Abstract
In 1979, a BALB/cJ mouse was identified with an exceptionally long body. This phenotype was found to be caused by a recessive mutation, designated longjohn (lgj), that mapped to the proximal region of chromosome 15. Several years later, a mouse with a similarly elongated body was identified in an outbred stock after chemical mutagenesis with ethylnitrosourea. This phenotype also was caused by a recessive mutation, designated strigosus (stri). The two mutations were found to be allelic. A third allele was identified in a DBA/2J mouse and was designated longjohn-2J (lgj2J). Analysis of skeletal preparations of stri/stri mice indicated that the endochondral ossification process was slightly delayed, resulting in an extended proliferation zone. A recent study reported that mice overexpressing brain natriuretic peptide, one of the members of the natriuretic peptide family, exhibit a skeletal-overgrowth syndrome with endochondral ossification defects. The Npr3 gene coding for type C receptor for natriuretic peptides (NPR-C), which is mainly involved in the clearance of the natriuretic peptides, mapped in the vicinity of our mouse mutations and thus was a candidate gene. The present study reports that all three mutations involve the Npr3 gene and provides evidence in vivo that there is a natriuretic-related bone pathway, underscoring the importance of natriuretic peptide clearance by natriuretic peptide type C receptor.
Keywords: skeletal development, endochondral ossification, intervertebral disc
The family of natriuretic peptides comprises at least three structurally related peptides: atrial natriuretic peptide (ANP), brain natriuretic peptide (BNP), and C-type natriuretic peptide (CNP) (1–3). ANP and BNP act mainly as cardiac hormones, produced respectively by the atrium and the ventricle (4, 5). CNP predominates in the central nervous system but also is detected in a wide variety of tissues where it acts as a neuropeptide as well as a local regulator (6–9).
Natriuretic peptides exert their biological functions by specific binding to cell-surface receptors (10). Three specific receptors have been identified in mammalian tissues. Two are guanylyl cyclase-coupled receptors [GC-A and GC-B, also called natriuretic peptide receptor (NPR)-A and NPR-B, respectively] that act through activation of the cGMP-dependent signaling cascade, whereas the third receptor (type C receptor or NPR-C) is not coupled to guanylyl cyclase but is mainly involved in the clearance of the peptides (11, 12). Natriuretic peptides bind selectively to these receptors (13). The rank order of ligand selectivity for GC-A is ANP ≥ BNP ≫ CNP, for GC-B is CNP ≫ ANP ≥ BNP, and for NPR-C is ANP > CNP > BNP.
Natriuretic peptides are implicated in the control of extracellular fluid volume, blood pressure, and electrolyte homeostasis by their natriuretic–diuretic activity. However, evidence in vitro has accumulated that indicates that some of these peptides act on the differentiation and proliferation of bone cells, i.e., osteoblasts, chondrocytes, and osteoclasts (14–17). Studies in vivo support the suggestion of a natriuretic-related bone pathway: mice lacking cGMP-dependent protein kinase II display a dwarf phenotype (18), and transgenic mice overexpressing BNP exhibit a skeletal overgrowth phenotype (19); however, neither transgenic mice overexpressing ANP (20) nor knockout mice for GC-A (21, 22) display any bone-growth abnormality. These studies suggest that the natriuretic bone action is mediated by BNP or CNP through GC-B or an as-yet-unknown GC-coupled receptor. Here we report evidence in vivo of a natriuretic-related bone action in that mice homozygous for mutations in the Npr3 gene coding for NPR-C receptor display a skeletal-overgrowth phenotype.
MATERIALS AND METHODS
Origin of Mutations.
The first mutant allele arose in 1979 in BALB/cJ mice and was designated longjohn (lgj) because affected mice displayed an exceptionally long body. The second allele, identified at the Institut Pasteur and provisionally designated strigosus (stri), which in Latin means “long and emaciated,” arose in 1989 in an outbred stock after chemical mutagenesis with ethylnitrosourea. Subsequently, stri was transferred onto the 129/Sv strain. The third allele arose in 1996 in DBA/2J mice and was designated longjohn-2J (lgj2J). Both lgj and lgj2J were spontaneous mutations that occurred at The Jackson Laboratory. The three mutations were proven to be allelic by using progeny testing.
