Abstract
The fossil record of marine gastropods has been used as evidence to support the operation of species selection; namely, that species with limited dispersal differentially increase in numbers because they are more likely to speciate than widely dispersing species. This conclusion is based on a tacit phylogenetic assumption that increases in species with limited dispersal are solely the result of speciation within monophyletic groups with low dispersal. To test this assumption, we reconstructed a phylogeny from nuclear sequence data for 70 species of the marine gastropod genus Conus and used it to map the evolution of developmental mode. All eight species without planktonic life history phases recently and independently evolved this characteristic from ancestors with planktonic larval phases, showing that transitions in developmental mode are common in this group. A simple model of species diversification shows that such shifts can control the relative numbers of species with and without dispersing larval stages, leading to apparent species selection. Such results challenge the conclusion that increases in the number of nonplanktonic species relative to species with planktonic larvae over geologic time is necessarily a result of higher rates of speciation of nonplanktonic lineages and show that demonstration of species selection requires a phylogenetic framework.
Species selection occurs when the extinction or diversification of a species is affected by fitness differences of heritable species-level traits (1). In particular, geographic range and potential for gene flow among populations are influenced by a species’ capacity for dispersal (1–5). Marine species with limited dispersal often have smaller geographic ranges (4–9) and more heterogeneous population structures (10, 11) than species with high levels of dispersal. Because local catastrophes are more likely to eliminate a species with a limited geographic range, extinction of species with nondispersing developmental modes is more likely than of species with planktonic larvae. For example, nonplanktonic gastropod lineages from the Late Cretaceous have significantly lower geologic durations [median age = 2 million years (my)] than lineages with planktonic larvae (median age = 6 my) (5). Because low levels of gene flow and low dispersal among populations are also likely to lead to divergence of lineages, nonplanktonic species are more likely to diversify (i.e., have greater speciation rates) than species with planktonic larvae. Among 16 genera of gastropods in the Late Cretaceous (5) and six families of gastropods in the Early Tertiary (6), the number of nonplanktonic species increases at a greater rate than the number of species with planktonic dispersal; these observations suggest that species with nonplanktonic developmental modes have greater speciation rates than species with planktonic modes.
Modern textbooks in evolution cite work on gastropod fossil patterns (6, 12, 13) as “… currently the best candidate for an evolutionary trend produced by species level selection” (ref. 14, p. 584; see also ref. 15, p. 693). This trend is the increase of nonplanktonic species in spite of potential enhanced susceptibility to extinction within several families in the Cenozoic (6). Hansen (6) used the observation that the number of nonplanktonic species increased relative to the number of species with a planktonic larval phase as evidence for greater speciation rates of nonplanktonic species. If species only give rise to species with the same life history (i.e., species with similar life histories are independent monophyletic groups), it follows that species with nonplanktonic developmental modes speciated at a higher rate than species with planktonic larvae. However if nonplanktonic lineages arise from lineages with planktonic larvae and transitions are prejudiced in this direction because of a strong “conversion bias” (see ref. 16, p. 102), then increases in the number of nonplanktonic species over time could be the result of these biased shifts in species’ developmental modes and not necessarily increased speciation rates.
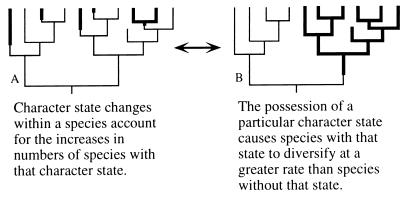
Without an understanding of phylogenetic relationships, species selection as it relates to origination rates is impossible to evaluate. There are two extreme interpretations of observations based on gastropods that illustrate the principles involved (Fig. 1). At one extreme, if a character (e.g., dispersal ability) has no effect on the diversification of lineages and frequently changes its character state (Fig. 1A), then evolutionary shifts are responsible for the prevalence of species with this character state, not species selection. At the other extreme, if character states change rarely and these different states have significant impacts on rates of speciation such that monophyletic groups are formed (Fig. 1B), then species selection might be shown to have operated. Thus, species selection requires monophyletic species groups whereas developmental transitions will generate de novo shifts of larval form many times during the evolutionary history of the group. Thus, as originally suggested by Stanley (ref. 17, p. 282), species selection is an intrinsically phylogenetic hypothesis that must be evaluated phylogenetically.
Figure 1.
Models of the evolutionary history of a group of species with interspecific variation in a character that may affect speciation and extinction rates of lineages. (A) No higher order effect. Transitions in the character state of this character occur commonly. Species with these traits (bold lines) do not form monophyletic groups. Species selection does not produce any observed pattern of trait variation among fossil assemblages. (B) Very few (one) transitions in character state. Different character states affect species’ origination rates. Species with similar traits form monophyletic groups. Species selection in this case could change the frequency of this trait among species over time.
