Abstract
Fifteen to twenty-five percent of sporadic colorectal carcinomas are replication error (RER) positive. Because the frequency of mutations in the mismatch repair genes (hMLH1 and hMSH2) is low in these tumors, we have investigated the role of mutational inactivation, methylation of the promoter region, and loss of heterozygosity (LOH) as a possible explanation for the mutator phenotype of RER+ colorectal cancer cell lines. Genomic DNA was extracted from a panel of 49 human colorectal cancer cell lines. The RER status was determined by amplification of BAT-26. All exons of hMLH1 and hMSH2 were amplified with the PCR and screened by using single-strand conformational polymorphism and direct sequencing. The methylation status was ascertained by methylation-specific PCR after bisulfite modification of DNA. Western blotting for hMLH1 was performed on methylated cell lines before and after the addition of the demethylating agent 5-azacytidine. LOH was sought by genescan analysis of amplified CA repeat markers and indirectly by determining the number of homozygotes in the cell lines and human random controls. Twelve cell lines from ten tumors (24%) were RER+. Hypermethylation of the hMLH1 promoter occurred in five of ten (50%) RER+ tumors, whereas three of thirty-two (6%) RER tumors showed partial methylation. None of the fully methylated cell lines expressed hMLH1, although all reexpressed hMLH1 after treatment with 5-azacytidine. There was no LOH in the RER+ tumors in either hMLH1 or hMSH2. Our results suggest that mutations of hMLH1 together with hypermethylation of the promoter region, but not LOH, are the cause of the mutator phenotype in the majority (70%) of RER+ tumors.
Hereditary nonpolyposis colorectal cancer (HNPCC) is caused by germline mutations in the DNA mismatch repair (MMR) genes (1). To date, inactivating mutations have been described in five MMR genes: hMSH2, hMLH1, hPMS1, hPMS2, and GTBP (hMSH6) (2–7). These cancers, from affected HNPCC kindreds, exhibit genomic instability that can be detected as changes in the length of microsatellite sequences (8). This microsatellite instability or replication error (RER) is seen in almost all colorectal cancers (CRC) removed from patients with HNPCC (8) and in approximately 15–20% of sporadic CRC from patients with no obvious family history (9). Approximately 70% of HNPCC patients with RER+ tumors are found to have germline mutations in one of the MMR genes (10). The majority of these mutations (95%) have been described in hMLH1 and hMSH2 (11), with mutations in other MMR genes being rare (11). Although some of the RER+-sporadic CRC have been shown to have mutations in one of the MMR genes (12), the majority of these cancers have no identifiable mutation (13, 14). This has also been the case in RER+ sporadic endometrial (15) and gastric cancers (16). Presumably, other nonmutational mechanisms or novel genes must be responsible for the microsatellite instability seen in these RER+-sporadic CRC. Nonmutational mechanisms that inactivate genes include epigenetic processes such as promoter region hypermethylation, which may result in transcriptional loss. This methylation has previously been described in a number of tumor-suppressor genes (17), and recently it has been shown that hypermethylation of the promoter region of the hMLH1 gene may cause a lack of expression of its protein and therefore may account for microsatellite instability in RER+sporadic CRC (10, 18–20). It has also been demonstrated that loss of hMLH1 protein expression in RER+ gastric cancers is associated with promoter methylation in 90% of cases (16). Expression of hMLH1 protein may be restored in methylated RER+ cell lines after treatment with 5-azacytidine (19, 21). As in other tumor-suppressor genes, loss of heterozygosity (LOH) may occur at MMR loci, thereby inactivating one allele of the MMR genes. Previous workers have shown LOH in RER+-sporadic CRC at the hMLH1 locus, but not at the hMSH2 locus (22, 23).
We have studied several possible mechanisms of inactivation of MMR genes in RER+-sporadic CRC, so as to ascertain the whole spectrum of events causing the mutator phenotype in RER+ tumors. Having examined a large panel of human CRC cell lines for the incidence of RERs, we then searched thoroughly for the possible mechanisms of inactivation of MMR genes and subsequent microsatellite instability in those RER+ cell lines. This study has included mutational screening, analysis of the methylation status of the promoter region, and LOH studies of the hMLH1 and hMSH2 genes in all 49 cell lines. The functional consequences of promoter methylation and RER+ cell lines were then further investigated.
MATERIALS AND METHODS
Cell Lines, Tissue Culture, and DNA Extraction.
The 49 human colorectal carcinoma cell lines (derived from 42 patients) used in this study are listed in Table 1. Cells were maintained in 75-cm2 sterile Falcon tissue culture flasks (Beckton Dickinson) in DMEM supplemented with 10% FCS at 37°C in 10% CO2. Genomic DNA was extracted by using standard techniques.
Table 1.
