Abstract
Crigler–Najjar syndrome type I is characterized by unconjugated hyperbilirubinemia resulting from an autosomal recessive inherited deficiency of hepatic UDP-glucuronosyltransferase (UGT) 1A1 activity. The enzyme is essential for glucuronidation and biliary excretion of bilirubin, and its absence can be fatal. The Gunn rat is an excellent animal model of this disease, exhibiting a single guanosine (G) base deletion within the UGT1A1 gene. The defect results in a frameshift and a premature stop codon, absence of enzyme activity, and hyperbilirubinemia. Here, we show permanent correction of the UGT1A1 genetic defect in Gunn rat liver with site-specific replacement of the absent G residue at nucleotide 1206 by using an RNA/DNA oligonucleotide designed to promote endogenous repair of genomic DNA. The chimeric oligonucleotide was either complexed with polyethylenimine or encapsulated in anionic liposomes, administered i.v., and targeted to the hepatocyte via the asialoglycoprotein receptor. G insertion was determined by PCR amplification, colony lift hybridizations, restriction endonuclease digestion, and DNA sequencing, and confirmed by genomic Southern blot analysis. DNA repair was specific, efficient, stable throughout the 6-month observation period, and associated with reduction of serum bilirubin levels. Our results indicate that correction of the UGT1A1 genetic lesion in the Gunn rat restores enzyme expression and bilirubin conjugating activity, with consequent improvement in the metabolic abnormality.
UDP-glucuronosyltransferases (UGTs) are a family of membrane-bound enzymes that catalyze the conjugation of numerous xenobiotics and endogenous substrates with glucuronic acid. Of the known isoforms, only UGT1A1 is physiologically relevant in bilirubin glucuronidation and biliary excretion of this potentially toxic metabolite (1, 2). Crigler–Najjar (CN) syndrome is the inherited deficiency of hepatic UGT1A1 activity and is characterized by elevated serum levels of unconjugated bilirubin (3). Of the two types of CN syndrome, type I is more severe and is characterized by a nearly complete absence of UGT1A1 activity, whereas incomplete deficiency of the enzyme is associated with the less severe type II form (4, 5).
The homozygous Gunn rat, a mutant strain of Wistar rat, is an accurate animal model for CN syndrome type I. Its liver lacks UGT1A1 activity because of the deletion of a single guanosine (G) base in UGT1A1 that results in a frameshift and a premature stop codon (6, 7). Recombinant adenoviral vectors have been used in vivo to correct the hyperbilirubinemia in the Gunn rat with persistent expression of the human bilirubin UGT1A1 enzyme for as long as 2 months (8, 9). Significant progress also has been made in overcoming the immunogenicity of the adenoviral-based vectors (9–11), but their use requires repeated treatments and immunomodulation of the host to maintain therapeutic levels of UGT1A1.
A novel approach, based on mechanisms of DNA repair (12), was reported to correct single nucleotide mutations in episomal and genomic DNA (13, 14). It was observed that an oligonucleotide (ON) composed of both DNA and RNA exhibited increased pairing efficiency with a genomic DNA target (15, 16). The chimeric RNA/DNA ON was designed for increased stability, resistance to nucleases, and improved localization to genomic target sites (17). In a typical duplex structure, the double-stranded region of the molecule is capped by single-stranded thymidine hairpins. The 5′ and 3′ ends of the molecule are juxtaposed and sequestered by using a 5-bp GC clamp at the 3′ end. The RNA residues are 2′-O-methylated to prevent RNase H degradation as well as to improve the formation of joint molecules (18). The homology segment between the RNA/DNA ON and its genomic target is designed with a single mismatch, which promotes the site-directed genomic alteration by endogenous repair pathways (17, 19).
We have used this technology previously to introduce site-specific missense mutations in genomic DNA in cultured human hepatoma cells (20) and in nonreplicating isolated rat hepatocytes (20, 21). In addition, >40% of the rat hepatic factor IX alleles were mutated in vivo by using a nonviral delivery system targeted to the hepatocyte via the asialoglycoprotein receptor (21, 22). Both the genomic and phenotypic changes were stable for more than 1 year in quiescent as well as regenerated livers.
Here, we demonstrate that chimeric RNA/DNA ONs can be used for site-directed insertion of a single G nt in genomic DNA from cultured hepatocytes and intact liver of the Gunn rat. The repair process is dose dependent and associated with restoration of the wild-type BstNI restriction endonuclease site in the UGT1A1 gene. In addition, the phenotypic change is characterized by the hepatic appearance of UGT1A1 protein, secretion of conjugated bilirubin in bile, and decreased serum bilirubin levels. This strategy of genomic alteration circumvents many of the disadvantages associated with viral vector-mediated gene transfer. Our results suggest that site-directed gene repair offers an attractive alternative to gene augmentation using recombinant viruses or hepatocyte transplantation (23) in the treatment of CN syndrome type I.
MATERIALS AND METHODS
Synthesis of the Chimeric ONs.
The chimeric RNA/DNA ONs were obtained from Kimeragen (Newtown, PA). They were synthesized by using DNA and 2′-O-methyl RNA phosphoramidite nucleoside monomers as described (13). After deprotection and purification by HPLC, more than 98% of the purified ONs were full length. Fluorescently labeled ONs were synthesized by using a fluorescein-modified deoxynucleotide at the initial 5′ position of the all-DNA strand.