The distinction between the rates of origination of nonplanktonic species can be tested through a phylogenetic reconstruction of species relationships and a concomitant analysis of dispersal mode. Using this phylogenetic approach, we investigated the transitions of developmental mode within the marine gastropod genus Conus. Based on recent phylogenetic investigations (18), veliger larvae among caenogastropods are homologous and possession of this life history phase is ancestral in this group. Lack of planktonically dispersing larvae among caenogastropods thus reflects a derived condition. Although there are examples of species of annelids and echinoderms that may have evolved a planktonic life history mode from a nonplanktonic ancestor (19–21), transitions in developmental mode among caenogastropods and other marine invertebrate groups presumably are biased from planktonic to nonplanktonic modes (22–31). If a nonplanktonic life history is selectively advantageous in certain circumstances and the transition from dispersing to nondispersing life history stages requires very few steps, then transitions of developmental mode may occur frequently. In Conus the length of the precompetent planktonic larval stage ranges from 0 to 30 days and has been estimated for 61 Indo-West Pacific species (ref. 8 and references therein). We specifically addressed the following questions: (i) How often have nonplanktonic lineages arisen in Conus? (ii) Are nonplanktonic species monophyletic? (iii) Is there any evidence of transition from species with nonplanktonic to planktonic developmental modes? (iv) Can observed shifts in development alter the proportions of nonplanktonic species over time? (v) Are nonplanktonic species morphologically more diverse than species with planktonic larvae? The results show that nonplanktonic developmental modes have evolved numerous times in Conus and that these developmental transitions occur frequently enough to resemble the effects of species selection.
METHODS
Specimens.
The specimens used in this study were largely from collections made throughout the Indo-West Pacific; one species was collected from the western Atlantic. Species designations were based on Walls (32) and Röckel et al. (33) and confirmed by A.J. Kohn (University of Washington, Seattle) or by comparisons to collections at the Bernice P. Bishop Museum (Honolulu). The lengths of the larval stages used in this study are those as summarized by Kohn and Perron (8) and Röckel et al. (33).
DNA Extraction, Amplification, and Sequencing.
DNA was isolated by using a modified cetyltrimethylammonium bromide extraction protocol (34) followed by phenol/chloroform extraction and alcohol precipitation methods (35). Although mitochondrial sequences adequately resolve relationships of recently diverged taxa (unpublished data), we targeted a nuclear sequence that is evolving at a slower rate than mtDNA to increase the resolution of relationships of basal lineages. Calmodulin primers (cal1 = 5′-GCCGAGCTGCARGAYATGATCAA-3′, cal2 = 5′-GTGTCCTTCATTTTNCKTGCCATCAT-3′) were designed from exon sequences flanking a conserved intron position in Aplysia californica (GenBank accession nos. X64653 and X64654), Drosophila melanogaster (X05949 and X05950), and Homo sapiens (X52608). These primers span a conserved intron position flanked by 52 bp of exon sequence. Amplifications were carried out under typical conditions (36). Amplification products subsequently were cloned in T-tailed plasmid vectors and sequenced by using solid-phase sequencing (36).
Sequence Alignment and Statistics.
Sequences were aligned by eye with the xesee program (37) and analyzed by using distance (mega, ref. 38) and parsimony (paup* 4.0b2; ref. 39) methods. The Kimura two-parameter algorithm was used to estimate genetic distances; neighbor-joining and heuristic parsimony searches were used for phylogeny reconstruction. Trees were rooted with the most basal Conus species, C. distans, as determined from phylogenies reconstructed from mitochondrial sequences (unpublished data). Levels of support for branches were estimated with bootstrapping methods in both mega and paup. The number of transitions from planktonic to nonplanktonic developmental modes was determined from the number of clades comprised of species with mixed developmental modes. Phylogenies were constrained for monophyly of all nonplanktonic species and monophyly of nonplanktonic species in clades with low bootstrap support. These trees were compared with the unconstrained tree, and significance of differences between constrained and unconstrained trees was determined with Templeton’s Wilcoxon signed-rank test (39).
Estimation of Transition Rate.
The proportion of transitions in developmental mode (T) was estimated from the number of branches in the tree (both internal and external) in which a transition in developmental mode has occurred (Nc) divided by the total number of branches in which the developmental mode of the lineage is known (Nt):
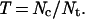 |
Calibration of Rate of Sequence Evolution.