Skeletal Development: Stainings and Histology.
All observations reported here were made by using stri/stri and normal sib controls produced by intercrossing F1 mice that were obtained by mating 129/Sv stri/stri mice with FVB/N mice. The genotype of all mice analyzed was verified by using two polymorphic markers flanking the mutation, D15Mit11 and D15Mit162. Combined alcian blue/alizarin red S skeletal stainings were made at postpartum (P) stages (P0.5–1, P2.5–3, P6.5–7, and P10.5–11), as described (23). A minimum of six control and eight mutant mice were studied at each stage. In addition, hematoxylin and eosin-stained serial sections of 10% formalin fixed forepaws, hindpaws, and axial skeletons of 21-day-old mice were analyzed.
Genetic Mapping.
The assignment of lgj to the proximal region of chromosome 15 was accomplished by using an interspecific backcross involving the BALB/cJ-lgj and the CAST/Ei inbred strains. Further refinement of the genetic position was accomplished by using stri and two interspecific backcrosses, one with the PWK/Pas strain and the other with the CAST/Ei strain. Microsatellite markers used were PCR-amplified under standard conditions (1.5 mM MgCl2; 55°C annealing; Taq polymerase, Eurobio, Paris). Electrophoresis was performed on 4% agarose gels (Nusieve 3:1, FMC). Npr3 gene was mapped by using single-strand conformation polymorphism with primers selected in the genomic 5′-flanking region of its cDNA sequence (GenBank accession no. D78176) as follows: 5′-GACATTTTAGGGGGCTCGTAA-3′ and 5′-GCTGCTGTTAAGTCATAGTTA-3′.
RNA Preparation and Reverse Transcription (RT)-PCR.
Total RNA was extracted from lung, kidney, and heart of normal and mutant (homozygous lgj, lgj2J, and stri) mice, 2 months of age, by using the RNAxel kit (Eurobio). One microgram was used as template for first-strand cDNA synthesis with oligo(dT) primer by using Moloney murine leukemia virus reverse transcriptase (GIBCO/BRL). Primers were designed from the Npr3 cDNA sequence (GenBank accession no. D78175): 5′-CGGTCCTTTCTGCTGTTCACTTTC-3′ and 5′-TTAAGCCACCGAAAAATGTGA-3′ (amplification of the whole cDNA, from base pair 4 to base pair 1,611). PCR amplification of cDNAs was performed with the Advantage cDNA PCR kit (CLONTECH). Amplification conditions were: 94°C for 1 min; 35 cycles, 94°C for 30 s; 58°C for 45 s; 68°C for 3 min; 68°C for 3 min. RT-PCR products were resolved on 1.2% agarose gels (STG, Eurobio). To obtain the 5′ flanking region of Npr3 (GenBank accession no. D78176), genomic DNA from the control and mutant animals was used as a template for PCR reactions by using primers 5′-CCGCTGCCACGCTATTTAAAC-3′ and 5′-AGCACGCACGCCGAGAAAGT-3′ (GenBank accession nos. D78176 and D78175, respectively).
DNA Sequencing. RT-PCR and PCR products were cloned (TA cloning kit, Invitrogen), and sequencing was performed by using the ABI Prism Dye Terminator Cycle Sequencing Ready Reaction kit (Perkin–Elmer). Sequencing was carried out by using an ABI 370A DNA sequencer (Perkin–Elmer).
The correct gene symbols for the three mutant alleles reported here are Npr3lgj, Npr3lgj-2J, and Npr3stri. For easier reading, however, we have used the simplified lgj, lgj2J, and stri.
RESULTS
Phenotype.
