⋅A transitions within CG dinucleotides. Tracing the pattern and direction of shared derived substitutions on each of the HERV trees revealed such a bias. Table 1 shows the distribution of C⋅G → T⋅A changes among the C/G-containing dinucleotides in the ancestral LTR sequence. In every case, the number of C⋅G → T⋅A substitutions per CG dinucleotide is 5- to 10-fold higher than for any of the six other dinucleotide contexts. Indeed, over 40% of the total C → T and G → A transitions are attributable to CG changes, despite the fact that CG is much less frequent than any other dinucleotide. This imbalance is consistent with methyl-CG deamination as a mechanism for generating C⋅G → T⋅A transitions (39); however, the issue of whether a mechanism for promoting CG deamination has evolved specifically as a defense against ERVs requires a careful comparison of the substitution patterns in ERV sequences with those of other nuclear DNA markers.
Table 1.
Distribution of inferred transitions as a function of dinucleotide
| Dinucleotide | RTVL-Ia
|
HERV-K(C4)
|
HERV-K(HML6.17)
|
HERV-K18
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of subst. | NNs per LTR | Ratio | No. of subst. | NNs per LTR | Ratio | No. of subst. | NNs per LTR | Ratio | No. of subst. | NNs per LTR | Ratio | |
| C → T occurring in | ||||||||||||
| CC | 18 | 52 | 0.35 | 13 | 49 | 0.27 | 11 | 45 | 0.24 | 1 | 79 | 0.01 |
| CA | 6 | 43 | 0.14 | 6 | 37 | 0.16 | 10 | 37 | 0.27 | 1 | 61 | 0.02 |
| CG | 15 | 8 | 1.88 | 11 | 11 | 1.00 | 12 | 12 | 1.00 | 6 | 17 | 0.35 |
| CT | 11 | 53 | 0.21 | 2 | 53 | 0.04 | 7 | 45 | 0.16 | 5 | 80 | 0.06 |
| G → A occurring in | ||||||||||||
| CG | 12 | 8 | 1.50 | 3 | 11 | 0.27 | 21 | 12 | 1.75 | 12 | 17 | 0.70 |
| AG | 6 | 34 | 0.18 | 1 | 38 | 0.03 | 6 | 31 | 0.19 | 2 | 66 | 0.03 |
| GG | 1 | 18 | 0.06 | 3 | 28 | 0.11 | 9 | 32 | 0.28 | 3 | 60 | 0.05 |
| TG | 6 | 34 | 0.18 | 6 | 47 | 0.13 | 9 | 35 | 0.23 | 1 | 82 | 0.01 |
NNs, dinucleotides.
Estimating the Time of Integration
The genetic distance between the 5′ and 3′ LTRs of an ERV reflects mutations accumulated since the time of integration and should therefore be proportional to the age of the provirus. HERV-KC4, HERV-KHML6.17, and RTVL-Ia are found in both OWMs and hominoids, which are estimated to have last shared a common ancestor over 31 million years ago. By contrast, HERV-K18, RTVL-Ha, and RTVL-Hb are found only in humans, chimpanzees, and gorillas, which are thought to have diverged around 5 million years ago (40–42). To estimate the age of each provirus the human/chimpanzee distances from each tree were used to calibrate the rate of molecular evolution at each locus (Table 2). The most recent common ancestor of humans and chimpanzees lived approximately 4.5 million years ago (40–42), so dividing the distance between the human and chimpanzee sequences (substitutions per site) by this number gives rates ranging from 2.3 to 5.0 × 10−9 substitutions per site per year. These numbers are similar to the estimated rates of evolution for pseudogenes and noncoding regions of mammalian genes (43–45). Applying each rate to the divergence between the 5′ and 3′ LTRs of the same locus gives integration times consistent with estimates based on species distribution (Table 2).
Table 2.