The ages of the oldest sister species, based on deepest fossil records, were used to estimate the rate of calmodulin intron sequence evolution. We can assume that if two lineages coexisted at a particular time, then the splitting of these lineages had to have taken place before that time. Because this estimate uses the time at which both lineages existed and not the actual date of divergence (which could be much earlier), it reflects the maximum rate of sequence evolution. For an independent estimate of the sequence evolution rate, we also used distances from the western Atlantic species to possible sister taxa in the eastern Pacific.
Diversity of Nonplanktonic Species.
Many Conus species have numerous ecotypes, morphotypes, or synonyms attributed to them. In some cases malacologists have grouped these into one species or split them into many (32, 33). Certainly it is unclear whether these types represent distinct species or different forms of the same species. As a proxy for morphological diversity, we counted the number of synonyms or names attributed to morphotypes and ecotypes of the species in our sample from information provided in Röckel et al. (33) and compared the average number for species of each developmental mode.
Sampling Bias.
The species used in this study are a fraction of the diversity of Conus (roughly 15% of the species in the genus), and represent about one-fifth of Conus’ diversity in the Pacific. Also, the taxa used may not reflect a purely random sample of species with each developmental mode because of potential differences in the distribution and geographic ranges of species with these modes. However, the selection of nonplanktonic species was over a broad geographic range (collected from Australia, Guam, Hawaii, Papua Niugini, and the Philippines).
RESULTS
Molecular Data.
Sequences were obtained from 70 species of Conus. The recovered sequences ranged in length from 261 to 300 nt. Because these sequences are from introns, they are largely free to vary and have a dense phylogenetic signal: 203 positions were variable and 127 were parsimoniously informative. The average pairwise distance (Kimura two-parameter corrected) was 9.72% and ranged from 0.36% to 17.50%. The average ratio of transition/transversion substitutions was 1.58 and ranged from 0 to 8.25. Variation within the exon was limited largely to silent sites, and distance estimates from exon sequences were on average one-third as large as distances from intron sequences.
Trees.
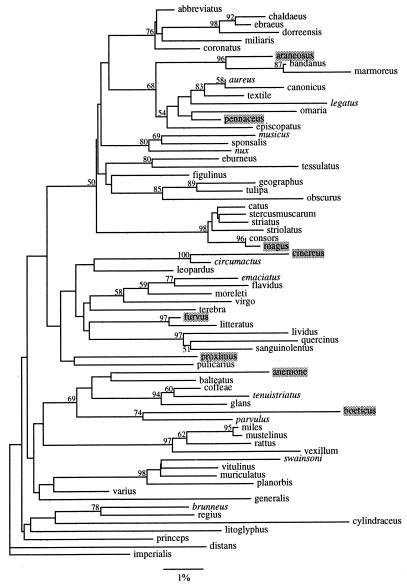
Among 70 Conus species, phylogenetic analyses of calmodulin sequences revealed numerous well-supported clades (Fig. 2). Clades with high bootstrap support were similarly identified in a phylogeny reconstructed from mitochondrial sequence data (unpublished data). Although the support for these clades is robust, the relationships among clades are poorly resolved, probably because of saturation of the phylogenetic signal. Nevertheless, the resolution of many nodes provides a powerful framework for interpreting the evolution of nonplanktonic developmental modes as well as for current studies of the evolution of feeding modes and conotoxins in this genus.
Figure 2.
Neighbor-joining tree reconstructed from calmodulin intron sequence data of Conus. Bootstrap values indicated on branches. Bootstrap values less than 50% were omitted. Names in italics are species whose developmental mode is unknown. Names shaded in gray are nonplanktonic species. All names in normal typeface are species with a planktonic larval phase.
Mapping of Developmental Modes. Mapping of developmental modes indicates that the nonplanktonic mode arose several times in this genus (Fig. 2). The eight species with nonplanktonic development fall out into eight different parts of the reconstructed phylogeny. The most similar species are C. araneosus and C. pennaceus and even this pair differs by 5% sequence divergence. That these eight species represent independent origins of nonplanktonic development is supported by several lines of evidence. First, a phylogeny that constrains all nonplanktonic species to be monophyletic is 50 steps longer and significantly different than the unconstrained tree (P < 0.0001). This result strongly suggests multiple origins of species with nonplanktonic development.
Second, four of the eight nonplanktonic species cluster with planktonic species with very strong bootstrap support (>95%). Because the remaining four nonplanktonic species cluster with planktonic species or species with unknown developmental mode with low or little bootstrap support (<75%), we forced these species to be monophyletic with the next closest nonplanktonic species and compared the lengths of the resulting trees. In three cases, the constrained trees were significantly longer than the unconstrained tree (P < 0.05). The tree that constrained the monophyly of C. anemone and C. boeticus was four steps longer than the unconstrained tree (P < 0.15). Thus, there were at least seven independent origins of nonplanktonic Conus species, though a parsimonious interpretation of our phylogenetic data suggests that all eight nonplanktonic species arose independently.