Homozygous mutant mice (lgj, lgj2J, and stri) are easily distinguished from their wild-type counterparts as early as 6 days of age by an increase in body length, longer digits, and a typical cone-shaped implantation of the tail. No skeletal anomalies are observed in heterozygotes, indicating that the mutations are fully recessive. When older, mutant mice are exceptionally thin and have arachnodactyly, thoracic kyphosis, and frequent tail and/or sacral kinks (Fig. 1). No gross abnormalities are observed in craniofacial structures. At necropsy, normal body fat deposits are absent. The lgj and lgj2J alleles cause a somewhat more severe phenotype than the stri allele in that homozygous lgj and lgj2J mice become humpbacked at younger age and have sacral kinks. Many homozygous lgj2J mice die suddenly in their adulthood.
Figure 1.
Phenotype. Wild-type sib of homozygous lgj2J mouse (Upper Left), lgj2J/lgj2J mutant (Upper Right), lgj/lgj mutant (Lower Left), and stri/stri mutant (Lower Right) mice at 3 months of age.
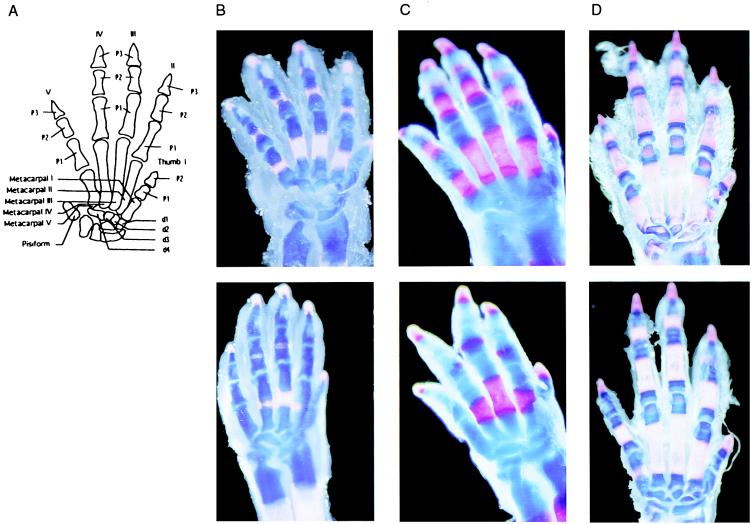
Skeletal Stainings and Histological Analysis. Analysis of skeletons by combined alcian blue/alizarin red S stainings revealed that a delay in the endochondral ossification occurs in the mutant mice. This delay was most noticeable in the hind- and forepaws, but it also was evident throughout the skeleton. It also was observed in neonates and presumably occurs in utero. We focused on forepaw ossification for this study. A schematic hand representation with bone denomination is given (Fig. 2A). P3 phalanges stained red in all preparations. At P0.5 (Fig. 2B), mutant mice exhibit small ossification centers only in metacarpals and P1 phalanges of digits III and IV, whereas there are already ossification centers in metacarpals as in P1 and P2 phalanges of digits II–V in wild-type mice. At P2.5 (Fig. 2C), only metacarpals and P1 phalanges for digits II, III, and IV exhibit ossification centers in mutant mice, whereas wild-type mice exhibit ossification centers in metacarpals, in P1 and P2 phalanges for digits II to V. At P6.5–7 (Fig. 2D), delayed ossification is most noticeable at the extremities of metacarpals and phalanges (especially in digits I and V) and in carpal bones. At this stage, digits are already longer in mutants as compared with controls.
Figure 2.
Alizarin red S/alcian blue staining of forepaw at various postpartum developmental stages. (A) Hand schematic representation with legends. (B) Forepaw of 0.5-day-old control (Upper) and stri/stri mutant (Lower) mice. (C) Forepaw of 2.5-day-old control (Upper) and stri/stri mutant (Lower) mice. (D) Forepaw of 6.5-day-old control (Upper) and stri/stri mutant (Lower) mice.
