Abstract
A rice Dwarf 1 gene was identified by using a map-based cloning strategy. Its recessive mutant allele confers a dwarf phenotype. Linkage analysis revealed that a cDNA encoding the α-subunit of GTP-binding protein cosegregated with d1 in 3,185 d1 segregants. Southern hybridization analysis with this cDNA as a probe showed different band patterns in several d1 mutant lines. In at least four independent d1 mutants, no gene transcript was observed by Northern hybridization analysis. Sequencing analysis revealed that an 833-bp deletion had occurred in one of the mutant alleles, which resulted in an inability to express GTP-binding protein. A transgenic d1 mutant with GTP-binding protein gene restored the normal phenotype. We conclude that the rice Dwarf 1 gene encodes GTP-binding protein and that the protein plays an important role in plant growth and development. Because the d1 mutant is classified as gibberellin-insensitive, we suggest that the GTP-binding protein might be associated with gibberellin signal transduction.
Dwarf mutants in plants are crucial for elucidating regulatory mechanisms for plant growth and development. This character is also favored in breeding. Dwarf mutants have been isolated in many species and have been extensively analyzed for their mode of inheritance and their response to plant hormones. There are various reasons for their dwarf phenotypes, associated with, for example, gibberellins (1–3), brassinosteroids (4, 5), abnormal cell walls (6), and abnormal cell elongation (5, 7).
In rice, at least 54 dwarf mutants are known, but only two, d18 and d35, are known to have a deficiency in their gibberellin biosynthetic pathway (8, 9). Rice dwarf mutant Daikoku, carrying the d1 gene, was first isolated as a spontaneous mutant that was not only short but had broad, dark green leaves, compact panicles, and short, round grains (Fig. 1). These phenotypes are all induced by a recessive allele (d1) of the Dwarf 1 (D1) gene and are thought to reflect aberrant physiological and biochemical pathways in plant growth and development. The rice d1 mutant was classified as gibberellin-insensitive (10). Gibberellins (GAs) are a large family of tetracyclic diterpenoid plant growth regulators and are associated with a number of plant growth and development processes such as seed germination, stem elongation, flowering, and fruit development (1–3) and regulation of gene expression in the cereal aleurone layer (11). Many GA-related mutants have been isolated from many plant species and have been classified. GA-insensitive mutants that did not respond to exogenous GAs were defined as dwarf mutants: GAI in Arabidopsis, D8 and D9 in maize, Rht3 in wheat (3), and d1 in rice (10).
Figure 1.
Phenotypes of d1 mutant, Daikoku. In each photograph, Daikoku is on the left and Nipponbare (wild-type variety) is on the right. (A) Plant height. (B) Panicle. (C) Seeds.
These mutants were thought to be associated with GA-signal transduction. The mechanisms of signal transduction triggered by GAs are still unknown. Isolation and characterization of genes defining the dwarf mutation caused by GA insensitivity can help to clarify not only the molecular mechanisms of plant growth and development but also signal transduction of GAs.
We report here the map-based cloning of the rice D1 gene. The amino acid sequence of D1 revealed that it encodes the α-subunit of GTP-binding protein (G protein). Although the α-subunit of G protein homologues have been isolated from several plants (12, 13), their functions are so far not clear. The most intriguing finding in this study is that the α-subunit of G protein plays an important role in plant growth and development.
MATERIALS AND METHODS
Plant Materials.
Six d1 mutant lines (HO532, HO533, HO537, HO538, HO541, and HO552) have been maintained for more than 60 years at Kyushu University (14). These mutants were all japonica and all spontaneous mutants. FL2 (d1) is a marker line derived from HO538. Two d1 mutant lines (CM392 and CM1729) induced by N-methyl-N-nitrosourea (mutants of Kinmaze) were provided by H. Satoh (Institute of Genetic Resources, Kyushu University, Japan).
Mapping Population.