The RER status of 49 human colorectal cancer cell lines derived from 42 patients
| RER+ | RER− | ||
|---|---|---|---|
| VACO 5 | HRA19 | SCKO-1 | CACO2 |
| SW48 | *VACO4S/A | PCJW | C99 |
| LS411 | VACO10MS | LS1034 | C84 |
| LS174T | T84 | LIM1863 | C80 |
| LOVO | SW948 | *HT29/WIDR | C75 |
| HCT116 | SW837 | HCA46 | C70 |
| HCA7 | *SW620/480 | COLO320DM | C32 |
| *DLD1/HCT15 | SW403 | *COLO201/205/206 | C10 |
| *GP2d/5d | SW1417 | CC20 | C106 |
| LS180 | SW1222 | CC07 | SW1116 |
| COLO741 | HT55 | ||
Lines beginning with the prefix C were established at the Cancer and Immunogenetics laboratory in Oxford, U.K.
Cell lines from the same patient.
PCR Amplification.
By using previously described intron-complementary primers, all 19 exons of the hMLH1 gene and 16 exons of the hMSH2 gene (including the intron–exon boundaries) were amplified specifically from genomic DNA (24). However, primers for exon 12 of hMLH1 were redesigned (5′-TAC CTC ATA CTA GCT TCT TTC TTA GT-3′ sense, 5′-CTG TAC TTT TCC CAA AAG GCC AT-3′ antisense) because of poor amplification with published primers. All reactions contained approximately 300 ng of template DNA in a total volume of 50 μl with final reaction concentrations of 1× standard PCR buffer (Promega)/200 mM dNTPs/1.5 mM Mg2+/0.2 mM of each primer/1 unit of Taq polymerase. Amplification was performed by using a protocol of 94°C for 4 min; 35 cycles of 94°C for 1 min, 50°C for 1 min, 72°C for 1 min; and finally 72°C for 10 min.
Single-Strand Conformational Polymorphism (SSCP) Analysis.
SSCP was performed as previously described (24). Electrophoresis was performed in 10% nondenaturing polyacrylamide gels and the PCR products visualized with silver staining by using standard methods. To confirm whether any observed SSCP band shifts in the cell lines were polymorphisms, we screened 89 samples of genomic DNA prepared from the peripheral blood of United Kingdom human random controls in the regions of interest (collected originally for an HLA allele frequency study and supplied by J. Bodmer, Oxford, U.K.).
Direct Sequencing of the hMLH1 and hMSH2 Genes.
The nucleotide sequences of the PCR products showing an abnormal electrophoretic mobility on SSCP analysis were determined by direct sequencing of purified PCR product in a thermocycle sequencing reaction with the dRhodamine Sequencing kit on a 377 prism sequencer (Applied Biosystems). The sequences obtained from our experiments were performed in duplicate and alongside samples with known wild-type genotypes and then compared with the published sequences of the hMLH1 and hMSH2 exons (GenBank accession nos. U17857-U17839 and U41221-U41206) by using sequencher 3.0 software.
Determination of the RER Status of the Cell Lines.
To determine the RER status of the cell lines, we amplified BAT-26, a single Poly(A) tract, previously shown to be highly sensitive and specific for microsatellite instability (25), by using fluorescently labeled primers and similar PCR conditions to those previously described. PCR products were loaded on a 377 prism sequencer (Applied Biosystems). Results were analyzed by using genescan software (version 2.0.2). All PCRs and analyses were repeated at least in duplicate. Any cell lines presenting ambiguous results were further investigated by using BAT-25, and all RER+ designations were confirmed by using the CA repeat marker D15S58.
Evaluation of Methylation Status of the hMLH1 and hMSH2 Promoter Regions.
The methylation status of the hMLH1 and hMSH2 promoter regions was evaluated by performing methylation-specific PCR (MSP) of genomic DNA that had been purified after chemical treatment with sodium bisulfite as described (26). Primer sequences for “unmethylated” and “methylated” MSP of hMLH1 and hMSH2 promoter regions and PCR conditions have been described (21, 26). The PCR products were stained with ethidium bromide and visualized under UV illumination after 3% agarose gel electrophoresis. Those cell lines with a positive result for methylation of the hMLH1 region were confirmed by using the PCR-based HpaII restriction enzyme assay as detailed previously (10). Unmethylated and methylated positive controls were included in all reactions.
Western Blotting and Demethylation with 5-Azacytidine.
Approximately 106 cells were lysed in RIPA buffer (150 mM NaCl/1% Nonidet P-40/0.5% deoxycholate/0.1% SDS/50 mM Tris, pH 7.5) and the lysate electrophoresed on a 20% SDS polyacrylamide gel. The proteins were transferred to Hybond, ECL nitrocellulose membrane (Amersham Pharmacia) and probed with 2 μg/ml anti-human MLH1 mab (PharMingen) and anti-human actin mab (Sigma) as an internal control. After incubation with horseradish peroxidase-coupled secondary antibody (Dako), immune complexes were visualized with enhanced chemiluminescence (Amersham Pharmacia). Cell lines with evidence of MMR gene promoter methylation were then cultured in the presence of 2 μg/ml 5-azacytidine (added to tissue culture flasks on day 2 and day 5/6 after passage, with a change of medium at 24 hr). After 1 week, a cell lysate was prepared for Western analysis and the cell lines kept in culture. Further lysate was prepared after 2 wk and 4 wk after the addition of 5-azacytidine.
LOH Studies for hMLH1 and hMSH2.