Truncal stainings in 10.5-day-old control and stri/stri mutant mice revealed that all truncal vertebrae were present in mutant mice but were enlarged (Fig. 3A). Both vertebral bodies and transverse processes were affected. Vertebral bodies were mainly enlarged, whereas transverse process abnormalities varied depending on the vertebrae topographic position, i.e., thoracic, lumbar, sacral, or caudal. Sacral region transverse processes are sharper and more rostrally oriented, especially in the last vertebrae that forms the pelvis (Fig. 3A, Lower Right). Two mutant mice (of 40 studied) exhibited special abnormalities in vertebrae: one had a partial thoracic supernumerary vertebrae, with a small rib, leading to 14 thoracic vertebrae and 7 intersternebrae spaces at sternum level whereas the other one had a small rib on the first lumbar vertebrae (data not shown).
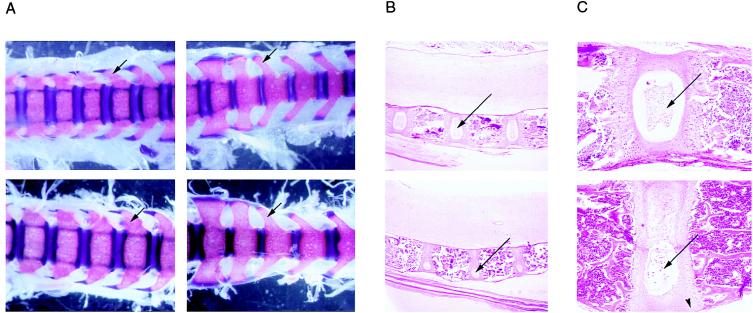
Figure 3.
Skeletal phenotype at the trunk level. (A) Alizarin red S/alcian blue staining of the trunk in 10.5-day-old control (Upper) and stri/stri mutant (Lower) mice. Dorsal view from lumbar and sacral region. Arrows denote the transverse processes. (B) Hematoxylin and eosin-stained histological sections of cervical-thoracic vertebrae in 21-day-old control (Upper) and stri/stri mutant (Lower) mice. Magnification ×2. Arrows denote the nucleus pulposus localized in the intervertebral disc. (C) Higher magnification (×40) of hematoxylin and eosin-stained histological sections of cervical–thoracic vertebrae in 21-day-old control (Upper) and stri/stri mutant (Lower) mice. Arrows denote the nucleus pulposus. Arrowhead denotes aberrant proliferation.
Histological analysis of 21-day-old homozygous stri and normal mice revealed no differences in membranous ossification, bone marrow, central nervous system, hypophysis, or skeletal-muscle development. In forepaws, the growth plates were normally organized (assessed at the junction between the digits and the carpal). Defects were observed in the trunk, in the intervertebral discs, and in the growth plates located at the extremities of vertebrae. Truncal vertebrae were enlarged, and intervertebral discs were very thin (Fig. 3B). The nucleus pulposus, a notochordal remnant normally present in the center of the intervertebral disc, was either absent or shrunken in the ventral part of the disc and included in a smaller hydrated poach. The dorsal part of the intervertebral disc was calcified (Fig. 3C). The growth plates presented a normal thickness but aberrant proliferations at their ventral and dorsal extremities invading the vertebral bodies.
Genetic Mapping.