Shifts in Developmental Mode.
There are 117 branches present on the phylogeny (Fig. 2) for which the developmental mode of the lineage is either known or can be reconstructed based on parsimony criteria. Of these, 109 branches represent lineages in which no transitions in development have occurred, whereas the eight remaining branches represent lineages in which developmental transitions have occurred. The probability of a change in development mode is 0.068 (8/117). In no case does it appear that a nonplanktonic lineage gave rise to a lineage with planktonic larvae.
Sequence Evolution Rate Calibration. Of the species sequenced, C. lividus and C. quercinus are the two most closely related species that have records dating back to the Miocene. Specimens of C. lividus were found in the Vigo Formation of the Philippines (40) which has faunal assemblages reflecting Lower, Middle, and Upper Miocene deposition spanning an age of 5–22.5 my (41). Specimens of C. quercinus were found in the Tjilanang Beds of Indonesia (42), which have been assigned to the junction of the Middle and Upper Miocene (43), approximately 10.4 million years ago (mya) (44). These species therefore diverged at least 10.4 mya. Genetic distance between C. lividus and C. quercinus is 6.1% (based on the Kimura two-parameter model) and so the maximum rate of calmodulin intron sequence divergence is ≈0.6%/my for this pair.
C. brunneus and C. regius are eastern Pacific and western Atlantic species, respectively and are sister species that likely diverged with the emergence of the Isthmus of Panama (3–3.5 mya) or somewhat earlier because of oceanographic changes associated with isthmus formation (45). The genetic distance between these species is 4.7%. Assuming a minimum time of divergence for these taxa (3 my), the rate of calmodulin sequence divergence is ≈1.6%/my.
If our phylogenetic model and temporal calibrations are reasonable, we can use these rate calibrations to estimate the recency of the origins of nonplanktonic species from known lineages with planktonic larvae within Conus. Nonplanktonic species are between 0.4 and 7.1% distant from their nearest relative with a dispersing life history mode and on average 3.8% distant. Using the rate calibrations, we estimated that some nonplanktonic lineages diverged from their common ancestor as recently as between 0.25 and 0.67 mya while the earliest diverged between 4.4 and 11.8 mya (Table 1).
Table 1.
Genetic distances and estimated times since divergence from rate calibrations from fossil data (0.6%/my) and Panamanian divergence (1.6%/my) between nonplanktonic species and their closest relative with a planktonic larval phase (see Fig. 2)
| Nonplanktonic species | Closest sister taxon | Genetic distance, % | Minimum estimate of time since divergence, mya
|
|
|---|---|---|---|---|
| Fossil rate | Panamanian rate | |||
| C. magus | C. consors | 0.4 | 0.67 | 0.25 |
| C. furvus | C. litteratus | 1.5 | 2.50 | 0.94 |
| C. cinereus | C. circumactus* | 3.0 | 5.00 | 1.88 |
| C. pennaceus | C. omaria | 3.4 | 5.67 | 2.13 |
| C. araneosus | C. bandanus | 3.5 | 5.83 | 2.19 |
| C. anemone | C. balteatus | 6.2 | 10.33 | 3.88 |
| C. proximus | C. pulicarius | 6.2 | 10.33 | 3.88 |
| C. cinereus | C. leopardus | 7.1 | 11.83 | 4.44 |
| C. boeticus | C. parvulus* | 7.3 | 12.17 | 4.56 |
Mode of development is unknown.
Diversity of Nonplanktonic Species.
On average, nonplanktonic species have more names attributed to them as synonyms or named ecotypes or morphotypes (7.5) than species with planktonic larvae (2.2). For example, the nonplanktonic species Conus anemone from Australia has eight other names that refer to a variety of morphologically distinct forms distributed throughout the nominal species’ range (33).
DISCUSSION
In the genus Conus, parallel developmental shifts are common. When the developmental mode of the sister species is known, all of the nonplanktonic species we analyzed can be shown to be independently derived from species with a planktonic larval phase. There are no clades composed entirely of nonplanktonic species, and most clades composed of species with both developmental modes are supported by high bootstrap values (Fig. 2). In this case, a robust phylogenetic framework allows us to identify convergent developmental modes and show that one of the requirements of species selection—the proliferation of monophyletic, nonplanktotrophic lineages—is not met in the genus.