Substitution lines that substitute chromosomes of Kasalath (indica) for that of Nipponbare (japonica) were used for crossing. Given that d1 was located on chromosome 5, substitution line SL18, which substitutes chromosome 5 of Kasalath (indica), was crossed with line FL2 (d1) (japonica). About 13,000 F2 seeds were sown in a nursery. At the seedling stage, 3,185 dwarf plants were selected and transplanted into a paddy field. One month after transplanting, two green leaves of each individual were collected, and leaves of five plants were combined to obtain 637 pooled samples (15).
DNA Extraction, Southern Hybridization, and Linkage Analysis.
The cetyltrimethylammonium bromide (CTAB) method (16) was used with minor modifications for extracting total DNA from rice leaves. The extracted DNA was digested with restriction enzymes. Three micrograms of each digested DNA was loaded on a 0.8% agarose gel and run for 16 h at 20 V and then blotted onto a nylon membrane. Southern hybridization (17) was done with horseradish peroxidase-labeled restriction fragment-length polymorphism (RFLP) markers according to the protocol for the Enhanced Chemiluminescent (ECL) direct nucleic acid labeling and detection system (Amersham Pharmacia). Linkage analysis was done by using the pooled-sampling method (15).
Screening of Yeast Artificial Chromosome (YAC) Library and P1-Derived Artificial Chromosome (PAC) Library. A rice YAC library containing 6,934 clones with an average insert size of 350 kilobases and covering six times the haploid genome was constructed previously (japonica cultivar Nipponbare; ref. 18). All YAC clones were blotted on five high-density filters, multiplied on many replica filters, and screened by colony hybridization with RFLP markers or a three-step PCR with YAC end-fragment DNA.
A rice PAC library (japonica cultivar Nipponbare) was kindly provided by the Rice Genome Research Program (RGP). From this library, 18,432 clones were screened by PCR with Sequence Tagged Site (STS) primers.
Three-step PCR Screening of Expressed Sequence Tags (ESTs).
A screening system using a three-step PCR to isolate YAC clones with STS markers (19) was modified. The 20-base primers were designed from the 3′-end sequences of rice ESTs. In the first screening, a PCR reaction was done with nine superpools of YAC DNA mixture (W pools), each coming from 9 × 96 clones, yeast host strain AB1380, and rice cultivar Nipponbare. In the second screening, 29 subpools (8 X pools, 12 Y pools, and 9 Z pools) of each W pool that was positive in the first screening were surveyed with the same primers. The YAC clones passing the second PCR were considered as candidates carrying ESTs, and the positive clones were finally confirmed by a third PCR using the individual candidate YAC DNAs as templates.
Isolation of YAC End DNA Fragment.
YAC end DNA fragments were amplified by using a slightly modified cassette-PCR method (20) and were cloned to the TA vector (Invitrogen).
RNA Extraction and Northern Hybridization.
Total RNA was extracted from 2-week-old seedlings by using a single-step method (21) with minor modifications. Three micrograms of poly(A)+RNA was recovered from total RNA by Oligotex-dT30 (Takara Shuzo, Kyoto) and was electrophoresed on 1.0% agarose containing 18% formaldehyde for 3 h at 75 V and then blotted onto a nylon membrane. Northern hybridization was done in Rapid hybridization buffer (Amersham Pharmacia) at 65°C overnight with 32P-labeled cDNA as a probe. The nylon membrane was washed twice in 1× SSPE (3M NaCl/173 mM NaH2PO4⋅2H2O/25 mM EDTA, pH 7.4) buffer containing 1% SDS at 65°C for 20 min then twice in 0.1× SSPE buffer containing 1% SDS at 65°C for 30 min. The banding patterns were detected on x-ray film after a week-long exposure at −80°C.
Sequence Analysis.
The primers were designed from the genomic sequences of the α-subunit of G protein in rice (IR36, indica) (22). Genomic regions of the α-subunit of G protein in Nipponbare (japonica) and a d1 mutant HO541 (japonica) were amplified by PCR with the primers. Amplified products were cloned to the TA vector (Invitrogen) and were sequenced.