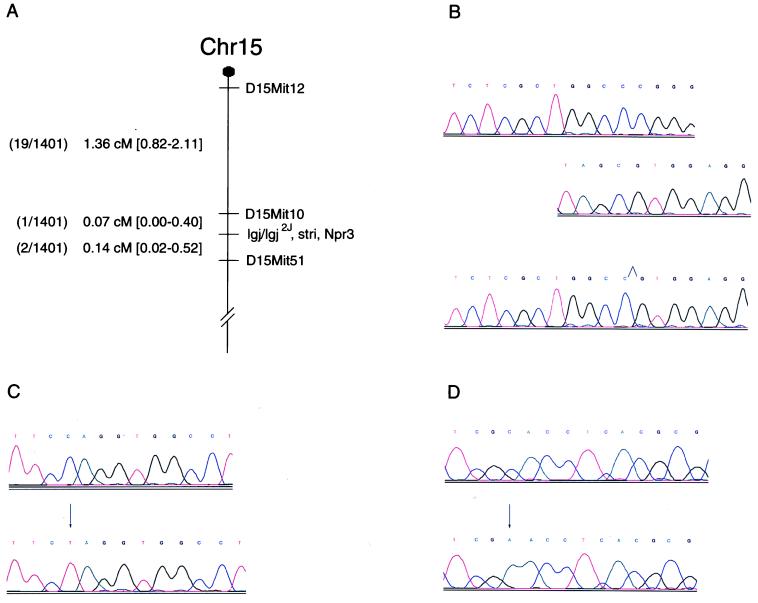
Details involving the mapping of lgj and stri to chromosome 15 will be presented elsewhere. Briefly, in the first cross involved in mapping lgj to chromosome 15, no recombination was noted in 104 backcross offspring for lgj, growth hormone receptor (Ghr) (24), and five microsatellite markers (D15Mit12, D15Mit11, D15Mit51, D15Mit162, and D15Mit52). In the two other crosses used for further refinement, a total of 1,401 mice were typed for stri and for nine microsatellite markers. This result positioned stri between D15Mit10 (proximal) and D15Mit51 (distal). A subset of 22 mice that inherited a recombinant chromosome between D15Mit12, which is proximal to D15Mit10, and D15Mit51 were genotyped for Npr3. No recombination was found between stri and Npr3, placing these two genes less than 1 centiMorgan apart (Fig. 4A).
Figure 4.
Genetic map and sequence chromatograms. (A) Two distinct interspecific backcrosses were made, and 1,401 backcross mice were analyzed. Markers were ordered by minimizing double recombinants. Number of recombinant animals is given for each interval. Genetic distances and confidence intervals (in brackets) were calculated at risk α = 0.05. (B) Sequence chromatograms for control (2 chromatograms, at the top) and lgj/lgj mutant (Lower) mice. Deletion site is indicated in the mutant chromatogram, the two chromatograms above are part of control sequence viewed as flanking the deletion. (C) Sequence chromatograms for control (Upper) and lgj2J/lgj2J mutant (Lower) mice. Arrow denotes mutation site. (D) Sequence chromatograms for control (Upper) and stri/stri mutant (Lower) mice. Arrow denotes mutation site.
RT-PCR and Sequencing.Npr3 attracted our attention as a candidate gene for stri, lgj, lgj2J for three reasons. (i) Npr3 mapped near D15Mit10 (25), the nearest proximal flanking marker we had identified in our crosses involving the stri allele. (ii) None of the 1,401 backcross mice we analyzed inherited a chromosome 15 containing recombination between stri and Npr3. (iii) A recent study (19) reported that mice overexpressing BNP, one of the natural ligands for NPR-C encoded by Npr3, presented a skeletal overgrowth syndrome with endochondral ossification defects similar to what we had observed in mutant animals. To determine whether any of the mutant alleles involved the Npr3 gene, primers were designed and used to prepare cDNA by using RT-PCR and RNA isolated from each of the mutant mice. The RT-PCR product from the lgj mutant was smaller than the 1,611 bp expected, whereas products from the other mutants and controls were of the expected size (data not shown). cDNA was sequenced from all three mutants and revealed three different mutations. The lgj allele contains an in-frame 36-bp deletion (Fig. 4B) between position 195 and 232, leading to a protein truncated by 12 aa. The lgj2J allele exhibits a C → T transition at position 283 (Fig. 4C), causing a stop codon. The stri allele has a C → A transversion at position 502 (Fig. 4D), leading to an asparagine-to-histidine substitution. All three mutations occurred in the predicted extracellular domain of the protein, affecting amino acids that are highly conserved among species (Fig. 5).
Figure 5.
Partial predicted amino acid sequence encoded by Npr3 gene compared among species. Amino acid sequence is numbered according to mouse sequence. The three mutation sites are boxed as follows: shaded box, lgj allele; normal box, lgj2J allele; striated box, stri allele. Bold underlined amino acids denote sequencing discrepancies with previous published cDNA sequence, which leads to three changes in the predicted amino acid sequence resulting in a protein that shares three more conserved residues among species.