Transitions of developmental modes appear to be common in other marine invertebrates as well. Molecular phylogenetic studies of echinoderms, bivalves, and gastropods have documented the occurrence of rampant shifts in developmental modes and life histories. Closely related sea urchin species have completely different developmental mechanisms giving rise to either nonplanktonic or planktonic developmental modes (25, 27). Lineages with nondispersing life history phases have been estimated to have arisen multiple times in asterinid starfish (29) and at least 14 times in sea urchins (46). O’Foighil and Smith (30) reported that lineages with nonplanktonic larvae arose twice during the evolutionary history of lasaeid clams. Nonplanktonic species also arose numerous times in both littorinid (47, 48) and turritellid gastropods (49).
There is no evidence of a transition from nondispersing to dispersing life history phases in Conus. Strathmann (22, 23, 50, 51) states that although this direction of transition is probably rare, it is nevertheless possible. However, the most parsimonious explanation for the distribution of nonplanktonic species throughout the phylogeny (Fig. 2) is that possessing a planktonic larval phase unanimously represents the ancestral condition and lacking a dispersing larval phase is derived.
Based on calibrations of sequence evolution and the fossil record (Table 1), all of the nonplanktonic Conus species in our sample appear to have arisen from lineages with planktonic larvae between 0.25 and 11.8 mya. These estimates represent minimum times of divergence from a sister species with a planktonic larval phase; this source of error perhaps is balanced by the assumption that the transition of developmental mode probably occurred sometime after the divergence of the two lineages. The rate calibrations are moreover best estimates from an incomplete understanding of the times of divergence of Conus lineages and are not likely to reflect actual times of origination of nonplanktonic species because of the unequal branch lengths between nonplanktonic species and their sister species. However, nonplanktonic species also tend to occur on branches near the tips of the tree (Fig. 2). This phylogenetic pattern is the expected one for traits that arise frequently but do not lead to proliferation of monophyletic species groups (Fig. 1). By contrast, lineages with a planktonic larval phase are more deeply rooted within the evolutionary history of this group and possession of planktonic larvae has likely been the common developmental mode for this genus for tens of millions of years (52).
Our results show that shifts in developmental mode have occurred at least eight times in Conus and that the nonplanktonic species we sampled have arisen only from lineages with planktonic larvae. These implications challenge the idea that greater increases in the number of nonplanktonic species relative to species with dispersing larvae are caused solely by greater speciation rates of the former. Hansen’s (6) data show that the number of nonplanktonic gastropod species rapidly increased from the Early Paleocene through the Middle Eocene relative to increases in the number of lineages with planktonic larvae. If nonplanktonic species only give rise to nonplanktonic species and species with planktonic larvae only give rise to species with planktonic larvae, then from Hansen’s (6) data, nonplanktonic species must have had higher rates of species diversification than species with planktonic larvae to account for their greater rates of appearance. However, if as we have shown for Conus, nonplanktonic species commonly originate from species with dispersing larvae, developmental transitions may contribute to increases in the number of nonplanktonic lineages over time.
We can estimate the potential impact of developmental shifts on apparent species selection with a simple model of species diversification that assumes as an initial condition that speciation and extinction rates are identical for species with each life history. Transitions of developmental mode occur from planktonic to nonplanktonic modes, and we use the model to estimate the probability of increases in nonplanktonic species relative to species with planktonic larvae in the complete absence of species selection. This model omits the potential increased extinction rates of nonplanktonic species (5) to gauge the impact of developmental shifts on species life history patterns without assuming the action of species selection. Instead, the model tests the effect transitions can have on the numbers of species with and without dispersing larvae over time.
When the phylogenetic relationships of lineages are unclear, transitions from planktonic to nonplanktonic developmental modes will appear to simultaneously decrease the speciation rate of lineages with planktonic larvae and increase the speciation rate of nonplanktonic lineages. Under this model, the number of lineages with planktonic larvae is determined by the probabilities of speciation, extinction, and developmental transitions. The number of nonplanktonic lineages will change as a function of speciation and extinction plus the probability of transitions from a planktonic developmental mode to a nonplanktonic one:
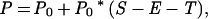 |
and
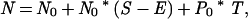 |
where P and N are the number of lineages with planktonic and nonplanktonic development after an opportunity for speciation, extinction, and transition, P0 and N0 are the number of lineages before that period, S is the probability of speciation, E is the probability of extinction, and T is the probability of transition.
