Abstract
DNA topoisomerase I is a nuclear enzyme involved in transcription, recombination, and DNA damage recognition. Previous studies have shown that topoisomerase I interacts directly with the tumor-suppressor protein p53. p53 is a transcription factor that activates certain genes through binding to specific DNA sequences. We now report that topoisomerase I can be stimulated by both latent and activated wild-type p53 as well as by several mutant and truncated p53 proteins in vitro, indicating that sequence-specific DNA-binding and stimulation of topoisomerase I are distinct properties of p53. These assays also suggest that the binding site for topoisomerase I on p53 is between amino acids 302 and 321. In living cells, the interaction between p53 and topoisomerase I is strongly dependent on p53 status. In MCF-7 cells, which have wild-type p53, the association between the two proteins is tightly regulated in a spatial and temporal manner and takes place only during brief periods of genotoxic stress. In marked contrast, the two proteins are constitutively associated in HT-29 cells, which have mutant p53. These findings have important implications for both cellular stress response and genomic stability, given the ability of topoisomerase I to recognize DNA lesions as well as to cause illegitimate recombination.
DNA topoisomerase I is a nuclear enzyme essential for most aspects of nucleic acid metabolism (1). The ability of topoisomerase I to relax supercoiled DNA through transient single-stranded DNA cleavage, strand passage, and religation is needed during transcription to relieve superhelical stress (2, 3). In addition, topoisomerase I serves as a modulator of transcriptional initiation through physical interaction with transcription factors such as the TATA box-binding protein, which is a constituent of the general transcription factor IID (TFIID) complex (4, 5). This property does not require DNA cleavage but is based on protein–protein interactions. Recent results show that topoisomerase I also possesses a protein-kinase activity, which is specific for serine residues of splicing factors containing an Arg-Ser motif. Phosphorylation of these splicing factors is believed to influence gene expression by altering the splice pattern (6, 7).
Eukaryotic topoisomerase I is structurally, functionally, and evolutionarily related to site-specific recombinases (8). During catalysis, topoisomerase I forms a covalent DNA–protein complex with the 3′ end of one DNA strand. This reaction intermediate can religate with either the 5′ hydroxyl end of the cleaved strand or with a 5′ hydroxyl end of a heterologous DNA molecule, resulting in recombinant DNA (9, 10). In yeast, increased topoisomerase I activity is accompanied by increased levels of illegitimate recombination, whereas treatment of mammalian cells with camptothecin and other topoisomerase I-directed antitumor drugs leads to sister chromatid exchange and chromosomal aberrations (11, 12). This is most likely because camptothecin treatment results in accumulation of covalent DNA–topoisomerase I complexes, which are potentially recombinogenic (for a recent review, see ref. 13).
Purified topoisomerase I is able to recognize endogenous DNA lesions such as mismatched bases, abasic sites, and deaminated cytosines. The binding of topoisomerase I is associated with nicking of DNA at the first phosphodiester bond 5′ to the DNA lesion and formation of stable DNA–topoisomerase I complexes similar to those that accumulate in the presence of camptothecin (14, 15). Covalent DNA–topoisomerase I complexes are also formed in the proximity of cyclopyrimidine dimer lesions as well as in living cells after UV irradiation (16, 17).
The tumor-suppressor gene product p53 is a key factor in the regulation of stress-induced pathways affecting DNA repair, cell-cycle progression, and apoptosis. p53 is a transcription factor that activates certain genes through binding to specific DNA sequences (18, 19). In addition, p53 is able to bind many cellular and viral proteins (for review, see ref. 20). p53 is altered by missense mutations in most human tumors (21). These mutations usually lead to expression of full-length proteins that are deficient for sequence-specific DNA binding. In contrast, mutant p53 proteins are not necessarily altered with respect to their ability to interact with other proteins (20, 22).
We have recently shown that p53 interacts directly with topoisomerase I in vitro as well as in carcinoma cells treated with genotoxic agents. The molecular interaction leads to activation of topoisomerase I both as measured by relaxation of supercoiled DNA and by phosphorylation of Ser-Arg-containing splicing factors (23, 24). A similar interaction has been observed between topoisomerase I from nontransformed keratinocytes and glutathione S-transferase (GST)–p53 fusion proteins (25).
We now report that there are no qualitative or quantitative differences in the way topoisomerase I is stimulated by p53 proteins with wild-type or mutant function in vitro. In contrast, the interaction between the two proteins is highly dependent on p53 status in living cells. The association between wild-type p53 and topoisomerase I is regulated in a spatial and temporal manner and takes place only during brief periods of genotoxic stress, whereas mutant p53 is constitutively associated with topoisomerase I. These findings have important implications for both cellular stress response and genomic stability, given the ability of topoisomerase I to recognize DNA lesions as well as to cause illegitimate recombination.
EXPERIMENTAL PROCEDURES
Chemicals.
Camptothecin and mitomycin C were purchased from Sigma. A polypeptide corresponding to amino acids 302–321 of human p53 was obtained from Neosystem (Strasbourg, France).
Purification of Topoisomerase I and p53.