Polyethylenimine (PEI) and Liposomal Formulations. The 25-kDa PEI (Fluka) was lactosylated by using sodium cyanoborohydride (Sigma) as described (22). For in vitro transfections, the chimeric ONs were combined with PEI at nine equivalents of PEI nitrogen per ON phosphate in 0.15 M NaCl. For in vivo delivery, the chimeric ONs were complexed with PEI at an ON phosphate/PEI amine ratio of 1:6 in 5% dextrose (22, 24).
Lipid films of dioleoyl phosphatidylcholine/dioleoyl phosphatidylserine/galactocerebroside (Avanti Polar Lipids) were prepared at a 1:1:0.16 molar ratio, hydrated, and extruded down to 0.5 μm as described (22). For in vitro transfections, 150 μg of UGT1A1/0.5 ml of 0.15 M NaCl was used to hydrate a 0.5-mg lipid film. For in vivo delivery, 600 μg of ON/ml of 5% dextrose was used to hydrate a 2-mg lipid film. Fluorescently labeled ONs were encapsulated in the anionic liposomes by using the same methods. The encapsulation efficiency of the RNA/DNA ONs was >80%.
Cell Culture and Transfections. Gunn rat hepatocytes immortalized by using the simian virus 40 temperature-sensitive large T-antigen were maintained at the permissive (33°C) temperature in supplemented DMEM (25). Cells were detached by using trypsin-EDTA and replated at a density of 2 × 105 cells per 35-mm Primaria (Becton–Dickinson) dish at the nonpermissive temperature (37°C) 24 h before transfection. Cells were transfected in 1 ml of the same medium supplemented with 2.5 mM CaCl2 by using a 100-μl aliquot of transfecting solution containing the chimeric ONs complexed to PEI or vehicle alone. After 18 h, 2 ml of medium was added, and the hepatocytes were maintained for an additional 30 h at 37°C before harvesting by scrapping. For repeat transfections, the medium was removed after 48 h and replaced, and the cells were transferred to 33°C for expansion. One week later, the cells were prepared, transfected, and harvested as outlined above.
Gunn rat hepatocytes were isolated by collagenase perfusion as described (25) and plated at a density of 1 × 106 cells/T25 flask in a chemically defined medium (hepatocyte growth medium, HGM) (26). Cells were transfected with 300 μl of the liposome-encapsulated chimeric ONs, or vehicle alone, in 3 ml of HGM supplemented with 10% heat-inactivated FBS and 2.5 mM CaCl2. Three milliliters of FBS-supplemented HGM was added 18 h after transfection, and the cultures were maintained an additional 30 h at 37°C. Parallel transfections of Gunn rat hepatocytes were done with the fluorescein-labeled chimeric ONs, and the cells were analyzed by confocal microscopy as described (20, 22).
In Vivo Delivery Systems. Male rats (≈65 g; Harlan Sprague–Dawley) received 200 μg of fluorescein-labeled chimeric ONs that were either naked, encapsulated in anionic liposomes, or complexed to PEI in 5% dextrose by tail vein injection. For asialoglycoprotein receptor competition, animals received bolus injections of 5 mg/100 g body weight of asialofetuin (ASF) in 0.15 M NaCl 1 min before and 3 min after injection of the fluorescently labeled ONs (27). Tissue samples were frozen in OCT, and the cryosections were fixed for 10 min with 4% paraformaldehyde (wt/vol) in PBS, pH 7.4. Tissue distribution of the fluorescently labeled ONs was determined by confocal microscopy as described (21).
Gunn rats (≈80 g; Harlan Sprague–Dawley) were administered aliquots of 200 μg of UGT1A1 either complexed to PEI or encapsulated in anionic liposomes, or an equal amount of vehicle alone by tail vein injection in 5% dextrose on 5 consecutive days. Seven days and 4 months postinjection, random liver tissue samples were removed for DNA isolation. At 6 months, bile samples were collected from the animals as described (10) as well as blood and liver tissue for enzyme activity, DNA, and Western blot analysis.
A separate group of Gunn rats (≈200 g) was injected on 5 consecutive days with either vehicle, or the chimeric ONs complexed to PEI or encapsulated in liposomes. A total dose of 3 mg/rat (600 μg/day × 5) was administered by tail vein injection in 5% dextrose. Rats treated a second time received the same dosing schedule. Blood was drawn under ether anesthesia for serum bilirubin levels and alanine aminotransferase activity (Sigma). Bile samples were collected by bile duct cannulation as described (10).
PCR Amplification, Cloning, and Analysis. Genomic DNA larger than 100 bp was isolated by using the high pure PCR template preparation kit (Boehringer Mannheim). DNA from liver tissue samples was isolated as described (21). PCR amplification (30 cycles of 94°C for 45 s, 55°C for 20 s, and 72°C for 45 s) of a 379-nt region of the rat UGT1A1 gene using the primers 5′-GGGATTCTCAGAATCTAGACATT-3′ (sense) and 5′-GTGTGTGGTATAAATGCTGTAGG-3′ (antisense) (28) was performed with 300 ng of the isolated DNA. To rule out PCR artifacts, 1 μg of UGT1A1 alone, or 300 ng of Gunn rat DNA incubated with up to 1.5 μg of the UGT1A1 chimeric ON, was subjected to PCR amplification. The amplification products were subcloned into the TA cloning vector pCR 2.1 (Invitrogen), and the ligated material was used to transform frozen competent Escherichia coli.