Allele loss could be studied only indirectly because there was an absence of constitutional DNA. Five microsatellite markers were chosen for both hMLH1 (D3S1448, D3S1100, D3S3564, D3S3605, and D3S3593) and hMSH2 (D2S119, D2S2294, D2S2306, D2S2259, and D2S2291). Because approximately 20% of individuals are homozygous at any one microsatellite, it is highly unlikely (P = 0.0003) that an individual will be homozygous in the germline for all five markers. PCR reactions were carried out in a total volume of 50 μl with final reaction concentrations of 1× standard PCR buffer (Promega)/dNTPs (200 mM)/1.5 mM Mg2+/0.2 mM of each primer/1 unit of Taq polymerase. The amplification protocol used an annealing temperature of 50–55°C (see PCR Amplification). Three microliters of diluted (1:10) PCR product was combined with 2 μl of formamide-loading buffer and 0.5 μl of Rox size standard. This mixture was denatured at 96°C for 5 min and quenched on ice before gel loading (4.25% denaturing acrylamide) on a 377 prism sequencer (Applied Biosystems). Results were analyzed by using genescan software (version 2.0.2). The likely incidence of LOH at hMLH1 and hMSH2 was further determined by choosing a common polymorphism within both genes and then sequencing the region of interest for the 49 cell lines and 89 human random controls. This method is particularly important in RER+ tumors because the data from CA repeat markers may be difficult to interpret.
RESULTS
RER Status of the Cell Lines.
The RER status of the cell lines is as previously described (Table 1) (27). There were 12 RER+ cell lines from 10 tumors, which make up 24% of our total collection.
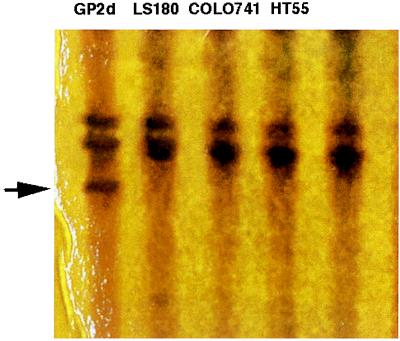
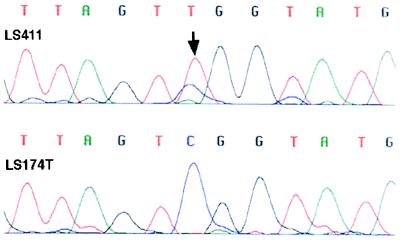
SSCP and Sequence Analysis. We detected four missense and one nonsense mutations in six (50%) RER+ cell lines from five tumors in hMLH1 (Table 2). There were no mutations detected in hMSH2. In LS180 (exon 4 at codon 117, ACG → ATG; Thr → Met), LS411 (exon 8 at codon 226, CGG → TGG; Arg → Trp), HCT116 (exon 9 at codon 252, TCA → TAA; Ser → STOP), and VACO5 (exon 12 at codon 409, CAG → CAT; Gln → His), mutations were caused by a single bp substitution. In GP2d and GP5d (exon 16 at codon 618, AAG → GCG; Lys → Ala), which display similar SSCP band shifts (Fig. 1) and come from the same tumor, this missense mutation was caused by a 2 bp substitution. In all missense cases, the presence of the wild-type sequence indicates that these cell lines are heterozygous (Fig. 2) for their respective mutations. Eighty-nine human random controls were examined for similar bandshifts in the relevant exons. One identical change was detected in exon 16, which was confirmed by sequencing.
Table 2.
Inactivating mutations in RER+ cell lines, and methylation of the hMLH1 promoter region
| Cell line | Mutation
|
hMLH1 Methylation | |||
|---|---|---|---|---|---|
| Gene | Codon | Nucleotide change | Consequence | ||
| VACO5 | hMLH1 | 409 | CAG → CAT | Gln → His | Yes |
| SW48 | Yes | ||||
| LS411 | hMLH1 | 226 | CGG → TGG | Arg → Trp | Yes |
| LS174T | No | ||||
| LOVO | * | No | |||
| HCT116 | hMLH1 | 252 | TCA → TAA | Ser → STOP | No |
| HCA7 | Yes | ||||
| DLD1/HCT15 | † | No | |||
| GP2d/5d | hMLH1 | 618 | AAG → GCG | Lys → Ala | No |
| LS180 | hMLH1 | 117 | ACG → ATG | Thr → Met | Yes |
There was no methylation of the hMSH2 promoter region in any of the cell lines. In only one cell line (LS174T) have we failed to explain the cause of microsatellite instability [LOVO (12) and DLD1/HCT15 (7) have mutations in hMSH2 and GTBP, respectively].
Homozygous deletion in exons 4–8, hMSH2.
Homozygous single-bp deletion in GTBP.
Figure 1.
A silver-stained SSCP polyacrylamide gel of exon 16 in hMLH1. The heterozygous band shift at GP2d is arrowed. Sequencing confirmed this to be a 2-bp substitution at codon 618 (AAG → GCG; Lys → Ala), previously described in HNPCC kindred. This sequence change was also found in one of the 89 United Kingdom human random controls, giving further weight to the association between subpolymorphic missense variants and CRC.