Integration time estimates
| Locus | Rate,* (subst./site/yr) × 109 | Avg distance between 5′ and 3′ LTRs, subst./site | Calc. age, 106 yr | Age of LCA, 106 yr |
|---|---|---|---|---|
| HERV-K(C4) | 2.28 ± 0.4 | 0.08 | 36.8 | 31 |
| HERV-K(HML6.17) | 3.71 ± 0.4 | 0.12 | 32.3 | 31 |
| RTVL-Ia | 2.28 ± 0.6 | 0.10 | 44.8 | 31 |
| HERV-K18 | 5.00 ± 0.8 | 0.03 | 5.7 | 4.5 |
| RTVL-Ha | 4.39 ± 1.5 | 0.03 | 6.7 | 4.5 |
A number of authors have pointed out that molecular clock calibrations are subject to a wide margin of error, and are usually based on imprecise estimates of divergence dates (46–48). The calculations in Table 2 are therefore only rough estimates of absolute time, but they are nonetheless useful for comparing the relative ages and rates of evolution of different HERV loci.
Conclusions
The study reported here is, to our knowledge, the first to take advantage of special properties of retroelements to provide insight into evolutionary mechanisms. The HERVs analyzed above include six unlinked loci, representing five unrelated HERV sequence families. Except where noted, these sequences gave trees that were consistent with the well established phylogeny of the old world primates, including OWMs, apes, and humans. Within this time scale genetic distances were less than 10% for all orthologous comparisons, and correction for multiple substitutions did not significantly alter branch lengths or tree topologies (data not shown). As with other nuclear DNA sequences, analyses of older phylogenetic relationships by using ERVs are likely to require such corrections.
One surprising result is the high frequency of conversion we observed. Indeed, only two of the six loci analyzed had suffered no such events in any lineage. Solo LTRs, which arise by recombinational deletion of the intervening viral genes, and which are found by the thousands in the genomes of many animal species, are further evidence for high frequency of recombination involving ERV sequences (1, 49–51). The mechanism that gives rise to such events is unlikely to be provirus-specific, but probably reflects the likelihood of conversion among any repeated, nuclear DNA sequences. Because many ERVs belong to multicopy families, it is also possible that interlocus recombination gives rise to concerted evolution among some of these loci. This latter mechanism may explain the rather confusing topology of the RTVL-Hb tree (Fig. 2G).
The use of LTR-to-LTR divergence to estimate insertion times has been reported previously (8, 15, 18, 25), but such studies invariably ignore the possibility of sequence conversion. Only one report (25) discussed the concern that sequence conversion between LTRs can result in an underestimate of insertion time, and suggested that conversion events should be detectable as deletions or alterations of the sequences flanking the LTRs. However, most of the loci analyzed in our study have clearly undergone conversion/recombination, yet none of these events resulted in loss or alteration of flanking sequences (data not shown). Phylogenetic analysis using HERV LTR sequences gives rise to trees with a predictable topology, on which is superimposed the phylogeny of the host taxa, and allows ready detection of conversion events. Once aberrant sequences are identified, they can be eliminated from an analysis, and the remaining sequences can be used to calculate insertion times, delineate substitution patterns, and decipher host phylogeny. Because ERVs are abundant within the genomes of many animal species, including (but not limited to) plants, insects, mollusks, fish, rodents, domestic pets, and livestock, the ERV approach can be applied to an endless variety of phylogenetic puzzles (1, 2).
Acknowledgments
We dedicate this paper to the memory of Igor Slobodkin, Ph.D. We thank Steve O’Brien for helpful comments and discussion. J.M.C. was supported by National Cancer Institute Grant R35CA44385 and is a Research Professor of the American Cancer Society. W.E.J. was supported by National Institutes of Health Training Grants T32GM07310 and T32AI07422.
ABBREVIATIONS
- ERV
endogenous retrovirus
- HERV
human ERV
- OWM
Old World monkey
- LTR
long terminal repeat
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. 172454–172500).