Isolation of Cosmid Clone Carrying the D1 Gene and Complementation Test. A cosmid vector (pPZP2CH-lac) for complementation testing was kindly provided by T. Fuse of the Bio-oriented Technology Research Advancement Institution, Japan. A PAC clone carrying the candidate D1 gene was partially digested with Sau3AI, and fragment DNAs were ligated into the BamHI cloning site of the cosmid vector. The cosmid clones were packaged in vitro with GigaPak III Gold (Stratagene) and infected into competent Escherichia coli XL1-BlueMRF′. Cosmid clones carrying the candidate D1 gene were screened by using PCR with STS primers.
Transformation was used for complementation testing with the Agrobacterium system (23). The cosmid DNA carrying the D1 gene was infected into callus of a d1 mutant line in darkness at 25°C for 3 days. The callus was regenerated in a growth chamber, and the regenerated plants were grown in an isolated greenhouse.
RESULTS
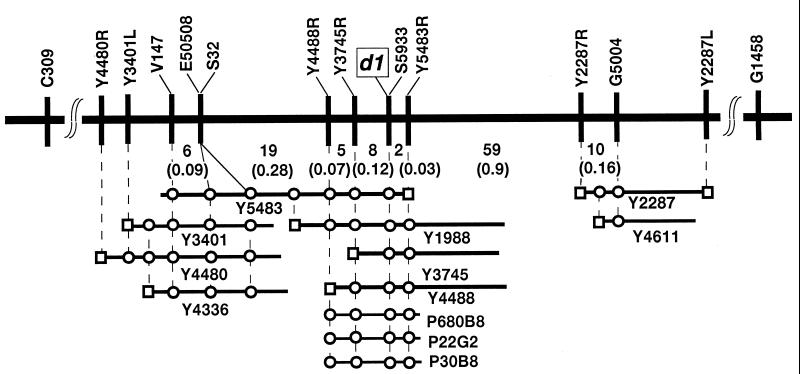
High-Resolution Mapping and Physical Mapping. In general, variations in genotypes of individual F2 plants derived from a cross between japonica and indica varieties create large variations in phenotypes. This variation causes difficulties in identifying true mutants among F2 plants, such as mutants for culm height. However, individual F2 plants derived from crosses between japonica marker line FL2 (d1) and SL18, which substitutes chromosomes of Kasalath (indica) outside the target region of chromosome 5 for those of Nipponbare (japonica), showed clear phenotypic differences for d1. We could identify dwarfism in individual plants with high reliability. Of about 13,000 F2 plants derived from the cross, 3,185 dwarf (d1 homozygous) plants were identified. DNA of the 637 pools was extracted for pooled-sample mapping. Southern hybridization and linkage analysis were done with 13 RFLP markers between C309 and G1458, which were mapped around d1 (24) on chromosome 5 (25). Of the 637 pools, 37 and 71 recombinant pools were identified with RFLP markers V147 and G5004, respectively, as the closest markers flanking either side of d1. d1 was located between these markers within 1.65 centiMorgan (cM) (Fig. 2). To select the individual recombinant plants from each pool, DNA was extracted from the 540 individual plants (5 plants × 108 recombinant pools), and each genotype was surveyed by using V147 and G5004 as probes. As a result, 37 and 71 recombinant plants were identified individually and used for fine mapping between V147 and G5004.
Figure 2.
High-resolution RFLP linkage map and physical map of the d1 locus. The vertical bar represents the RFLP markers, and the numbers of recombinants are indicated under the linkage map. Genetic distances between adjacent markers are shown in parentheses. □, End-fragment DNA of YAC clone; ○, RFLP markers or YAC end-fragment DNA contained in the YAC clones (Y number) and PAC clones (P number).