After plating, the colonies were lifted onto Micron Separations MagnaGraph nylon filters, replicated, and processed for hybridization with 32P-end-labeled 17-mer ON probes 1206A (5′-ATGTCCTGAAATGACTG-3′) or 1206G (5′-ATGTCCTGGAAATGACT-3′). Hybridizations were performed at 37°C for 24 h and the filters were washed as described (20). Plasmid DNA prepared from colonies hybridizing with 1206A or 1206G was sequenced on an ABI 370A sequencer (Perkin–Elmer) by using the mp13 forward and reverse primers as well as a gene-specific primer 5′-CCCATGGTATTTATGAAGGAATATGC-3′ corresponding to nucleotides 1071–1106 of the rat UGT1A1 cDNA (7). The PCR amplicons from DNA samples isolated after 6 months were subjected to BstNI restriction endonuclease digestion and separated by using 1% agarose gel electrophoresis to distinguish the wild type from the mutant UGT1A1 Gunn rat gene sequence (29).
Southern and Western Blot Analyses. Genomic DNA from the 6-month liver samples was digested sequentially with EcoRI then BstNI, and the fragments were resolved by electrophoresis through a 1% agarose gel. After capillary transfer to nitrocellulose membrane, the blots were hybridized for 24 h at 65°C in 6× SSC containing 1% SDS, 5× Denhardt’s, and 200 μg/ml denatured sonicated fish sperm DNA with 32P-labeled probe corresponding to the 379-nt PCR-amplified fragment of the rat UGT1A1 gene. After hybridization, the filters were washed in 1× standard saline phosphate/EDTA (0.154 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA; SSPE), 0.5% SDS and then 0.1× SSPE, 0.5% SDS at room temperature and 37°C, respectively, and analyzed by phosphoimaging.
Total homogenate and microsomes were isolated from the flash-frozen liver tissue samples by using the buffers and procedures outlined (30). Protein concentrations were determined with the Bio-Rad protein assay kit. Aliquots of 100 μg of total or microsomal proteins were separated by using 7.5% SDS/PAGE. After electrophoretic transfer onto nitrocellulose membranes, immunoblots were incubated sequentially with H2O2, 5% milk blocking solution, primary antibody (1:5,000) to rat UGT1A1 (28), and horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody. UGT1A1 protein was detected by using the Ultra chemiluminescent system (Pierce).
Serum Bilirubin, UGT1A1 Activity, and HPLC Analysis.
Serum bilirubin concentrations were determined in blood drawn from the Gunn rats by using a Sigma diagnostic kit. The UGT1A1 enzyme activity was assayed in digitonin-activated liver homogenates with bilirubin as the acceptor aglycone, as described (1, 2). Bile pigments collected from the cannulated bile ducts of each animal group were evaluated for bilirubin glucuronidation by HPLC analysis as described (11). Authentic pigments were used as standards, and pigments were identified by retention times.
RESULTS
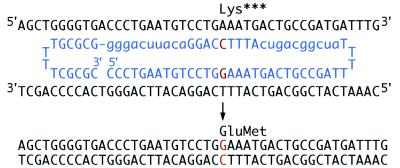
UGT1A1 Correction in Cultured Hepatocytes. We designed the chimeric ONs with the hybrid RNA/DNA strand targeting the nontranscribed DNA sequence of the UGT1A1 gene (Fig. 1). The sequence of the RNA/DNA molecule was identical to that of the mutant gene with one change. An additional G was placed as the center nt within the stretch of nine DNA residues, flanked on both sides by blocks of modified RNA. The genomic target site corresponded to nucleotide 1206 of the complementary strand of the mutant cDNA (7).
Figure 1.
Targeting strategy to correct the UGT1A1 frameshift mutation in the Gunn rat. The 2′-O-methylated RNA residues of the targeting RNA/DNA ON (blue) are indicated in lowercase and the DNA residues in capital letters. Blocks of 10 modified RNA residues flank both sides of a 9-residue stretch of DNA, which contains the base change required for correction. The ON sequence is complementary to 28 residues of genomic DNA spanning the site of mutation with the exception of a G base (orange) targeted for position 1206. The cell’s endogenous DNA repair process mediates insertion of G at the target site, thereby correcting the frameshift mutation and restoring UGT1A1 activity. The folded double-hairpin structure containing four T residues in each loop, a 5-bp GC clamp, and the modified RNA residues significantly improve resistance to nuclease degradation.
Both the liposomal and PEI delivery systems were targeted to the hepatocyte asialoglycoprotein receptor (22, 31). Gunn rat hepatocytes initially were transfected with the fluorescently labeled ONs at 150, 180, and 300 nM concentrations. There was significant cell uptake and nuclear localization of the labeled chimeric molecules in both the immortalized and primary Gunn rat hepatocytes (data not shown). These cells then were transfected with unlabeled UGT1A1 ONs, which were either complexed to PEI or encapsulated in the anionic liposomes. The frequency of G nt insertion at position 1206 was determined by hybridization of duplicate colony lifts of the PCR-amplified and cloned 379-nt stretch of exon 4 of the rat UGT1A1 gene (28).