While sequencing the normal wild-type allele, we found two discrepancies with the published cDNA sequence: between position 172 and 175, ATTA should read GATT and in position 395, G should read A (data not shown; submitted to GenBank). These differences lead to three changes in the predicted amino acid sequence resulting in a protein that shares three more conserved residues among species (Fig. 5).
DISCUSSION
In this report we found that three allelic mice exhibiting skeletal overgrowth are mutated in the Npr3 gene encoding for NPR-C receptor. Npr3 knockout mice also exhibit the same skeletal-overgrowth phenotype (26). We therefore assert that Npr3 gene is the causative gene for the skeletal abnormalities reported here.
NPR-C is mainly involved in the clearance of the natriuretic peptides. The peptides bind to it and are internalized and enzymatically degraded, after which NPR-C returns to the cell surface (10). NPR-C is a protein with extracellular and transmembrane domains and a short cytoplasmic segment. All three mutations reported here are in the predicted extracellular domain of the protein, affecting amino acids that are highly conserved among species and presumably important in the ligand-binding process. The most probable mechanism involved is a ligand-binding defect and lack of clearance by NPR-C. The resulting accumulation of unbound natriuretic peptides would mediate the skeletal-overgrowth phenotype, as first observed in BNP-transgenic mice (19).
Bone formation follows two distinct ossification pathways: membranous and endochondral. Both mechanisms occur simultaneously in developing mammals, but at different locations, i.e., craniofacial bone structures form via the membranous process whereas long bones, some short, flat, and irregular bones (such as the carpal and tarsal bones), and vertebrae form via the endochondral process (27). In this paper, we report that the three mutants exhibit a skeletal-overgrowth phenotype characterized by an elongated body, thoracic kyphosis, arachnodactyly, and sacral or tail kinks, but no significant changes in craniofacial bones. This correlates with a previous BNP-transgenic study (19) suggesting that natriuretic peptides are involved in the process of endochondral ossification but not in the process of membranous ossification.
In endochondral ossification, a cartilaginous structure is previously formed and subsequently replaced by bone tissue (ossification). In this process, the cartilaginous structure, composed of proliferative and hypertrophic chondrocytes, serves both to drive the growth of skeletal elements and to form a scaffold for the osteoblasts implicated in ossification. Growth plates, localized at the end of each bone, are implicated in the longitudinal growth of bones by the endochondral ossification process. Previous studies in vitro (17, 19) suggested that natriuretic peptides act on longitudinal bone growth by activating, via their GC-coupled receptors, the proliferation and differentiation of the growth-plate chondrocytes. Our skeletal and histologic data that showed a delay in the forepaw ossification and aberrant proliferations in the vertebrae growth plates in stri/stri mutant mice support this suggestion and underscore the physiological role of the NPR-C receptor in modulating the bone action of the natriuretic peptides.
The three mutants displayed phenotypic differences. Whether these differences are caused by differences in genetic background, differences in the protein coded by the mutant allele, or a combination of the two remains to be determined. The sudden-death phenotype in homozygous lgj2J mice may be caused by a cardiovascular action of the natriuretic peptides, as was observed in mice lacking the Npr1 gene coding for the GC-A receptor (22).
Although most ethylnitrosourea-induced germ-line mutations in the mouse are point mutations that occur at A/T sites (28), there are some reports of mutations at G/C sites (29). Thus, the stri mutation, which is a point mutation but not at an A/T site, is possibly caused by a less common type of ethylnitrosourea action.
In summary, our results provide evidence in vivo for a role of natriuretic peptides in the control of bone growth and underscore the importance of NPR-C as a down-regulator of the natriuretic peptide stimuli. Whether Npr3 mutant mice are useful models for human disease remains to be determined, given the differences in growth-plate development known to exist between these species (30). Nevertheless, the three mutations we have described here, ranging from a single amino acid substitution to a protein truncated at the extracellular domain, are useful models to further investigate physiological and pharmacological aspects of bone growth per se.