A YAC library was screened for construction of YAC contigs covering the d1 region. Four YACs (Y5483, Y3401, Y4480, and Y4336) were identified with RFLP marker V147, and two (Y2287 and Y4611) were identified with G5004 (Fig. 2). Both end sequences of these YACs were amplified by using the cassette-PCR method, and the products were also used for linkage mapping by RFLPs. Fragment Y5483R was mapped between d1 and G5004 and linked to d1 within 0.03 cM. By using Y5483R as a probe, three YACs (Y1988, Y3745, and Y4488) were also identified. End-fragment DNAs of Y3745R and Y4488R were both mapped between d1 and V147 with genetic distances from d1 of 0.12 and 0.19 cM, respectively. As a result, d1 was located between Y3745R and Y5483R within a 0.15-cM genetic distance (Fig. 2).
EST Mapping.
We used large-scale EST mapping on a YAC physical map. Three ESTs (E50508, S32, and S5933) were mapped onto the YAC clones in the d1 region. YACs Y5483, Y3401, Y4336, and Y4480 carried the sequences of E50508 and S32. YACs Y5483, Y1988, Y4488, and Y3745 carried the sequence of S5933. The three ESTs were used for genetic mapping in the same population as above. As a result, S5933, E50508, and S32 were mapped to within 0, 0.5, and 0.5 cM, respectively, of d1 in the 3,185 d1 segregants (Fig. 2). The cosegregated cDNA S5933 (350-bp insert) was derived from green shoots, and its translated amino acid sequence completely coincided with the sequence from Phe-332 to Thr-380 of the α-subunit of rice G protein RGA1 (22, 26).
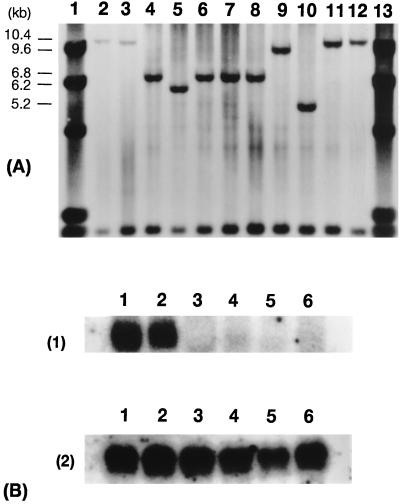
Mutant Analysis. Allelic tests were used to confirm the d1 allelism among the nine d1 mutant lines (FL2, HO532, HO533, HO537, HO538, HO541, HO552, CM392, and CM1729). F1 hybrids from crosses between FL2 and each of the other eight lines were all d1 dwarf. Allelic testing confirmed that these lines were caused by the mutation at the D1 locus. Genomic Southern hybridization analysis of these d1 mutant lines and two wild-type varieties, Nipponbare (japonica) and Kasalath (indica), was done with S5933 as a probe. No polymorphism was observed between the wild types, but five different band patterns were observed among the nine mutants (Fig. 3A). These results strongly suggested that several types of structural variations occurred in the region probed by S5933 in the nine d1 mutant lines.
Figure 3.
(A) Genomic Southern hybridization analysis of nine d1 mutants using S5933 as a probe. Lanes: 1, λ/HindIII; 2, Nipponbare; 3, Kasalath; 4, FL2; 5, HO532; 6, HO533; 7, HO537; 8, HO538; 9, HO541; 10, HO552; 11, CM382; 12, CM1792; 13, λ/HindIII. Three micorgrams of total DNA were digested with HindIII. (B) Northern hybridization analysis using S5933 (1) or S14002 (2) (actin) as a probe. Lanes: 1, Nipponbare; 2, Kasalath; 3, FL2; 4, HO532; 5, HO541; 6, HO552.
Poly(A)+RNA was extracted from four d1 mutants (FL2, HO532, HO541, and HO552) that showed different types of banding patterns in genomic Southern hybridization (Fig. 3A) and from Nipponbare and Kasalath. Northern analysis was done with S5933 as a probe. A 1.8-kilobase translated product that coincided well with the reported size for mRNA of the α-subunit of rice G protein (22, 26) was observed in Nipponbare and Kasalath but not in the independent four d1 mutant lines (Fig. 3B).