Figure 2.
Sequence chromatogram of exon 8 in hMLH1. The heterozygous single-bp substitution at codon 226 (CGG → TGG; Arg → Trp) in LS411 is demonstrated (arrow). LS411 was also shown to be methylated in the hMLH1 promoter region and did not express hMLH1 protein on Western blotting.
We detected various polymorphisms in a number of our cell lines (Table 3). In hMLH1, these genetic variants included a missense change in exon 8 at codon 219 (ATC → GTC; Ile → Val) and an intron 9 change (a → g). In hMSH2, these included intronic changes at intron 9 (a → t) and intron 10 (a → g) and a silent change in exon 10 at codon 526 (ACC → ACA; Thr → Thr). The polymorphic variants at codon 219 of hMLH1 and introns 9 and 10 of hMSH2 have been described by other groups (28, 29).
Table 3.
Polymorphisms in hMLH1 and hMSH2
| Polymorphism | Codon | Cell line |
|---|---|---|
| hMLH1 | ||
| ATC → GTC; Ile → Val | 219 | 18/42 tumors |
| a → g* | Intron 9 + 10 | LIM 1863 |
| hMSH2 | ||
| a → t | Intron 9-9 | 8/42 tumors |
| ACC → ACA*; Thr → Thr | 526 | DLD1, HCT15 |
| a → g | Intron 10 + 11 | 29/42 tumors |
Indirect evidence of LOH was sought by sequencing for two of the common polymorphisms of interest in all the 49 cell lines and 89 United Kingdom human random controls: in exon 8 of hMLH1 at codon 219 (ATC → GTC; Ile → Val) and intron 10 of hMSH2 (a → g).
Newly described sequence changes.
Methylation Status of hMLH1 and hMSH2 Promoter Regions.
In hMLH1, five RER+ cell lines from five tumors (50%) were fully methylated in the promoter region (Table 2) (Fig. 3). In three of these cell lines, we had also detected a heterozygous missense mutation in hMLH1 (LS180, LS411, and VACO5). There were no fully methylated RER− cell lines, although three cell lines (HT29, WIDR, and SW403) from two RER− tumors (6%) were found to be partially methylated, i.e., a PCR product was seen with both “unmethylated” and “methylated” primers. Fisher’s exact value was calculated for RER+ vs. RER− cell lines, methylated or unmethylated (P = 0.015), and for RER+ vs. RER− cell lines, fully methylated or unmethylated (P = 0.0004). We did not detect methylation in the promoter region of hMSH2 in any of the cell lines.
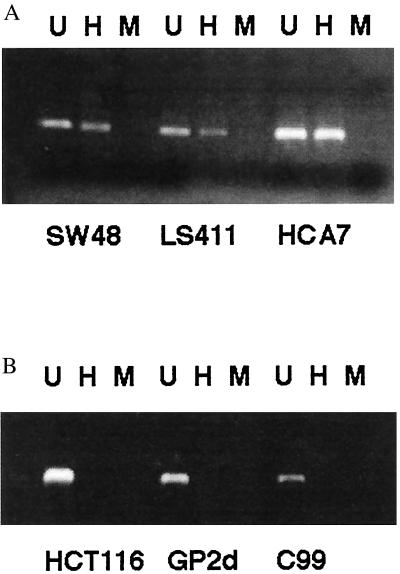
Figure 3.
PCR-based HpaII restriction enzyme assay demonstrating (a) a methylated hMLH1 promoter region in three RER+ cell lines (SW48, LS411, and HCA7), and (b) an unmethylated hMLH1 promoter region in two RER+ (HCT116 and GP2d) and one RER− cell lines (C99). (U, undigested; H, incubated with HpaII; M, incubated with MspI).
Western Blotting and Demethylation with 5-Azacytidine.
Cell lines with methylation (full or partial) of the promoter region of hMLH1 (HCA7, SW48, LS411, VACO5, SW403, and HT29) were analyzed, together with RER+ and RER− unmethylated controls (HCT116, GP2d, C70, and SW480). None of the fully “methylated” cell lines and HCT116 (homozygous hMLH1 nonsense mutation, but not methylated) expressed hMLH1 protein. The partially methylated cell lines SW403 and HT29, the RER+ cell line GP2d (heterozygous hMLH1 missense mutation, but not methylated), and the RER− controls all expressed hMLH1 protein. After treatment with 5-azacytidine, all fully “methylated” cell lines demonstrated reexpression of hMLH1 protein (Fig. 4). At 4 wk after the original treatment with 5-azacytidine, none of these fully methylated cell lines continued to express hMLH1 protein. The RER+ double mutant control, HCT116, did not reexpress hMLH1 protein after treatment with 5-azacytidine.
Figure 4.