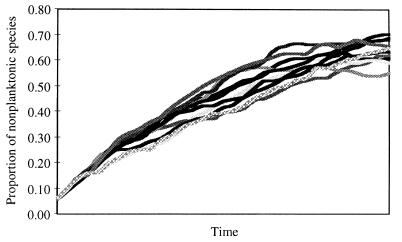
The model shows that the probability of developmental transition does not have to be large for nonplanktonic lineages to increase in numbers relative to species with planktonic larvae. We simulated the trajectories of the number of lineages with each life history over time by using the above formulae. This model assumes that probabilities for extinction and speciation of a lineage are equal and identical for each developmental mode; these probabilities were allowed to vary randomly between 0.225 and 0.275. This model also assumes a low probability of transition from planktonic to nonplanktonic developmental modes, which was set to vary randomly between 0.045 and 0.055. Initial numbers of lineages with each life history were set to 50 and 1, respectively. In 1,000 simulations in which each emulated 100 opportunities for lineages to either speciate, go extinct, change developmental mode, or remain intact, nonplanktonic species outnumber species with planktonic larvae in the end 853 times. Ten random examples of the trajectories from the simulations are presented in Fig. 3. Additional simulations were performed with the extinction rate of nonplanktonic species three times larger than the extinction rate of species with a planktonic larval phase. Even under these rigors nonplanktonic species can increase at a greater rate than species with planktonic development (when speciation rates are the same for each developmental type and transitions occur at a frequency of 0.05), although the parameter range in which this is true is more limited.
Figure 3.
Developmental shifts can mimic the effects of species selection. Simulated trajectories of the numbers of nonplanktonic species relative to the total number of species. Each run started with 50 species with planktonic larvae and one nonplanktonic species and consisted of 100 opportunities for speciation, extinction, or transition of developmental mode. Speciation and extinction probabilities (0.25) are identical for species with each developmental mode and were allowed to independently vary randomly within 10% of the mean probability. Transition probability was set to 0.05 and was allowed to vary randomly within 10% of this value.
These results show that biased transitions in developmental mode can account for greater increases in the number of nonplantkonic species relative to dispersing forms. Under this model, the proportions of lineages that undergo a transition of developmental mode (T) required to completely account for the greater increases in the number of nonplanktonic species relative to species with planktonic larvae range between 0.10 and 0.50 for the six gastropod groups studied by Hansen (6) (Table 2). Although these values are greater than the proportion we estimated from Conus, they suggest that developmental shifts could have a role in apparent species selection (Table 2). Probably both transitions and higher species accumulations of nonplanktonic taxa occur in most groups, but a focus on species selection has in the past only considered the latter explanation.
Table 2.
Proportions of transition per lineage (T) necessary to account for the greater increases in number of nonplanktonic species relative to species with planktonic larvae as estimated from Hansen’s (6) data
| Gastropod group | T | CT, % |
|---|---|---|
| Buccinidae | 0.10 | 68.0 |
| Fasciolariidae | 0.23 | 29.6 |
| Mitridae | 0.50 | 13.6 |
| Nassariidae | 0.30 | 22.7 |
| Olividae | 0.40 | 17.0 |
| Volutidae | 0.18 | 37.8 |
Contribution of developmental transitions (CT) to increase in numbers of nonplanktotrophs if T = 0.068 (see Discussion).
A phylogenetic perspective allows us to reject the hypothesis that species selection alone occurs in Conus. Yet it is impossible to assert that species selection is purely inoperative unless no monophyletic groups of nonplanktonic Conus occur. From a rough estimate of the number of names attributed to nonplanktonic species relative to the species with planktonic larvae used in this study, nonplanktonic species have on average 7.5 different names attributed to them while species with planktonic larvae only have 2.2. If all of the different types attributed to these species represent distinct or diverging species and if they are monophyletic, then this could provide some evidence for the operation of species selection in Conus. The morphologically distinct ecotypes of C. anemone in Australia (33) and diversity of nonplanktonic species in the Cape Verde Islands (8, 53) both may represent cases of heightened speciation rates among lineages of Conus with nondispersing developmental modes.
Developmental shifts and species selection represent two extremes (Fig. 1) that can be used to explain why there have been increases in the numbers of nonplanktonic species relative to species with planktonic larvae within prosobranch gastropods. There exists a broad continuum between these extremes in which evolutionary transitions plus species selection impact the rates of origination of nonplanktonic species. Conus probably represents one such intermediate case. Because all gastropod families that have been investigated phylogenetically also show evidence of parallel developmental shifts to nonplanktonic life history modes (47–49), the fossil taxa previously studied probably also fall somewhere in between. Among the gastropods studied by Hansen (6), for example, 14–68% of the species selection signal could be the result instead of developmental shifts if the rate of developmental evolution per lineage is similar to Conus (Table 2). As pointed out by Stanley (ref. 17, p. 282), even volutid gastropods, a family in which all species with planktonic larvae have gone extinct, could be made up of species with independent origins of nonplanktonic development and owe their current domination by low dispersal forms at least partially to developmental shifts rather than strict species selection.