The genomic region of the α-subunit of rice G protein in both Nipponbare and HO541 was cloned and sequenced. Deletion of 833 bp was detected in HO541 between the first exon and intron.
Complementation Test. A PAC library was screened for covering the d1 gene. Three PACs (P680B8, P22G2, and P30B8) were identified by PCR with STS primers of EST S5933 (Fig. 2). The flanking markers on both sides of the d1 gene, Y3745R and Y5483R, were contained in these PACs. A PAC clone (P22G2) was subcloned into cosmid vector (pPZP2CH-lac), and a cosmid clone carrying the α-subunit of rice G protein (C6) was screened by PCR with STS primers of EST S5933. Because this cosmid clone had the flanking markers on both sides of the d1 gene Y3745R and Y5483R, the D1 gene was included in the cosmid clone.
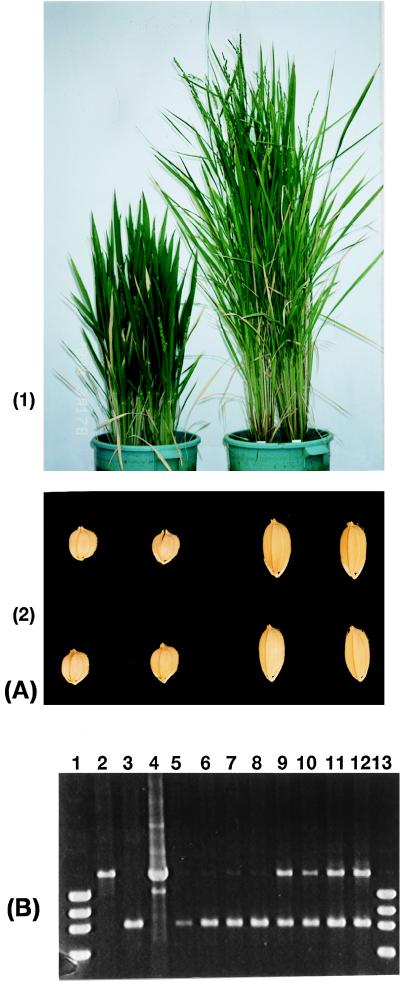
The cosmid clone was transformed into a d1 mutant line (HO541). Twenty-one transgenic plants with C6 clone in HO541 were obtained. Of 21 plants, 14 plants showed a normal phenotype and 7 plants showed a dwarf phenotype. Eleven transgenic plants with only vector in HO541 were obtained as control, and all showed a dwarf phenotype (Fig. 4A, 1 and 2). Those transgenic plants that showed a normal phenotype were tested if they carried the cosmid clone. Primers were designed between the first exon and the second exon of the region of α-subunit of rice G protein, because HO541 has an ∼833-bp deletion here. DNA of transgenic plants was extracted, and PCR amplification was performed with the primers. Transgenic plants showing a normal phenotype had the band of cosmid clone. Transgenic plants with the vector did not have the bands of cosmid clones (Fig. 4B). To test inheritance of transgenic plants, the segregation of the second generations of the transgenic plants was observed. The segregations of the second generation of the three normal transgenic plants were 60 normal and 20 dwarf, 46 normal and 18 dwarf, and 40 normal and 10 dwarf; all segregations were close to a 3:1 ratio, as expected.
Figure 4.
Complementation test. (A) Transgenic plants of d1 mutant (HO541). (1) Left, transgenic plant with cosmid vector; right, transgenic plant with cosmid clone (C6) containing D1 genomic region. (2) Left, seeds of transgenic plant with cosmid vector; right, seeds of transgenic plant with cosmid clone (C6). (B) Detection of integrated D1 gene in transgenic plants. Lanes: 1, marker (ϕX174/HaeIII); 2, Nipponbare (normal); 3, HO541 (d1 mutant); 4, cosmid clone (C6); 5–8, transgenic plants with cosmid vector in HO541 (phenotypes were all dwarf); 9–12, transgenic plants with cosmid clone C6 (phenotypes were all normal); 13, marker (ϕX174/HaeIII).