Western blot of cell lines: 1 wk after treatment with 5-azacytidine (VACO5, HCA7, and HCT116) and 2 wk after treatment with 5-azacytidine (HCA7, LS411 and SW48) and positive controls (C70, GP2d). Reexpression of hMLH1 can been seen 1 wk after treatment with 5-azacytidine in the methylated VACO5 and HCA7, but not in the unmethylated RER+ control, HCT116. Two weeks after treatment, expression of hMLH1 is virtually absent (faint band in HCA7, LS411, and SW48), suggesting that this epigenetic mechanism of gene silencing is driven by a dynamic process.
LOH at hMLH1 and hMSH2.
genescan analysis of the PCR products of CA repeat markers close to the hMLH1 locus suggested that six cell lines (HRA19, CC07, VACO4S/4A, C32, SW1116) from five tumors (12%) had LOH at all five markers. genescan analysis of five markers close to the hMSH2 locus suggested that three cell lines (T84, LIM1863, C32) had LOH (7%). All cell lines with LOH at hMLH1 and hMSH2 were RER−. Indirect evidence of LOH at these two sites was sought by sequencing for two common polymorphisms. Two single bp substitutions found on SSCP and direct sequencing were chosen, in exon 8 of hMLH1 at codon 219 (ATC → GTC; Ile → Val) and intron 10 of hMSH2 (a → g). In hMLH1, Fisher’s exact value was calculated for RER+ cell line homozygotes vs. human random control heterozygotes (P = 0.5) and for RER− cell line homozygotes vs. human random control heterozygotes (P = 0.035). In hMSH2, Fisher’s exact value (P > 0.5) was not significant for RER+ or RER− cell lines.
DISCUSSION
In this study, we screened 49 human CRC cell lines derived from 42 tumors for mutations in hMLH1 and hMSH2. Twelve cell lines from ten tumors (24%) were RER+, which is slightly more than expected and may reflect a relative advantage that RER+ sporadic CRC have in their ability to be established as cell lines from fresh surgical specimens. We found five mutations in hMLH1 in RER+ cell lines. There was only one homozygous mutation, the previously described nonsense mutation at codon 252 (5), and four heterozygous missense mutations, including a mutation at codon 409 in the well studied VACO5 cell line. Two of the other three missense mutations (codon 117 and 618) have been described in HNPCC kindreds (30, 48). DNA from a lymphoblastoid cell line was available from LS180 (missense mutation at codon 117), but this did not contain the sequence change, implying that this was a somatically derived mutation. The sequence change at codon 226 has also been described in an HNPCC kindred (31), but was reported as a nonsense mutation (CGG → TGG vs. CGA→TGA). Thirty-one percent of hMLH1 germline mutations are reported as missense mutations (32), which is less than our four of five somatic mutations being missense (80%). This is also in contrast to hMSH2 germline mutations, which are nonsense, frameshift, and splice site mutations in over 90% of cases. The pathogenic effects of a missense mutation may be difficult to interpret, but functional analysis of such mutations in Saccharomyces cerevisiae has shown the codon 117 and 618 changes (and many others) to alter normal hMLH1 function (the codon 226 change has not been studied) (33). It has been shown that hMLH1 is the most frequently altered protein (95%) in sporadic RER+ CRC (34), and it may be that the role of hMSH2 is more significant in HNPCC patients where mutations occur in roughly equal proportions in the two genes (30, 35). This difference suggests that the selective advantage of nonsense or frameshift mutations in hMSH2 is not enough at the somatic level to influence substantially the probability of successful tumor outgrowth. The pathogenic missense change at codon 618 was also found in one of the 89 human random controls. This gives further weight to the association between subpolymorphic missense variants, in this case in hMLH1, and the risk of CRC (36).
Five RER+ cell lines (50%) were fully methylated in the hMLH1 promoter region, although others have reported up to 84% of sporadic RER+ tumors showing promoter methylation (18). We did not detect promoter methylation in hMSH2, and this is in keeping with other workers (18, 20). Three of our five methylated cell lines also contained heterozygous missense mutations in hMLH1, which presumably confer a selective advantage to subsequent methylation of both wild-type and mutant sequences. Jones and Laird (37) have recently pointed out that Knudson’s two-hit hypothesis should extend to include epigenetic mechanisms of gene inactivation, such as methylation. In the case of methylation, it is not clear which event occurs first. Presumably, a missense change precedes methylation or one allele is methylated before the other, otherwise there is no basis for selection for the missense mutation. None of the methylated cell lines expressed hMLH1 protein, although this was restored transiently in all of these cell lines after treatment with 5-azacytidine. It is interesting that this effect was reversible in all of these cell lines at 4 wk, so that this epigenetic mechanism of gene silencing is driven by a dynamic process, presumably by the continuing action of the methyl transferase which is inhibited by 5-azacytidine. The underlying event that leads to methylation must therefore be a change, for example in some aspect of chromatin structure possibly connected with histone acetylation (38), which enables the methyl transferase to have access to the DNA. Previous work on an endometrial carcinoma cell line has shown that it is unlikely that the cell lines are overgrown by a subpopulation of cells resistant to 5-azacytidine (19). It has also been shown that cells reexpressing hMLH1 protein are able to perform strand-specific MMR of bp and insertion/deletion mismatches (21). It may be possible to suppress the RER+ mutator phenotype by reversing promoter methylation or the conditions that enable methylation, and this is particularly important as promoter methylation of hMLH1 has recently been associated with drug resistance in ovarian cancer (39). Others have described how tumor initiation may be suppressed in Min mice by the inhibition of DNA methylation (40) and antisense oligodeoxynucleotides that may inhibit methyltransferase and decrease carcinogenesis (41). However, these treatments do not address the underlying mechanism, such as chromatin confirmation, which enables access to methyl transferase and subsequent promoter hypermethylation (38). Ruschoff et al. (42) have shown that aspirin, although not a demethylating agent, may induce a genetic selection for microsatellite stability in a subset of MMR-deficient cells.