References
- 1.Boeke J D, Stoye J P. In: Retroviruses. Coffin J M, Hughes S H, Varmus H E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 343–436. [PubMed] [Google Scholar]
- 2.Herniou E, Martin J, Miller K, Cook J, Wilkinson M, Tristem M. J Virol. 1998;72:5955–5966. doi: 10.1128/jvi.72.7.5955-5966.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coffin J M. In: RNA Tumor Viruses. Weiss R, Teich N, Varmus H, Coffin J M, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. pp. 1109–1204. [Google Scholar]
- 4.Coffin J M. In: Fundamental Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott-Raven; 1996. pp. 763–844. [Google Scholar]
- 5.Benveniste R E, Todaro G J. Nature (London) 1976;261:101–108. doi: 10.1038/261101a0. [DOI] [PubMed] [Google Scholar]
- 6.Atchley W R, Fitch W M. Science. 1991;254:554–558. doi: 10.1126/science.1948030. [DOI] [PubMed] [Google Scholar]
- 7.Hillis D M, Bull J J. Science. 1991;254:528. doi: 10.1126/science.1948027. [DOI] [PubMed] [Google Scholar]
- 8.Shih A, Coutavas E E, Rush M G. Virology. 1991;182:495–502. doi: 10.1016/0042-6822(91)90590-8. [DOI] [PubMed] [Google Scholar]
- 9.van der Kuyl A C, Dekker J T, Goudsmit J. J Virol. 1995;69:7877–7887. doi: 10.1128/jvi.69.12.7877-7887.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atchley W R, Fitch W. Mol Biol Evol. 1993;10:1150–1169. doi: 10.1093/oxfordjournals.molbev.a040070. [DOI] [PubMed] [Google Scholar]
- 11.Frankel W N, Rudy C, Coffin J M, Huber B T. Nature (London) 1991;349:526–528. doi: 10.1038/349526a0. [DOI] [PubMed] [Google Scholar]
- 12.Wilkinson D A, Mager D L, Leong J C. In: The Retroviridae. Levy J, editor. Vol. 3. New York: Plenum; 1994. pp. 465–536. [Google Scholar]
- 13.Mariani-Costantini R, Horn T M, Callahan R. J Virol. 1989;63:4982–4985. doi: 10.1128/jvi.63.11.4982-4985.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maeda N, Kim H S. Genomics. 1990;8:671–683. doi: 10.1016/0888-7543(90)90254-r. [DOI] [PubMed] [Google Scholar]
- 15.Dangel A W, Baker B J, Mendoza A R, Yu C Y. Immunogenetics. 1995;42:41–52. doi: 10.1007/BF00164986. [DOI] [PubMed] [Google Scholar]
- 16.Tassabehji M, Strachan T, Anderson M, Campbell R D, Collier S, Lako M. Nucleic Acids Res. 1994;22:5211–5217. doi: 10.1093/nar/22.24.5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steinhuber S, Brack M, Hunsmann G, Schwelberger H, Dierich M P, Vogetseder W. Human Genet. 1995;96:188–192. doi: 10.1007/BF00207377. [DOI] [PubMed] [Google Scholar]
- 18.Mager D L, Freeman J D. Virology. 1995;213:395–404. doi: 10.1006/viro.1995.0012. [DOI] [PubMed] [Google Scholar]
- 19.Medstrand P, Mager D L, Yin H, Dietrich U, Blomberg J. J Gen Virol. 1997;78:1731–1744. doi: 10.1099/0022-1317-78-7-1731. [DOI] [PubMed] [Google Scholar]
- 20.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swofford D L. paup*, Phylogenetic Analysis Using Parsimony (*and other methods), version 4. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 22.Varmus H E, Swanstrom R. In: RNA Tumor Viruses. Weiss R, Teich N M, Varmus H E, Coffin J M, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1984. pp. 369–512. [Google Scholar]
- 23.Brown P O. In: Retroviruses. Coffin J M, Hughes S H, Varmus H E, editors. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 24.Withers-Ward E S, Kitamura Y, Barnes J P, Coffin J M. Genes Dev. 1994;8:1473–1487. doi: 10.1101/gad.8.12.1473. [DOI] [PubMed] [Google Scholar]