DISCUSSION
Linkage analysis revealed that a cDNA encoding the α-subunit of G protein cosegregated with d1 in 3,185 d1 segregants. Southern hybridization analysis using this cDNA as a probe showed different band patterns in several d1 mutant lines. In at least four independent d1 mutants, no gene transcript was observed by Northern hybridization analysis. Sequence analysis revealed that an 833-bp deletion had occurred in one of the mutant alleles (HO541). This result coincides well with the genomic Southern hybridization pattern using S5933 as a probe, in which an ∼800-bp difference in length was observed between Nipponbare (10.4 kilobase) and HO541 (9.6 kilobase) (Fig. 3A). Based on these results, we conclude that the rice D1 gene encodes the α-subunit of G protein. Transgenic plants restored the normal phenotype. This result finally confirmed the responsibility of the α-subunit of G protein for the d1 dwarf mutation in rice.
It is well known that G protein plays an important role in signal transduction in animals and microbes (27, 28). Heterotrimeric G protein has also been identified in several plant species (12, 13), and evidence of relatedness of G protein to signal transduction, such as in the K+ channel, has been shown (29).
Rice dwarf mutant d1 has dark green leaves, compact panicles, and short, round grains. Because these forms are all induced by d1 as pleiotropic effects of this gene, the heterotrimeric G protein in rice that D1 encodes is likely to play a key role in controlling rice growth and development. The abnormal phenotype was exhibited as a result of the lack of G protein as a signal associated with cellular growth, differentiation, and development in d1 mutants.
GAs are well known as regulators of plant growth and development, including seed germination, stem elongation, flowering, and fruit development (1–3), and regulation of α-amylase expression in the cereal aleurone layer (11). Recently, it has been shown that heterotrimeric G proteins are implicated in inducing the expression of α-amylase in oat aleurone by GA (32). This result is very significant. The rice mutant d1 was characterized as GA-insensitive, because while production of α-amylase by application of GA3 was saturated at 10−8 M GA3 in Nipponbare (normal cultivar) and Tan-ginbouzu dwarf (d18-gibberellin sensitive), no production of α-amylase in the d1 mutant was detectable at that concentration (10). Recently, putative G protein-coupled receptors were isolated from Arabidopsis (30), and one of these G protein-coupled receptors that influences sensitivity to cytokinins, one of the plant hormones, has been identified in Arabidopsis (31).
The G protein that the D1 gene encodes in rice might be associated with GA signal transduction. Many GA-associated mutants have been isolated and classified (3). GA biosynthetic pathways are well studied, and several genes coding enzymes catalyzing steps in GA biosynthesis have been isolated from GA-deficient mutant plants (33–37). For example, GA-sensitive dwarf mutant le in pea, first identified by Gregor Mendel, is reversed by GA1 and was shown to be deficient in GA-3-β-hydroxylase, which converts GA20 to GA1 in the GA biosynthetic pathway (38, 39). Even in this case, however, the molecular mechanism of signal transduction after triggering by GA is so far unknown. However, GA-insensitive and GA-constitutive mutants are useful for elucidating the mechanism of signal transduction of GAs; analysis of these mutants can help to clarify the molecular mechanisms. Several mutants have been defined as unresponsive to GA, for example, GAI in Arabidopsis, D8 and D9 in maize, Rht3 in wheat, or as constitutive GA-responsive mutants, such as spy and rga in Arabidopsis and sin in barley (3). GAI, spy, and rga were isolated in Arabidopsis (40–42). GAI works as a repressor of GA responses. spy contains a tetratricopeptide repeat region, which suppresses GA signal transduction and is epistatic to gai (41), and RGA is also a negative regulator of GA signalling (42). Regulators of GA signal transduction have been accumulating, but the mechanisms of signal components in GA-signal pathway triggered by GAs are still not clear. Our identification in rice of the G protein gene responsible for plant growth and development might help elucidate the signal-transduction pathway related to GA if the dwarfism is caused by a deficiency in this pathway. It is necessary to present genetical and biochemical evidence to prove direct participation of G protein to GA signaling in rice. We have several rice elongated mutants showing phenotype similar to that of Spy mutants in Arabidopsis. These rice mutants may associate with GA-signal transduction. Genetic analysis of epistasis with d1 and cloning of the spy-like gene are required. Also, further analysis of the structure of a plausible GA receptor and of the biochemical interactions between the receptor and G proteins is required to clarify the signal-transduction system mediated by the G protein in plants.