The mechanism enabling methylation is not understood at present, although mammalian methyltransferases have recently been described (43). Others have shown that sporadic RER+ CRC may be a subgroup of tumors that have increased promoter region methylation in a number of genes, including p16 and IGFII (44). Thus, the inactivation of hMLH1 by promoter methylation may, in turn, enhance the rate of further genetic events in sporadic RER+ cancers (37). Three RER- cell lines (SW403, WIDR, and HT29) were partially methylated, but expressed hMLH1 protein. WIDR and HT29 are derived from the same original tumor, and both cell lines are partially methylated after many passages. This concordance is strong indirect evidence that the mechanisms enabling methylation are very stable. It is unclear whether partial methylation is caused by methylation of one allele, incomplete methylation of both alleles, or some other phenomenon. However, it has been demonstrated that when hMLH1 is inactivated by an epigenetic mechanism, such as methylation, this is a biallelic event and is not accompanied by LOH (19). Nevertheless, it is not clear whether the changes on each chromosome that enable methylation are independent or correlated events.
We have not demonstrated LOH associated with RER+ tumors in either hMLH1 or hMSH2. This is in contrast to previous reports of LOH at hMLH1 (22, 23) but in agreement for hMSH2. The lack of LOH at hMLH1 seen in our study may be peculiar to cell lines. We also previously failed to detect LOH in a smaller number of cell lines (22). These data were confirmed with both genescan analysis of CA repeat markers and comparing sequence data for cell lines that were homozygote for common polymorphisms. The LOH that was seen in RER-tumors for hMLH1 may reflect LOH at other tumor-suppressor gene loci on chromosome 3, such as β-catenin. In tumors from HNPCC patients with germline mutations, promoter methylation in hMLH1 was seen in only 22% of cases (21). It may be that LOH plays a more important role as the “second hit” in those HNPCC patients who inherit a MMR gene mutation, whereas methylation is responsible for knocking out the hMLH1 gene in sporadic RER+ CRC. This would certainly explain our finding of methylation in 50% of the RER+ cell lines together with no apparent LOH and further supports our argument that tumors from HNPCC patients, which almost always acquire a raised mutation rate, mostly follow a different pathway from sporadic RER+ tumors (45, 46). In the latter, we believe that a single mutation initially will be selected for and that the subsequent event leading to the RER+ phenotype does not occur particularly early in the adenoma-to-carcinoma sequence. At this late stage, the mutation rate may be less limiting because of increased population size and possibly other constraints on tumor growth. This prediction is supported by experimental evidence (47).
The lack of LOH and methylation at hMSH2 is not altogether surprising, because alterations in the hMLH1 gene have been shown to be responsible for microsatellite instability in the majority of sporadic RER+ cancers (34). The situation for hMLH1 may be analogous to APC, where germline mutations occur much more often outside the mutation cluster region than do the somatic changes in sporadic cancers. This is presumably because the selective advantage needed to promote tumor outgrowth may be much less for germline mutations, which occur in all cells, than for somatic mutations, which necessarily occur in only a small proportion of cells. Dosage effects, presumably because of hMLH1 methylation, may give a sufficient advantage for tumor outgrowth, whereas for hMSH2 they do not. If gene dosage to half function alone does not give a sufficient selective advantage, a dominant-negative effect is needed to bring the level significantly below half. The codon 618 heterozygous missense change (with no methylation) may result in a dominant-negative effect beyond simply halving the level of activity.
In conclusion, inactivating mutations and/or hypermethylation of hMLH1 were found in 7 of 10 (70%) of RER+ tumors. In only one cell line (LS174T) have we failed to explain the cause of microsatellite instability [LOVO (12) and DLD1/HCT15 (7) have mutations in hMSH2 and GTBP, respectively]. Demethylation of all methylated RER+ cell lines resulted in reexpression of hMLH1 protein. Our results suggest that mutations of hMLH1 together with hypermethylation of the promoter region, but not LOH, are the underlying cause of the mutator phenotype in the vast majority of sporadic RER+ CRC cell lines.