Thus, parallel developmental shifts to nonplanktonic life history modes should be added with species selection as explanations of patterns of differential species accumulation in the fossil record. Further understanding of the relative roles of species selection and developmental shifts will depend on an evaluation of phylogenetic relationships and evolution of life history features both in fossil and extant species. Demonstrating species selection and verifying its operation require an explicit phylogenetic framework so that parallel evolutionary shifts can be identified and accounted for.
Acknowledgments
We thank the individuals and institutions that have contributed or facilitated in the collection of specimens for this study, especially H. Conley and G. C. Fiedler, and also P. S. Armstrong, R. Cowie, K. A. del Carmen, A. Kay, A. J. Kohn, C. Meyer, F. Moretzohn, G. Paulay, S. Romano, B. Smith, D. Strang, the Bernice P. Bishop Museum (Honolulu), the Christensen Research Institute (Madang, Papua Niugini), and the University of Guam. We also thank F. Cipriano, A. J. Kohn, C. Marshall, D. McHugh, G. Paulay, R. R. Strathmann, and two anonymous reviewers for input and criticisms of earlier drafts of this manuscript. This work was supported by the Research Council of the University of Hawaii and Organismic and Evolutionary Biology Department of Harvard University and grants from the Conchologists of America, Hawaiian Malacological Society, Lerner-Gray fund for marine research, Sigma-Xi, and the Western Society of Malacologists, and grants to S.R.P. from the National Science Foundation.
ABBREVIATIONS
- my
million years
- mya
million years ago
Footnotes
References
- 1.Grantham T A. Annu Rev Ecol Syst. 1995;26:301–321. [Google Scholar]
- 2.Jackson J B C. Am Nat. 1974;198:541–560. [Google Scholar]
- 3.Scheltema R S. In: Concepts and Methods of Biostratigraphy. Kaufman E G, Hazel J E, editors. Hutchinson, and Ross, Stroudsburg, PA: Dowden; 1977. [Google Scholar]
- 4.Jablonski D, Lutz R A. Biol Rev. 1983;58:21–89. [Google Scholar]
- 5.Jablonski D. Bull Mar Sci. 1986;39:565–587. [Google Scholar]
- 6.Hansen T A. Paleobiology. 1982;8:367–377. [Google Scholar]
- 7.Scheltema R S. In: Reproduction, Genetics and Distribution of Marine Organisms. Ryland J S, Tyler P A, editors. Fredensborg: Olsen and Olsen; 1989. pp. 183–188. [Google Scholar]
- 8.Kohn A J, Perron F E. Life History and Biogeography Patterns in Conus: Oxford Biogeography Series No. 9. Oxford: Clarendon; 1994. [Google Scholar]
- 9.Emlet R B. Evolution. 1995;49:476–489. doi: 10.1111/j.1558-5646.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 10.McMillan W O, Raff R A, Palumbi S R. Evolution. 1992;46:1299–1312. doi: 10.1111/j.1558-5646.1992.tb01125.x. [DOI] [PubMed] [Google Scholar]
- 11.Hellberg M E. Evolution. 1996;50:1167–1175. doi: 10.1111/j.1558-5646.1996.tb02357.x. [DOI] [PubMed] [Google Scholar]
- 12.Hansen T A. Science. 1978;199:885–887. doi: 10.1126/science.199.4331.885. [DOI] [PubMed] [Google Scholar]
- 13.Hansen T A. Paleobiology. 1980;6:193–207. [Google Scholar]
- 14.Ridley M. Evolution. Boston: Blackwell; 1993. [Google Scholar]
- 15.Futuyma D J. Evolutionary Biology. 3rd Ed. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 16.Gould S J. In: Perspectives on Evolution. Milkman R, editor. Sunderland, MA: Sinauer; 1982. pp. 83–104. [Google Scholar]
- 17.Stanley S M. Macroevolution. San Francisco: Freeman; 1979. [Google Scholar]
- 18.Ponder W F, Lindberg D R. Zool J Linn Soc. 1997;119:83–265. [Google Scholar]
- 19.McEdward L R. Biol J Linn Soc. 1995;54:299–327. [Google Scholar]
- 20.McEdward L R, Janies D A. Biol J Linn Soc. 1997;60:381–400. [Google Scholar]