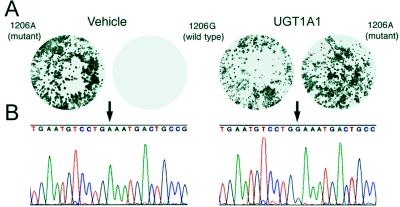
The filter lifts were hybridized with the 32P-end-labeled ON probes 1206A and 1206G (Fig. 2A). The overall frequency of conversion of the targeted nt was calculated by dividing the number of clones hybridizing with the 1206G probe by the total number of clones hybridizing with both probes. G insertion was observed only in hepatocytes transfected with UGT1A1, and not in cells transfected with vehicle or nonspecific chimeric ONs. Additionally, no hybridization of the 1206G probe occurred in clones derived from DNA isolated from untreated hepatocytes and PCR-amplified in the presence of 0.5–1.5 μg of the UGT1A1 ON. Nucleotide insertion was dose dependent and was as high as 15.3%. In addition, the frequency of G insertion increased to 23.7% after a second transfection of the immortalized Gunn rat hepatocytes.
Figure 2.
Filter lift hybridizations and sequence analysis of DNA from isolated hepatocytes. (A) Representative hybridization patterns of duplicate filter lifts of the cloned PCR amplicons with either 32P-labeled mutant 1206A or wild-type 1206G 17-mer probes. Hepatocytes were transfected with vehicle (Left) or UGT1A1 ON (Right). (B) The nt sequence of plasmid DNA isolated from clones hybridized with probes to mutant 1206A or wild-type 1206G displaying either A (arrow, Left) or G (arrow, Right), respectively.
We confirmed our results from the filter hybridizations by direct sequencing of at least 12 independent clones of the wild-type and mutant genes (Fig. 2B). The results indicated that colonies hybridizing to only 1206A exhibited the mutant sequence. In contrast, those colonies derived from UGT1A1-transfected Gunn rat hepatocytes hybridizing to the wild-type 1206G ON probe displayed a G at position 1206. The entire 379-nt PCR-amplified region of the UGT1A1 gene was sequenced for all of the clones and no alterations other than the directed change at the target site was observed. Finally, the start and end points of the 379-nt PCR-amplified genomic DNA samples corresponded exactly to those of the primers used for the amplification process, indicating that the clones sequenced were derived from genomic DNA, rather than nondegraded chimeric ONs.
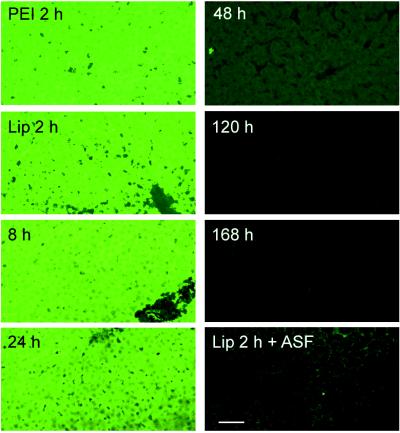
In Vivo Characterization of the Anionic Liposome and PEI Delivery Systems. The fluorescently labeled ONs, using either PEI or liposomes, were distributed homogeneously throughout the liver as early as 2 h after tail vein injection (Fig. 3). In contrast, there was only minimal uptake in lung, heart, and kidney. Coadministration of ASF, which binds avidly to the asialoglycoprotein receptor (27), almost totally inhibited the hepatic uptake of the ONs. This finding was associated with increased levels of the labeled molecules in the other organs. Interestingly, no detectable fluorescence was present in the testis even with coadministration of ASF. In animals injected with naked fluorescein-labeled ONs, there was almost no detectable fluorescence in the liver at 2 h. However, distribution to the other organs was similar to that observed when ASF was coadministered to inhibit liver uptake (data not shown).
Figure 3.
In vivo hepatic distribution of fluorescently labeled ONs. Rats received 200 μg of 5′ fluorescein-labeled chimeric ONs encapsulated in anionic liposomes or complexed with PEI by single bolus tail vein injection. At the indicated times, their livers were processed and examined by confocal microscopy. Lip, liposomes. (Bar = 100 μm.)
We then characterized the time course for hepatic disappearance of the fluorescein label in rats injected with the liposome-encapsulated ONs (Fig. 3). No significant change was observed until 24 h postinjection when the fluorescence began to decline. There was a dramatic decrease throughout the liver by 48 h, and by 120–168 h there was only background fluorescence. Disappearance of the fluorescein label in the other tissues mirrored that observed in the liver. The same pattern of distribution and disappearance was observed when the ONs were complexed to PEI.
In Vivo Correction of the Hepatic UGT1A1 Mutation.
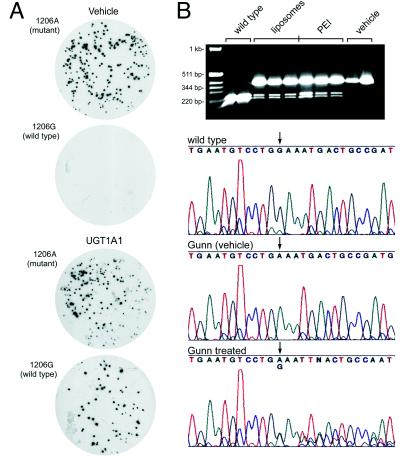
Chimeric ONs complexed with PEI or encapsulated in the anionic liposomes were administered in vivo by tail vein injection. Random samples of liver were harvested at 7 days and 4 and 6 months postinjection. Liver DNA was isolated, and the 379-nt sequence spanning the target site was PCR-amplified. Duplicate filter lifts of the transformed colonies were hybridized with the 17-mer ONs to either wild-type 1206G or mutant 1206A. Insertion frequency of G at the genomic target site was ≈20% with either delivery system (Fig. 4A) and was undetectable in the control groups. The frequency remained stable at ≈20% even when the same livers were analyzed 4 and 6 months postinjection (Table 1). The PCR amplicons from the 6-month samples were subjected to restriction endonuclease digestion with BstNI. Agarose gel analysis indicated partial cleavage at the wild-type BstNI site, whereas DNA from the vehicle controls remained resistant (Fig. 4B, Top). Finally, the 379-nt PCR-amplified DNA fragments were sequenced to confirm G insertion. Amplicons from the UGT1A1 livers exhibited a mix of wild-type G and mutant A at position 1206 (Fig. 4B, Bottom), whereas the control groups displayed only the mutant A (Middle).