ABBREVIATIONS
- HNPCC
hereditary nonpolyposis colorectal cancer
- MMR
mismatch repair
- RER
replication error
- CRC
colorectal cancer
- LOH
loss of heterozygosity
- SSCP
single-strand conformational polymorphism
References
- 1.Peltomaki P. J Pathol. 1995;176:329–330. doi: 10.1002/path.1711760402. [DOI] [PubMed] [Google Scholar]
- 2.Fishel R, Lescoe M K, Rao M R, Copeland N G, Jenkins N A, Garber J, Kane M, Kolodner R. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 3.Leach F S, Nicolaides N C, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen L A, Nystrom-Lathi M, et al. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 4.Bronner C E, Baker S M, Morrison P T, Warren G, Smith L G, Lescoe M K, Kane M, Earabino C, Lipford J, Lindblom A, et al. Nature (London) 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 5.Papadopoulos N, Nicolaides N C, Wei Y F, Ruben S M, Carter K C, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, Adams M D, et al. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 6.Nicolaides N C, Papadopoulos N, Liu B, Wei Y F, Carter K C, Ruben S M, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, et al. Nature (London) 1995;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 7.Papadopoulos N, Nicolaides N C, Liu B, Parsons P, Lengauer C, Palombo F, D’Arrigo A, Markowitz S, Willson J K V, Kinzler K W, et al. Science. 1995;268:1915–1917. doi: 10.1126/science.7604266. [DOI] [PubMed] [Google Scholar]
- 8.Aaltonen L A, Peltomaki P, Leach F, Sistonen P, Pylkkanen S M, Mecklin J P, Jarvinen H, Powell S, Jen J, Hamilton S R, et al. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 9.Thibodeau S N, Bren G, Scaid D. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 10.Kane M, Loda M, Gaida G, Lipman J, Mishra R, Goldman H, Jessup J M, Kolodner R. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 11.Papadopoulos N, Lindblom A. Hum Mutat. 1997;10:89–99. doi: 10.1002/(SICI)1098-1004(1997)10:2<89::AID-HUMU1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 12.Liu B, Nicolaides N C, Markowitz S, Willson J K V, Parsons R E, Jen J, Papadopoulos N, Peltomaki P, de la Chapelle A, Hamilton S R, et al. Nat Genet. 1995;9:48–55. doi: 10.1038/ng0195-48. [DOI] [PubMed] [Google Scholar]
- 13.Thibodeau S N, French A J, Roche P C, Cunningham J M, Tester D J, Lindor N M, Moslein G, Baker S M, Liskay R M, Burgart L J, et al. Cancer Res. 1996;56:4836–4840. [PubMed] [Google Scholar]
- 14.Borresen A L, Lothe R A, Meling G I, Lystad S, Morrison P, Lipford J, Kane M F, Rognum T O, Kolodner R D. Hum Mol Genet. 1995;4:2065–2072. doi: 10.1093/hmg/4.11.2065. [DOI] [PubMed] [Google Scholar]
- 15.Katabuchi H, van Rees B, Lambers A R, Ronnett B M, Blazes M S, Leach F S, Cho K R, Hedrick L. Cancer Res. 1995;55:5556–5560. [PubMed] [Google Scholar]
- 16.Leung S Y, Yeun S T, Chung L P, Chu K M, Chan A S Y, Ho J C I. Cancer Res. 1999;59:159–164. [PubMed] [Google Scholar]
- 17.Herman J G, Graff J R, Myohanen S, Nelkin B D, Baylin S B. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herman J G, Umar A, Polyak K, Graff J R, Ahuja N, Issa J J, Markowitz S, Willson J K V, Hamilton S R, Kinzler K W, et al. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veigl M L, Kasturi L, Olechnowicz J, Ma A, Lutterbaugh J D, Periyasamy S, Li G, Drummond J, Modrich P L, Sedwick W D, et al. Proc Natl Acad Sci USA. 1998;95:8698–8702. doi: 10.1073/pnas.95.15.8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cunningham J M, Christensen E R, Tester D J, Kim C, Roche P C, Burgart L J, Thibodeau S N. Cancer Res. 1998;58:3455–3460. [PubMed] [Google Scholar]
- 21.Herman J G, Umar A, Polyak K, Graff J R, Ahuja N, Issa J J, Markowitz S, Willson J KV, Hamilton S R, Kinzler K W, et al. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomlinson I P M, Ilyas M, Bodmer W F. Br J Cancer. 1996;74:1514–1517. doi: 10.1038/bjc.1996.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hemminki A, Peltomaki P, Mecklin J P, Jarvinen H, Salovaara R, Nystrom-Lahti M, de la Chapelle A, Aaltonen L A. Nat Genet. 1994;8:405–410. doi: 10.1038/ng1294-405. [DOI] [PubMed] [Google Scholar]
- 24.Beck N E, Tomlinson I P M, Homfray T, Frayling I, Hodgson S V, Haracopos C, Bodmer W F. Hum Genet. 1997;99:219–224. doi: 10.1007/s004390050343. [DOI] [PubMed] [Google Scholar]