Acknowledgments
We are very grateful to Drs. Patricia Baldacci and Xavier Montagutelli for discussions and critical reading of the manuscript. We thank Drs. Susan Ackerman, Leah Rae Donahue, and Kenneth Johnson for their thoughtful review of this manuscript. The technical assistance of Gérard Pivert with histology and Leona Gagnon with genotyping is most appreciated. We are indebted to Richard Samples for identifying the DBA/2J mouse carrying the lgj2J allele. This work was supported part by the Association Française contre les Myopathies (AFM) and by National Institutes of Health Grants GM20919 and RR01183 (to E.M.E.).
ABBREVIATIONS
- ANP
atrial natriuretic peptide
- BNP
brain natriuretic peptide
- CNP
C-type natriuretic peptide
- GC
guanylyl cyclase, NPR-A, -B, or -C, natriuretic peptide receptor type A, B, or C
Footnotes
References
- 1.Kangawa K, Matsuo H. Biochem Biophys Res Commun. 1984;118:131–139. doi: 10.1016/0006-291x(84)91077-5. [DOI] [PubMed] [Google Scholar]
- 2.Sudoh T, Kangawa K, Minamino N, Matsuo H. Nature (London) 1988;332:78–81. doi: 10.1038/332078a0. [DOI] [PubMed] [Google Scholar]
- 3.Sudoh T, Minamino N, Kangawa K, Matsuo H. Biochem Biophys Res Commun. 1990;168:863–870. doi: 10.1016/0006-291x(90)92401-k. [DOI] [PubMed] [Google Scholar]
- 4.De Bold A J. Science. 1985;230:767–770. doi: 10.1126/science.2932797. [DOI] [PubMed] [Google Scholar]
- 5.Mukoyama M, Nakao K, Hosoda K, Suga S-I, Saito Y, Ogawa Y, Shirakami G, Jougasaki M, Obata K, Yasue H, et al. J Clin Invest. 1991;87:1402–1412. doi: 10.1172/JCI115146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komatsu Y, Nakao K, Suga S, Ogawa Y, Mukoyama M, Arai H, Shirakami G, Hosoda K, Nakagawa O, Hama N, et al. Endocrinology. 1991;129:1104–1106. doi: 10.1210/endo-129-2-1104. [DOI] [PubMed] [Google Scholar]
- 7.Furuya M, Yoshida M, Hayashi Y, Ohnuma N, Minamino N, Kangawa K, Matsuo H. Biochem Biophys Res Commun. 1991;177:927–931. doi: 10.1016/0006-291x(91)90627-j. [DOI] [PubMed] [Google Scholar]
- 8.Suga S, Nakao K, Itoh H, Komatsu Y, Ogawa Y, Hama N, Imura H. J Clin Invest. 1992;90:1145–1149. doi: 10.1172/JCI115933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Middendorff R, Muller D, Paust H J, Davidoff M S, Mukhopadhyay A K. J Clin Endocrinol Metab. 1996;81:4324–4328. doi: 10.1210/jcem.81.12.8954035. [DOI] [PubMed] [Google Scholar]
- 10.Levin E R, Gardner D G, Samson W K. N Engl J Med. 1998;339:321–328. doi: 10.1056/NEJM199807303390507. [DOI] [PubMed] [Google Scholar]
- 11.Maack T, Suzuki M, Almeida F A, Nussenzveig D, Scarborough R M, McEnroe G A, Lewicki J A. Science. 1987;238:675–678. doi: 10.1126/science.2823385. [DOI] [PubMed] [Google Scholar]
- 12.Almeida F A, Suzuki M, Scarborough R M, Lewicki J A, Maack T. Am. J. Physiol. 1989. R469–R475. [DOI] [PubMed] [Google Scholar]