Figure 4.
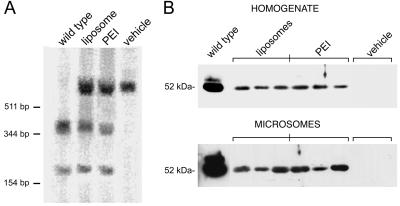
Filter lift hybridizations, restriction fragment length polymorphism, and sequence analysis of DNA isolated from liver. (A) Hybridization patterns of duplicate filter lifts of the cloned PCR products from liver DNA of Gunn rats 6 months postinjection with vehicle (Upper) or UGT1A1 ONs (Lower). (B) PCR amplicons were subjected to BstNI restriction enzyme digestion and analyzed by agarose gel electrophoresis and ethidium bromide staining (Top). Direct DNA sequencing of the PCR-amplified UGT1A1 gene surrounding the targeted G insertion site at position 1206 (arrow) is shown for wild-type (G, top sequence), vehicle (A, middle), and UGT1A1-treated Gunn rats (A and G, bottom). The size of the DNA standards is indicated at top left.
Table 1.
In vivo G insertion at nucleotide 1206 of the UGT1A1 gene in Gunn rat livers
| Vehicle | UGT1A1 dosage, mg | Insertion, %
|
||
|---|---|---|---|---|
| 1 week | 4 mos | 6 mos | ||
| Liposomes | 1 | 20.5 ± 6.1 | 17.3 ± 5.1 | 19.9 ± 3.0 |
| PEI | 1 | 23.0 ± 1.4 | 19.3 ± 2.1 | 20.7 ± 0.3 |
| PEI control | 0 | n.d. | n.d. | n.d. |
The data represent the mean percentage ±SD of G insertion from random liver tissue samples determined by filter lift hydridizations as described in Materials and Methods. Each treatment group contained at least three animals. n.d., not detectable.
Southern and Western Blot Analyses. DNA was isolated from a variety of liver tissue samples for genomic Southern blot analysis. In fact, DNA isolated from animals that were administered the UGT1A1 ON showed partial restoration (≈25%) of the BstNI restriction site in exon 4 of the UGT1A1 gene (29) (Fig. 5A). In contrast, the control samples showed no cleavage with BstNI at this site, whereas the wild-type DNA was completely cleaved. The results from the Southern blot analyses were similar for both the PEI and liposomal delivery systems.
Figure 5.
Southern and Western blot analyses of Gunn rat livers. Liver tissue was harvested for DNA and protein analysis 6 months after in vivo administration of the UGT1A1 ONs as described in Materials and Methods. (A) Southern blot analysis after sequential digestion of genomic DNA with EcoRI and BstNI. (B) Western blot analysis of total liver homogenate and microsomal extracts from the UGT1A1- and vehicle-treated Gunn rats. DNA size markers and protein molecular mass are indicated at left.
Total and microsomal proteins were isolated from liver tissue samples and subjected to Western blot analysis for detection of the 52-kDa UGT1A1 protein. The results (Fig. 5B) indicated that repair of the UGT1A1 gene sequence was associated with appearance of the bilirubin-conjugating enzyme. In contrast, there was no detectable UGT1A1 protein in control samples. The protein was enriched in the microsomal fraction and expressed at 8–15% of wild-type levels, in agreement with the observed enzyme activity in these samples.
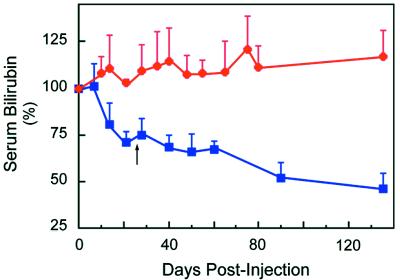
Effect of UGT1A1 Gene Correction on Serum Bilirubin Levels. The serum bilirubin levels of the Gunn rats were monitored after tail vein injection and indicated that a single dosing regimen of the UGT1A1 molecule, using either PEI or liposomes, resulted in an ≈25% decrease in serum bilirubin levels (Fig. 6). In contrast, rats administered vehicle or nonspecific ON showed no change, or even an increase in their serum bilirubin levels. A repeat dosing with UGT1A1 resulted in a further drop in serum bilirubin to <50% of the pretreatment levels, whereas no significant change was observed in the control rats. Blood studies for routine liver enzymes were performed with both delivery systems, and no changes were detected. Moreover, histologic examination of the livers 6 months after administration indicated that neither PEI, anionic liposomes, nor the chimeric ONs altered liver morphology (data not shown).
Figure 6.
Effect of UGT1A1 gene correction on serum bilirubin levels in Gunn rats. Animals were administered UGT1A1 (blue squares) or nonspecific (red circles) ONs complexed to PEI or encapsulated in anionic liposomes as described in Materials and Methods. The dosage was repeated for all groups 30 days after the final injection of the first series (arrow). Each data point is the mean ± SD of 11 animals. There was no significant difference between the PEI and anionic liposome groups. P < 0.001 ≥14 days for UGT1A1 ONs.