- 25.Zhou X P, Hoang J M, Li Y J, Seruca R, Carneiro F, Sobrinho-Simoes M, Lothe R A, Gleeson C M, Hilary Russell SE, Muzeau F, et al. Genes Chromosomes Cancer. 1998;21:101–107. doi: 10.1002/(sici)1098-2264(199802)21:2<101::aid-gcc4>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 26.Herman J G, Graff J R, Myohanen S, Nelkin B D, Baylin S B. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Efstathiou J A, Liu D, Wheeler J M D, Kim H C, Beck N E, Ilyas M, Karayiannakis A J, Mortensen N J McC, Kmiot W, Playford R J, et al. Proc Natl Acad Sci USA. 1999;96:2316–2321. doi: 10.1073/pnas.96.5.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tannergard P, Lipford J R, Kolodner R, Frodin J E, Nordenskjold M, Lindblom A. Cancer Res. 1995;55:6092–6096. [PubMed] [Google Scholar]
- 29.Borresen A L, Lothe R A, Meling G I, Lystad S, Morrison P, Lipford J, Kane M F, Rognum T O, Kolodner R D. Hum Mol Genet. 1995;4:2065–2072. doi: 10.1093/hmg/4.11.2065. [DOI] [PubMed] [Google Scholar]
- 30.Liu B, Parsons R, Papadopoulos N, Nicolaides N C, Lynch H T, Watson P, Jass J R, Dunlop M, Wyllie A, Peltomaki P, et al. Nat Med. 1996;2:169–174. doi: 10.1038/nm0296-169. [DOI] [PubMed] [Google Scholar]
- 31.Moslein G, Tester D J, Lindor N M, Honchel R, Cunningham J M, French A J, Halling K C, Schwab M, Goretzki P, Thibodeau S N. Hum Mol Genet. 1996;5:1245–1252. doi: 10.1093/hmg/5.9.1245. [DOI] [PubMed] [Google Scholar]
- 32.Peltomaki P, Vasen H F A. Gastroenterology. 1997;113:1146–1158. doi: 10.1053/gast.1997.v113.pm9322509. [DOI] [PubMed] [Google Scholar]
- 33.Shimodaira H, Filosi N, Shibata H, Suzuki T, Radice P, Kanamaru R, Friend S H, Koodner R D, Ishioka C. Nat Genet. 1998;19:384–389. doi: 10.1038/1277. [DOI] [PubMed] [Google Scholar]
- 34.Thibodeau S N, French A J, Cunningham J M, Tester D, Burgart L J, Roche P C, McDonnell S K, Schaid D J, Vockley C W, Michels V V, et al. Cancer Res. 1998;58:1713–1718. [PubMed] [Google Scholar]
- 35.Peltomaki P, de la Chapelle A. Adv Cancer Res. 1997;71:93–119. doi: 10.1016/s0065-230x(08)60097-4. [DOI] [PubMed] [Google Scholar]
- 36.Frayling I M, Beck N E, Ilyas M, Dove-Edwin I, Goodman P, Pack K, Bell J A, Williams C B, Hodgson S V, Thomas H J W, et al. Proc Natl Acad Sci USA. 1998;95:10722–10727. doi: 10.1073/pnas.95.18.10722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones P A, Laird P W. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 38.Cameron E E, Bachman K E, Myohanen S, Herman J, Baylin S B. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 39.Strathdee G, MacKean M J, Illand M, Brown R. Oncogene. 1999;18:2335–2341. doi: 10.1038/sj.onc.1202540. [DOI] [PubMed] [Google Scholar]
- 40.Laird P W, Jackson-Grasby L, Fazeli A, Dickinson S L, Jung W E, Li E, Weinberg R A, Jaenisch R. Cell. 1995;81:197–205. doi: 10.1016/0092-8674(95)90329-1. [DOI] [PubMed] [Google Scholar]
- 41.Ramchandani S, MacLeod A R, Pinard M, von Hofe E, Szyf M. Proc Natl Acad Sci USA. 1997;94:684–689. doi: 10.1073/pnas.94.2.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruschoff J, Wallinger S, Dietmaier W, Bocker T, Brockhoff G, Hofstadter F, Fishel R. Proc Natl Acad Sci USA. 1998;95:11301–11306. doi: 10.1073/pnas.95.19.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okano M, Xie S, Li E. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 44.Ahuja N, Mohan A, Li Q, Stolker J, Herman J, Hamilton S, Baylin S, Issa J P. Cancer Res. 1997;57:3370–3374. [PubMed] [Google Scholar]
- 45.Homfray T F R, Cottrell S E, Ilyas M, Rowan A, Talbot I C, Bodmer W F, Tomlinson I P M. Hum Mutat. 1998;11:114–120. doi: 10.1002/(SICI)1098-1004(1998)11:2<114::AID-HUMU3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 46.Tomlinson I P M, Novelli M, Bodmer W F. Proc Natl Acad Sci USA. 1996;93:14800–14803. doi: 10.1073/pnas.93.25.14800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young J, Leggett B, Gustafson C, Ward M, Searle J, Thomas L, Buttenshaw R, Chenevix-Trench G. Hum Mutat. 1993;2:351–354. doi: 10.1002/humu.1380020505. [DOI] [PubMed] [Google Scholar]
- 48.Wijnen J, Khan P M, Vasen H, van der Klift H, Mulder A, van Leeuwen-Cornelisse I, Bakker B, Losekoot M, Moller P, Fodde R. Am J Hum Genet. 1997;61:329–335. doi: 10.1086/514847. [DOI] [PMC free article] [PubMed] [Google Scholar]