Acknowledgments
We thank Dr. H. Satoh for providing plant materials, RGP for providing rice PAC library, and Dr. T. Fuse for providing cosmid vector.
ABBREVIATIONS
- D1,Dwarf 1
GA, gibberellin
- G protein
GTP-binding protein
- YAC
yeast artificial chromosome
- EST
expressed sequence tag
- PAC
P1-derived artificial chromosome
- RFLP
restriction fragment-length polymorphism
- cM
centiMorgan
Footnotes
References
- 1.Reid J B. J Plant Growth Regul. 1993;12:207–226. [Google Scholar]
- 2.Hooley R. Plant Mol Biol. 1994;26:1529–1555. doi: 10.1007/BF00016489. [DOI] [PubMed] [Google Scholar]
- 3.Ross J J, Murfet I C, Reid J B. Physiol Plant. 1997;100:550–560. [Google Scholar]
- 4.Li J, Chory J. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 5.Azpiroz R, Wu Y, LoCascio J C, Feldmann K A. Plant Cell. 1998;10:219–230. doi: 10.1105/tpc.10.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reiter W D, Chapple C C S, Somerville C R. Science. 1993;261:1032–1035. doi: 10.1126/science.261.5124.1032. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi T, Gasch A, Nishizawa N, Chua N-H. Genes Dev. 1995;9:97–107. doi: 10.1101/gad.9.1.97. [DOI] [PubMed] [Google Scholar]
- 8.Murakami Y. In: Science of Rice Plant. Matsuo T, Kumazawa K, Ishihara R, Hirata H, editors. Vol. 2. Tokyo: Food and Agriculture Policy Research Center; 1995. pp. 182–189. [Google Scholar]
- 9.Futsuhara Y, Kikuchi F. In: Science of Rice Plant. Matsuo T, Kumazawa K, Ishihara R, Hirata H, editors. Vol. 3. Tokyo: Food and Agriculture Policy Research Center; 1995. pp. 300–308. [Google Scholar]
- 10.Mitsunaga S, Tashiro T, Yamaguti J. Theor Appl Genet. 1994;87:705–712. doi: 10.1007/BF00222896. [DOI] [PubMed] [Google Scholar]
- 11.Nolan R C, Ho T-H D. Planta. 1988;174:551–560. doi: 10.1007/BF00634486. [DOI] [PubMed] [Google Scholar]
- 12.Terryn N, Montagu M V, Inze D. Plant Mol Biol. 1993;22:143–152. doi: 10.1007/BF00039002. [DOI] [PubMed] [Google Scholar]
- 13.Ma H. Plant Mol Biol. 1994;26:1611–1636. doi: 10.1007/BF00016493. [DOI] [PubMed] [Google Scholar]
- 14.Konishi N, Satoh . Catalogue of Rice Germplasm in Kyushu University. 1994. [Google Scholar]
- 15.Churchill G A, Giovannoni J J, Tanksley S D. Proc Natl Acad Sci USA. 1993;90:16–20. doi: 10.1073/pnas.90.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray M G, Thompson W F. Nucleic Acid Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Southern E M. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 18.Umehara Y, Inagaki A, Tanoue H, Yasukouchi Y, Nagamura Y, Saji S, Otsuki Y, Fujimura T, Kurata N, Minobe Y. Mol Breed. 1995;1:79–89. [Google Scholar]
- 19.Jones M H, Khwaja O S A, Briggs H, Lambson B, Davey P M, Chalmers J, Zhou C-Y, Walker E M, Zhang Y, Todd C, et al. Genomics. 1994;24:266–275. doi: 10.1006/geno.1994.1615. [DOI] [PubMed] [Google Scholar]
- 20.Isegawa Y, Sheng J, Sokawa Y, Yamanishi K, Nakagomi O, Ueda S. Mol Cell Probes. 1992;6:467–475. doi: 10.1016/0890-8508(92)90043-w. [DOI] [PubMed] [Google Scholar]