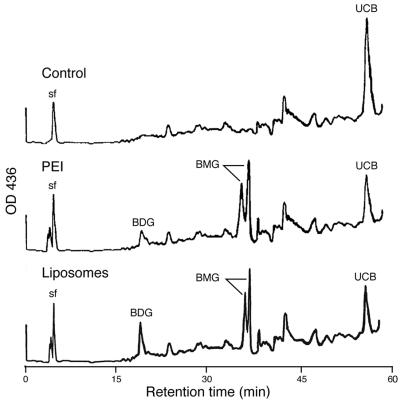
Hepatic UGT1A1 enzyme activity was confirmed by bile duct cannulation and HPLC analysis of bilirubin glucuronidation. In fact, bilirubin mono- and diglucuronides were detected only in those Gunn rats that were administered the UGT1A1 chimeric ONs (Fig. 7). No significant differences were detected between the PEI and liposomal delivery systems, and in both groups the bilirubin was conjugated primarily as the monoglucuronidated species. Only unconjugated bilirubin was present in the bile of the control Gunn rats.
Figure 7.
HPLC analysis of bile pigments from Gunn rat livers. Bile ducts were cannulated and bile collected for HPLC analysis from both PEI- and liposome-treated Gunn rats as described in Materials and Methods. BMG, bilirubin monoglucuronide; BDG, bilirubin diglucuronide; UCB, unconjugated bilirubin. The HPLC profiles are representative of four animals in each experimental group.
DISCUSSION
Chimeric RNA/DNA ONs have been used successfully for single nt substitution in episomal and genomic DNA of replicating cells (13, 14, 20, 32). They also have mediated efficient genomic site-specific nt exchange in isolated nonreplicating as well as quiescent hepatocytes in vivo (21, 22). The purpose of this study was to establish whether chimeric ONs could effect site-specific replacement of a G residue to correct the frameshift mutation in exon 4 of UGT1A1 in Gunn rats. Our results demonstrate efficient correction of the genetic lesion in both immortalized and primary Gunn rat hepatocytes, as well as the liver in situ. In addition, the long-term change together with our previous results with mutation of the rat factor IX gene (22) suggest that correction of the UGT1A1 gene was permanent.
The genomic insertion of G at the targeted site was not an artifact of PCR amplification, as recently suggested (33). Specifically, neither the control groups nor DNA samples spiked with the UGT1A1 ONs yielded wild-type clones. Also, despite the almost complete hepatic disappearance of the ONs from the liver by 48 h, the frequency of G insertion at 1 week was comparable with that observed 4 and 6 months later in the same livers. Furthermore, hepatic correction of the UGT1A1 gene mutation was confirmed by genomic Southern blot analysis, expression of the 52-kDa UGT1A1 enzyme, and a significant reduction in serum bilirubin levels that has been maintained as long as 10 months without additional treatment. In contrast, serum bilirubin remained unchanged, and in some cases increased in control animals. Finally, UGT1A1 enzyme activity was confirmed by the appearance of both mono- and diglucuronidated bilirubin in the Gunn rat bile (1).
The reduction in serum bilirubin levels was gradual and more closely resembled that observed with hepatocyte transplantation (23) rather than whole organ transplantation or overexpression of UGT1A1 transgenes (8, 10, 11). This finding may be explained by partial correction of the enzyme defect and slow release of bilirubin from the body stores of the Gunn rats, as well as zonal differences in hepatic UGT1A1 expression. The greater proportion of bilirubin monoglucuronide relative to the diglucuronide in bile is also reminiscent of partial bilirubin UGT deficiency states, including CN syndrome type II, Gilbert syndrome, and heterozygous Gunn rats (34, 35). The presence of a higher concentration of bilirubin relative to the number of UGT molecules favors the generation of bilirubin monoglucuronide over the formation of the diglucuronide (36). Also, even with partial gene correction, increased enzyme expression could be achieved by transcriptional induction of UGT1A1 with several different agents, including phenobarbital (5).
Based on the in vivo fluorescent studies, we estimate that ≈100,000 ONs were delivered to hepatocyte nuclei with each tail vein injection. If both alleles are equally amenable to gene repair and only a minority of them are repaired, it is more probable that a single allele would be corrected than both alleles in a given cell. Consistent with this notion, it was reported recently that repair of the tyrosine missense mutation in albino melanocytes appears to occur in a single allele in clonal isolates passaged as many as 10 times (32). This occurrence could be important in some diseases such as α1-antitrypsin deficiency, in which a codominant mutant protein may interfere with the function of the wild-type gene product (37).
The incorporation of 2′-O-methylated RNA residues in the structure of the chimeric ON increases the efficiency of nt exchange compared with all-DNA ONs (13, 14, 16). It appears that the RNA/DNA strand of the duplex is responsible for the initial pairing event, whereas the mismatch within the all-DNA homology strand activates the endogenous DNA repair process (17). In fact, the human recombinase HsRec2 protein, which facilitates homologous pairing (38), significantly increased joint molecule formation between the RNA/DNA ONs and complementary single-stranded DNA compared with the all-DNA ONs (18).