- 21.Bhaud M, Duchêne J C. Oceanologica Acta. 1996;19:335–346. [Google Scholar]
- 22.Strathmann R R. Evolution. 1978;32:894–906. doi: 10.1111/j.1558-5646.1978.tb04642.x. [DOI] [PubMed] [Google Scholar]
- 23.Strathmann R R. Evolution. 1978;32:907–914. doi: 10.1111/j.1558-5646.1978.tb04643.x. [DOI] [PubMed] [Google Scholar]
- 24.Strathmann R R, Eernisse D J. Am Zool. 1994;34:502–512. [Google Scholar]
- 25.Wray G A. Science. 1995;267:1115–1116. doi: 10.1126/science.267.5201.1115. [DOI] [PubMed] [Google Scholar]
- 26.Wray G A. Syst Biol. 1996;45:308–322. [Google Scholar]
- 27.Wray G A, Raff R A. Trends Evol Ecol. 1991;6:45–50. doi: 10.1016/0169-5347(91)90121-D. [DOI] [PubMed] [Google Scholar]
- 28.Hart M W. Evolution. 1996;50:174–187. doi: 10.1111/j.1558-5646.1996.tb04484.x. [DOI] [PubMed] [Google Scholar]
- 29.Hart M W, Byrne M, Smith M J. Evolution. 1997;51:1848–1861. doi: 10.1111/j.1558-5646.1997.tb05108.x. [DOI] [PubMed] [Google Scholar]
- 30.O’Foighil D, Smith M J. Evolution. 1995;49:140–150. doi: 10.1111/j.1558-5646.1995.tb05966.x. [DOI] [PubMed] [Google Scholar]
- 31.O’Foighil D, Smith M J. Mol Phylogenet Evol. 1996;6:134–142. doi: 10.1006/mpev.1996.0065. [DOI] [PubMed] [Google Scholar]
- 32.Walls J G. Cone Shells: A Synopsis of the Living Conidae. Neptune City, NJ: T.F.H. Publications; 1979. [Google Scholar]
- 33.Röckel D, Korn W, Kohn A J. Manual of the Living Conidae. Vol. 1. Berlin: Verlag; 1995. [Google Scholar]
- 34.Winnepenninckx B, Backeljau T, de Wachter R. Trends Genet. 1993;9:407. doi: 10.1016/0168-9525(93)90102-n. [DOI] [PubMed] [Google Scholar]
- 35.Palumbi S R. In: Molecular Systematics. 2nd Ed. Hillis D M, Moritz C, Mable B K, editors. Sunderland, MA: Sinauer; 1996. pp. 205–247. [Google Scholar]
- 36.Palumbi S R. In: Molecular Zoology: Advances, Strategies, and Protocols. Ferraris J R, Palumbi S R, editors. New York: Wiley; 1996. pp. 101–117. [Google Scholar]
- 37.Cabot E L, Beckenbach A T. Comp Appl Biosci. 1989;5:233–234. doi: 10.1093/bioinformatics/5.3.233. [DOI] [PubMed] [Google Scholar]
- 38.Kumar S, Tamura K, Nei M. mega: Molecular Evolutionary Genetics Analysis, Version 1.01. University Park: Pennsylvania State Univ.; 1993. [Google Scholar]
- 39.Swofford D L. paup*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. Sunderland, MA: Sinauer; 1999. [Google Scholar]
- 40.Dickerson R E. Proc Calif Acad Sci. 1921;11:1–26. [Google Scholar]
- 41.Ministry of Natural Resources. Geology and Mineral Resources of the Philippines. Manila: Bureau of Mines and Geo-Sciences; 1981. [Google Scholar]
- 42.Verk I M van der. Leidsche Geol Mededeel. 1931;5:209–296. [Google Scholar]
- 43.Shuto T. Geol Paleont Southeast Asia. 1975;15:289–301. [Google Scholar]
- 44.Harland W B, Armstrong R L, Cox A V, Craig L E, Smith A G, Smith D G. A Geologic Time Scale 1989. Cambridge: Cambridge Univ. Press; 1990. [Google Scholar]
- 45.Duque-Caro H. Palaeogeogr Paleoclimatol Palaeoecol. 1990;77:203–234. [Google Scholar]
- 46.Emlet R B. Adv Invert Reprod. 1990;5:329–335. [Google Scholar]
- 47.Reid D G. Philos Trans R Soc London B. 1989;324:1–110. [Google Scholar]
- 48.Reid D G, Rumbak E, Thomas R H. Philos Trans R Soc London B. 1996;351:877–895. doi: 10.1098/rstb.1996.0082. [DOI] [PubMed] [Google Scholar]
- 49.Lieberman B S, Allmon W D, Eldredge N. Paleobiology. 1993;19:205–215. [Google Scholar]
- 50.Strathmann R R. Annu Rev Ecol Syst. 1985;16:339–361. [Google Scholar]
- 51.Strathmann R R. Annu Rev Evol Syst. 1993;24:89–117. [Google Scholar]
- 52.Kohn A J. Malacologia. 1990;32:55–67. [Google Scholar]
- 53.Röckel D, Rolán E, Monteiro A. Cone Shells from Cape Verde Islands. Vigo, Spain: Feito; 1980. [Google Scholar]