- 21.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 22.Seo H-S, Kim H-Y, Jeong J-Y, Lee S-Y, Cho M-J, Bahk J-D. Plant Mol Biol. 1995;27:1119–1131. doi: 10.1007/BF00020885. [DOI] [PubMed] [Google Scholar]
- 23.Toki S. Plant Mol Biol Rep. 1997;15:16–21. [Google Scholar]
- 24.Ashikari M, Ideta O, Yoshimura A, Iwata N. Rice Genet Newslett. 1996;13:76–78. [Google Scholar]
- 25.Harushima Y, Yano M, Shomura A, Sato M, Shimano T, Kuboki Y, Yamamoto T, Lin S Y, Antonio B A, Parco A, et al. Genetics. 1998;148:479–494. doi: 10.1093/genetics/148.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishikawa A, Tsubouchi H, Iwasaki Y, Asahi T. Plant Cell Physiol. 1995;36:353–359. doi: 10.1093/oxfordjournals.pcp.a078767. [DOI] [PubMed] [Google Scholar]
- 27.Neer E J. Cell. 1995;80:249–257. doi: 10.1016/0092-8674(95)90407-7. [DOI] [PubMed] [Google Scholar]
- 28.Hamm H E. J Biol Chem. 1998;273:669–672. doi: 10.1074/jbc.273.2.669. [DOI] [PubMed] [Google Scholar]
- 29.Assman S M. Trends Plant Sci. 1996;1:73–74. [Google Scholar]
- 30.Josefsson L G, Rask L. Eur J Biochem. 1997;249:415–420. doi: 10.1111/j.1432-1033.1997.t01-1-00415.x. [DOI] [PubMed] [Google Scholar]
- 31.Plakidou-Dymock S, Dymock D, Hooley R. Curr Biol. 1998;8:315–324. doi: 10.1016/s0960-9822(98)70131-9. [DOI] [PubMed] [Google Scholar]
- 32.Jones H D, Smith S J, Desikan R, Plakidou-Dymock S, Lovegove A, Hooley R. Plant Cell. 1998;10:245–253. doi: 10.1105/tpc.10.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun T-P, Goodman H M, Ausubel F M. Plant Cell. 1992;4:119–128. doi: 10.1105/tpc.4.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun T-P, Kamiya Y. Plant Cell. 1994;6:1509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Y-L, Li L, Wu K, Peeters A J M, Gage D A, Zeevaart J A D. Proc Natl Acad Sci USA. 1995;92:6640–6644. doi: 10.1073/pnas.92.14.6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiang H-H, Hwang I, Goodman H M. Plant Cell. 1995;7:195–201. doi: 10.1105/tpc.7.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winkler R G, Helentjaris T. Plant Cell. 1995;7:1307–1317. doi: 10.1105/tpc.7.8.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lester D R, Ross J J, Davies P J, Reid J B. Plant Cell. 1997;9:1435–1443. doi: 10.1105/tpc.9.8.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin D N, Prpebsting W M, Hedden P. Proc Natl Acad Sci USA. 1997;94:8907–891. doi: 10.1073/pnas.94.16.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng J, Carol P, Richards D E, King K E, Cowling R J, Murphy G P, Harberd N P. Genes Dev. 1997;11:3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jacobsen S E, Binkowski K A, Olszewski N E. Proc Natl Acad Sci USA. 1996;93:9292–9296. doi: 10.1073/pnas.93.17.9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silverstone A L, Ciampaglio C N, Sun T-P. Plant Cell. 1998;10:155–169. doi: 10.1105/tpc.10.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]