With a novel bacterial test system, the functional capacity of these chimeric molecules to promote targeted nt conversion was shown to require both RecA recombinase and MutS, a mismatch repair enzyme (19). The human MutS homolog MSH2 also was required for nt conversion in a mammalian cell-free assay system (39). A two-step process was proposed in which RecA mediates strand pairing and formation of a double D-loop, whereas MutS mediates genomic repair (19, 39). In fact, MutS is active in mismatch repair pathways rather than in homologous recombination (40–42). It recently has been reported that the MutSα and MutSβ heterodimeric complexes of the mammalian MSH2 mismatch repair pathway are differentially expressed in cultured cells, and that the MSH2 protein is involved in modulating their levels (43, 44). Thus, cell lines with varying concentrations of these repair molecules may respond differently to genomic alteration by RNA/DNA ONs (45).
The use of RNA/DNA ONs to correct genetic diseases of the liver offers significant advantages over viral-mediated transgene expression. In particular, it overcomes the random genomic integration associated with certain viral vectors, and the observed immunogenicity and lack of persistent gene expression in others. However, the approach does require the use of ONs that are designed specifically to each genetic mutation. The percent decrease in serum bilirubin levels achieved in this study would be sufficient to convert potentially lethal CN syndrome type I to a manageable CN syndrome type II phenotype. Additionally, the cumulative effect of the repeated treatments coupled with the ability of this technology to induce site-specific nt alteration of genomic DNA without selection offers a potentially powerful technique for both ex vivo and in vivo gene therapy.
Acknowledgments
We acknowledge the expert technical assistance of Ms. Xiaoming Ma, Ms. Cheryle Linehan-Stieers, and Mr. Stefan Kren. This work was supported by a grant (to C.J.S.) from Kimeragen, Inc., Newtown, PA; and by Grants RO1-DK 39137 (to N.R.C.), RO1-DK 46057 (to J.R.C.), and P30-DK 41296 (Liver Research Core Center, Albert Einstein College of Medicine) from the National Institutes of Health.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: ASF, asialofetuin; CN, Crigler–Najjar; ON, oligonucleotide; PEI, polyethylenimine; UGT, UDP-glucuronosyltransferase.
References
- 1.Roy Chowdhury N, Gross F, Moscioni A D, Kram M, Arias I M, Roy Chowdhury J. J Clin Invest. 1987;79:327–334. doi: 10.1172/JCI112816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosma P J, Seppen J, Goldhoorn B, Bakker C, Oude Elferink R P J, Roy Chowdhury J, Roy Chowdhury N, Jansen P L M. J Biol Chem. 1994;269:17960–17964. [PubMed] [Google Scholar]
- 3.Crigler J F, Najjar V A. Pediatrics. 1952;10:169–172. [PubMed] [Google Scholar]
- 4.Arias I M. J Clin Invest. 1962;41:2233–2245. doi: 10.1172/JCI104682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roy Chowdhury J, Wolkoff A W, Roy Chowdhury N, Arias I M. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw-Hill; 1995. pp. 2161–2208. [Google Scholar]
- 6.Iyanagi T, Watanabe T, Uchiyama Y. J Biol Chem. 1989;264:21302–21307. [PubMed] [Google Scholar]
- 7.Roy-Chowdhury J, Huang T, Kesari K, Lederstein M, Arias I M, Roy-Chowdhury N. J Biol Chem. 1991;266:18294–18298. [PubMed] [Google Scholar]
- 8.Askari F K, Hitomi Y, Mao M, Wilson J M. Gene Ther. 1996;3:381–388. [PubMed] [Google Scholar]
- 9.Li Q, Murphree S S, Willer S S, Bolli R, French B A. Hum Gene Ther. 1998;9:497–505. doi: 10.1089/hum.1998.9.4-497. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi M, Ilan Y, Roy Chowdhury N, Guilda J, Horwitz M, Roy Chowdhury J. J Biol Chem. 1996;271:26536–26542. doi: 10.1074/jbc.271.43.26536. [DOI] [PubMed] [Google Scholar]
- 11.Ilan Y, Prakash R, Davidson A, Jona V, Droguett G, Horwitz M S, Roy Chowdhury N, Roy Chowdhury J. J Clin Invest. 1997;99:1098–1106. doi: 10.1172/JCI119238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kmiec E B, Cole A, Holloman W K. Mol Cell Biol. 1994;14:7163–7172. doi: 10.1128/mcb.14.11.7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoon K, Cole-Strauss A, Kmiec E B. Proc Natl Acad Sci USA. 1996;93:2071–2076. doi: 10.1073/pnas.93.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole-Strauss A, Yoon K, Xiang Y, Byrne B C, Rice M C, Gryn J, Holloman W K, Kmiec E B. Science. 1996;273:1386–1389. doi: 10.1126/science.273.5280.1386. [DOI] [PubMed] [Google Scholar]