- 13.Suga S-I, Nakao K, Hosoda K, Mukoyama M, Ogawa Y, Shirakami G, Arai H, Saito Y, Kambayashi Y, Inouye K, Imura H. Endocrinology. 1992;130:229–239. doi: 10.1210/endo.130.1.1309330. [DOI] [PubMed] [Google Scholar]
- 14.Hagiwara H, Sakaguchi H, Itakura M, Yoshimoto T, Furuya M, Tanaka S, Hirose S. J Biol Chem. 1994;269:10728–10733. [PubMed] [Google Scholar]
- 15.Inoue A, Hiruma Y, Hirose S, Yamaguchi A, Hagiwara H. Biochem Biophys Res Commun. 1995;215:1104–1110. doi: 10.1006/bbrc.1995.2577. [DOI] [PubMed] [Google Scholar]
- 16.Suda N, Tanaka K, Yasoda A, Ogawa Y, Itoh H, Nakao K. Biochem Biophys Res Commun. 1996;223:1–6. doi: 10.1006/bbrc.1996.0836. [DOI] [PubMed] [Google Scholar]
- 17.Yasoda A, Ogawa Y, Suda M, Tamura N, Mori K, Sakuma Y, Chusho H, Shiota K, Tanaka K, Nakao K. J Biol Chem. 1998;273:11695–11700. doi: 10.1074/jbc.273.19.11695. [DOI] [PubMed] [Google Scholar]
- 18.Pfeifer A, Aszodi A, Seidler U, Ruth P, Hofmann F, Fässler R. Science. 1996;274:2082–2086. doi: 10.1126/science.274.5295.2082. [DOI] [PubMed] [Google Scholar]
- 19.Suda M, Ogawa Y, Tanaka K, Tamura N, Yasoda A, Takigawa T, Uehira M, Nishimoto H, Itoh H, Saito Y, et al. Proc Natl Acad Sci USA. 1998;95:2337–2342. doi: 10.1073/pnas.95.5.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steinhelper M E, Cochrane K L, Field L J. Hypertension. 1990;16:301–307. doi: 10.1161/01.hyp.16.3.301. [DOI] [PubMed] [Google Scholar]
- 21.Lopez M J, Wong S K-F, Kishimoto I, Dubois S, Mach V, Friesen J, Garbers D L, Beuve A. Nature (London) 1995;378:65–68. doi: 10.1038/378065a0. [DOI] [PubMed] [Google Scholar]
- 22.Oliver P M, Fox J E, Kim R, Rockman H A, Kim H S, Reddick R L, Pandey K N, Milgram S L, Smithies O, Maeda N. Proc Nat Acad Sci USA. 1997;94:14730–14735. doi: 10.1073/pnas.94.26.14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimmel C A, Trammell C. Stain Technol. 1981;56:271–273. doi: 10.3109/10520298109067325. [DOI] [PubMed] [Google Scholar]
- 24.Eicher E M, Lee B K. Mamm Genome. 1991;1:57–58. doi: 10.1007/BF00350847. [DOI] [PubMed] [Google Scholar]
- 25.Savinova O V, Matsukawa N, Smithies O, John S W M. Mamm Genome. 1997;8:788. doi: 10.1007/s003359900571. [DOI] [PubMed] [Google Scholar]
- 26.Matsukawa, N., Grzesik, W. J., Takahashi, N., Pandey, K. N., Pang, S., Yamauchi, M. & Smithies, O. (1999) Proc. Natl. Acad. Sci., in press. [DOI] [PMC free article] [PubMed]
- 27.O’Rahilly R, Gardner E. In: Bones and Joints. Ackerman L V, Spjut H J, Abell M R, editors. Baltimore: Williams & Wilkins; 1976. pp. 1–15. [Google Scholar]
- 28.Favor J, Neuhäuser-Klaus A, Ehling U H, Wulff A, van Zeeland A A. Mutat Res. 1997;374:193–199. doi: 10.1016/s0027-5107(96)00229-1. [DOI] [PubMed] [Google Scholar]
- 29.Provost G S, Short J M. Proc Natl Acad Sci USA. 1994;91:6564–6568. doi: 10.1073/pnas.91.14.6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howell D S, Dean D D. In: Disorders of Bone and Mineral Metabolism. Coe F L, Favus M J, editors. New York: Raven; 1992. pp. 313–353. [Google Scholar]