- 25.SanMiguel P, Gaut B, Tikhonov A, Nakajima Y, Bennetzen J L. Nat Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 26.Purvis A. Philos Trans R Soc London B. 1995;348:405–421. doi: 10.1098/rstb.1995.0078. [DOI] [PubMed] [Google Scholar]
- 27.Napier J R, Napier P H. The Natural History of the Primates. Cambridge, MA: MIT Press; 1985. [Google Scholar]
- 28.Fleagle J G. Primate Adaptation and Evolution. San Diego: Academic; 1988. [Google Scholar]
- 29.Ruvolo M. Mol Biol Evol. 1997;14:248–265. doi: 10.1093/oxfordjournals.molbev.a025761. [DOI] [PubMed] [Google Scholar]
- 30.Ruvolo M, Disotell T R, Allard M W, Brown W M, Honeycutt R L. Proc Natl Acad Sci USA. 1991;88:1570–1574. doi: 10.1073/pnas.88.4.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ono M. J Virol. 1986;58:937–944. doi: 10.1128/jvi.58.3.937-944.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ono M, Kawakami M, Takezawa T. Nucleic Acids Res. 1987;15:8725–8737. doi: 10.1093/nar/15.21.8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meese E, Gottert E, Zang K D, Sauter M, Schommer S, Mueller-Lantzsch N. Cytogenet Cell Genet. 1996;72:40–42. doi: 10.1159/000134157. [DOI] [PubMed] [Google Scholar]
- 34.Löwer R, Löwer J, Kurth R. Proc Natl Acad Sci USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein J, O’hUigin C, Figueroa F, Mayer W E, Klein D. Mol Biol Evol. 1993;10:48–59. doi: 10.1093/oxfordjournals.molbev.a039999. [DOI] [PubMed] [Google Scholar]
- 36.Mager D L, Henthorn P S. Proc Natl Acad Sci USA. 1984;81:7510–7514. doi: 10.1073/pnas.81.23.7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mager D L, Freeman J D. J Virol. 1987;61:4060–4066. doi: 10.1128/jvi.61.12.4060-4066.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoder J, Walsh C, Bestor T. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 39.Krawczak M, Cooper D N. In: Human Genome Evolution. Jackson M S, Strachan T, Dover G, editors. Oxford: BIOS Scientific; 1996. pp. 1–33. [Google Scholar]
- 40.Gingerich P D. Yearbook Phys Anthropol. 1984;27:57–72. [Google Scholar]
- 41.Pilbeam D R. Sci Am. 1984;250(3):84–96. doi: 10.1038/scientificamerican0384-84. [DOI] [PubMed] [Google Scholar]
- 42.Takahata N, Satta Y. Proc Natl Acad Sci USA. 1997;94:4811–4815. doi: 10.1073/pnas.94.9.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li W-H, Wu C-I, Luo C-C. Mol Biol Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- 44.Miyata T. In: Molecular Evolution, Protein Polymorphism, and the Neutral Theory. Kimura M, editor. Tokyo: Scientific Societies Press; 1982. pp. 233–266. [Google Scholar]
- 45.Nei M. Molecular Evolutionary Genetics. New York: Columbia Univ. Press; 1987. [Google Scholar]
- 46.Hillis D M, Mable B K, Moritz C. In: Molecular Sytematics. Hillis D M, Moritz C, Mable B K, editors. Sunderland, MA: Sinauer; 1996. pp. 515–543. [Google Scholar]
- 47.Avise J C. Molecular Markers, Natural History and Evolution. New York: Chapman & Hall; 1994. [Google Scholar]
- 48.Arnason U, Gullberg A, Janke A. J Mol Evol. 1998;47:718–727. doi: 10.1007/pl00006431. [DOI] [PubMed] [Google Scholar]
- 49.Copeland N G, Hutchison K W, Jenkins N A. Cell. 1983;33:379–387. doi: 10.1016/0092-8674(83)90419-1. [DOI] [PubMed] [Google Scholar]
- 50.Stoye J P, Fenner S, Greenoak G E, Moran C, Coffin J M. Cell. 1988;54:383–391. doi: 10.1016/0092-8674(88)90201-2. [DOI] [PubMed] [Google Scholar]
- 51.Mager D L, Goodchild N L. Am J Human Genet. 1989;45:848–854. [PMC free article] [PubMed] [Google Scholar]