- 15.Kotani H, Kmiec E B. Mol Cell Biol. 1994;14:6097–6106. doi: 10.1128/mcb.14.9.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotani H, Kmiec E B. Mol Cell Biol. 1994;14:1949–1955. doi: 10.1128/mcb.14.3.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kmiec E B, Kren B T, Steer C J. In: The Development of Human Gene Therapy. Friedmann T, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 643–670. [Google Scholar]
- 18.Havre P A, Kmiec E B. Mol Gen Genet. 1998;258:580–586. doi: 10.1007/s004380050771. [DOI] [PubMed] [Google Scholar]
- 19.Kren B T, Metz R, Kumar R, Steer C J. Sem Liver Dis. 1999;19:93–104. doi: 10.1055/s-2007-1007101. [DOI] [PubMed] [Google Scholar]
- 20.Kren B T, Cole-Strauss A, Kmiec E B, Steer C J. Hepatology. 1997;25:1462–1468. doi: 10.1002/hep.510250626. [DOI] [PubMed] [Google Scholar]
- 21.Kren B T, Bandyopadhyay P, Steer C J. Nat Med. 1998;4:285–290. doi: 10.1038/nm0398-285. [DOI] [PubMed] [Google Scholar]
- 22.Bandyopadhyay P, Ma X, Linehan-Stieers C, Kren B T, Steer C J. J Biol Chem. 1999;274:10163–10172. doi: 10.1074/jbc.274.15.10163. [DOI] [PubMed] [Google Scholar]
- 23.Fox I J, Roy Chowdhury J, Kaufman S S, Goertzen T C, Roy Chowdhury N, Warkentin P I, Dorko K, Sauter B V, Strom S C. N Engl J Med. 1998;338:1422–1426. doi: 10.1056/NEJM199805143382004. [DOI] [PubMed] [Google Scholar]
- 24.Abdallah B, Hassan A, Benoist C, Goula D, Behr J P, Demeneix B A. Hum Gene Ther. 1996;7:1947–1954. doi: 10.1089/hum.1996.7.16-1947. [DOI] [PubMed] [Google Scholar]
- 25.Fox I J, Roy Chowdhury N, Gupta S, Kondapalli R, Schilsky M L, Stockert R J, Roy Chowdhury J. Hepatology. 1995;21:837–846. [PubMed] [Google Scholar]
- 26.Block G D, Locker J, Bowen W C, Petersen B E, Katyal S, Strom S C, Riley T, Howard T A, Michalopoulos G K. J Cell Biol. 1996;132:1133–1149. doi: 10.1083/jcb.132.6.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hara T, Aramaki Y, Tsuchiya S, Hosoi K, Okada A. Biopharmaceutics Drug Dispos. 1987;8:327–339. doi: 10.1002/bdd.2510080404. [DOI] [PubMed] [Google Scholar]
- 28.Ilan Y, Roy Chowdhury N, Prakash R, Jona V, Attavar P, Guha C, Tada K, Roy Chowdhury J. Transplantation. 1997;64:8–13. doi: 10.1097/00007890-199707150-00003. [DOI] [PubMed] [Google Scholar]
- 29.Huang T-J, Roy Chowdhury J, Lahiri P, Yerneni P C, Bommineni V R, Arias I M, Roy Chowdhury N. Hepatology. 1992;16:756–762. doi: 10.1002/hep.1840160323. [DOI] [PubMed] [Google Scholar]
- 30.Liu L, Jang G-F, Farnsworth C C, Yokoyama K, Glomset J A, Gelb M H. Methods Enzymol. 1995;250:189–206. doi: 10.1016/0076-6879(95)50072-3. [DOI] [PubMed] [Google Scholar]
- 31.Bandyopadhyay P, Kren B T, Ma X, Steer C J. BioTechniques. 1998;25:282–292. doi: 10.2144/98252gt03. [DOI] [PubMed] [Google Scholar]
- 32.Alexeev V, Yoon K. Nat Biotech. 1998;16:1343–1346. doi: 10.1038/4322. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, Eriksson M, Falk G, Graff C, Presnell S C, Read M S, Nichols T C, Blombäck M, Anvret M. Antisense Nucleic Acid Drug Dev. 1998;8:531–536. doi: 10.1089/oli.1.1998.8.531. [DOI] [PubMed] [Google Scholar]
- 34.Blanckaert N. Biochem J. 1980;185:115–128. doi: 10.1042/bj1850115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fevery J, Blanckaert N, Heirwegh K P M, Préaux A-M, Berthelot P. J Clin Invest. 1977;60:970–979. doi: 10.1172/JCI108877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordon E R, Goresky C A. Can J Biochem. 1980;58:1302–1310. doi: 10.1139/o80-175. [DOI] [PubMed] [Google Scholar]
- 37.Teckman J H, Qu D, Perlmutter D H. Hepatology. 1996;24:1504–1516. doi: 10.1002/hep.510240635. [DOI] [PubMed] [Google Scholar]
- 38.Rice M C, Smith S T, Bullrich F, Havre P, Kmeic E B. Proc Natl Acad Sci USA. 1997;94:7417–7422. doi: 10.1073/pnas.94.14.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cole-Strauss A, Gamper H, Holloman W K, Muñoz M, Cheng N, Kmiec E B. Nucleic Acids Res. 1999;27:1323–1330. doi: 10.1093/nar/27.5.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yáñez R J, Porter A C G. Gene Ther. 1998;5:149–159. doi: 10.1038/sj.gt.3300601. [DOI] [PubMed] [Google Scholar]
- 41.Kolodner R D. Trends Biochem Sci. 1995;20:397–401. doi: 10.1016/s0968-0004(00)89087-8. [DOI] [PubMed] [Google Scholar]
- 42.Modrich P. J Biol Chem. 1997;272:24727–24730. doi: 10.1074/jbc.272.40.24727. [DOI] [PubMed] [Google Scholar]
- 43.Genschel J, Littman S J, Drummond J T, Modrich P. J Biol Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 44.Umar A, Risinger J I, Glaab W E, Tindall K R, Barrett J C, Kunkel T A. Genetics. 1998;148:1637–1646. doi: 10.1093/genetics/148.4.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santana E, Peritz A E, Iyer S, Uitto J, Yoon K. J Invest Dermatol. 1998;111:1172–1177. doi: 10.1046/j.1523-1747.1998.00403.x. [DOI] [PubMed] [Google Scholar]