Human DNA topoisomerase I was prepared from insect cells as described (6, 26). This purification procedure resulted in a topoisomerase I protein that migrated as a single 100-kDa polypeptide band on SDS/PAGE. Human p53 and GST–p53 fusion proteins were purified from bacteria as described, leading to pure protein preparations as judged by Coomassie blue staining of SDS/PAGE gels (23, 27).
Sequence-Specific DNA Binding.
Electrophoretic mobility-shift assays were carried out with oligonucleotides containing the p53-binding site of GADD45 or with mutant GADD45 that had lost the p53-binding site (28). Oligonucleotides 5′-AATTCTCGAGCAGAACATGTCTAAGCATGCTGGGCTCGAG-3′ and 5′-AATTCTCGAGCAGAAAATTTCTAAGAATTCTGGGCTCGAG-3′ were phosphorylated in the presence of [γ-32P]ATP and T4 polynucleotide kinase and annealed with the complementary oligonucleotides in the presence of 100 mM NaCl. Protein–DNA binding was carried out for 30 min at 4°C in 20 μl of reaction buffer [50 mM Hepes, pH 7.0/50 mM KCl/0.1 mM DTT/1 mg/ml BSA/0.001% Triton X-100/20% glycerol/120 ng/μl double-stranded poly(dI,dC)] containing ≈3 ng of end-labeled double-stranded oligonucleotides and p53 that had been preincubated with PAb 421 anti-p53 monoclonal antibodies (Oncogene Science) or with topoisomerase I for 30 min on ice. Samples were analyzed in 4% native polyacrylamide gels, prepared, and prerun in 0.5× TBE buffer containing 0.01% Triton X-100. Electrophoresis was carried out at 4°C at 200 V, and the gels were dried and autoradiographed.
DNA Relaxation.
Different concentrations of topoisomerase I and p53 were mixed on ice with 0.5 μg of pBR322 DNA in 20 mM Tris⋅HCl (pH 7.5), 150 mM KCl, 0.5 mM EDTA, and 0.5 mM DTT (20 μl final volume), and relaxation assays were performed as described (29).
DNA Cleavage.
Different concentrations of topoisomerase I were mixed on ice with 300 ng of GST–p53 and 20,000 dpm of 3′ end-labeled pBR322 DNA in 20 mM Tris⋅HCl (pH 7.5), 60 mM KCl, 0.5 mM EDTA, and 0.5 mM DTT (20 μl final volume). Samples were incubated at 37°C for 10 min, and the reactions were terminated by the addition of 2 μl of 2.5% SDS/2.5 mg/ml proteinase K and incubated for 30 min at 50°C. The samples were denatured, and DNA fragments were separated by agarose gel electrophoresis followed by autoradiography as described (29).
Cell Culture and Preparation of Nuclear Extracts.
MCF-7 human mammary adenocarcinoma cells and HT-29 human colon carcinoma were grown in DMEM supplemented with 10% fetal calf serum and antibiotics (0.1 μg/ml streptomycin and 100 units/ml penicillin). M1 murine myeloid leukemia cell lines were grown in RPMI 1640 medium supplemented with 10% fetal calf serum and antibiotics. Nuclear extracts were prepared from ≈5 × 106 cells in exponential growth phase and partly purified by ammonium sulfate precipitation (23). For comparison of catalytic activity, purified extracts from different cell lines were adjusted to the same protein concentration followed by serial dilutions.
Immunolocalization of p53.
Cells were attached to circular slides overnight and exposed to 10 μg/ml mitomycin C for 4 hours followed by postincubation in drug-free medium for 20 hours. After drug exposure, cells were fixed with 3.7% formaldehyde, permeabilized in 0.25% Triton X-100, and blocked with 1% BSA. Cells were then incubated for 1 hour with anti-p53 mAbs (PAb 1801, Oncogene Science) at 1:50 followed by secondary anti-mouse FITC-conjugated antibodies (Amersham Pharmacia). Coverslips were mounted in Vectashield (Vector Laboratories) and analyzed with an epifluorescence microscope Axiovert 100M equipped with appropriate filters and laser confocal scanning system LSM 510 by using a plan Apochromat ×63 objective (Zeiss).
Western Blot Analysis.
Nuclear extracts were prepared from drug-treated cells as described above. Proteins were separated on a 4–12% polyacrylamide SDS gel and transferred to a nitrocellulose filter. The presence of p53 and topoisomerase I was revealed by PAb 421 (Oncogene Science) and Scl70 (TopoGen, Columbus, OH), respectively, followed by incubation with peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch) and detection by enhanced chemiluminescence (Amersham Pharmacia).
Immunoprecipitation of p53.
Nuclear extracts were prepared from ≈3 × 107 cells and partly purified by ammonium sulfate precipitation. Four hundred microliters of each nuclear extract was incubated with 10 μl of anti-p53 antibodies (PAb 421) for 1 hour on ice under gentle agitation followed by addition of 100 μl of protein A-Sepharose (CL-4B, Amersham Pharmacia) prepared in 50 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, and 10 mM 2-mercaptoethanol for an additional 1 hour on ice. After washing, 10 μl of each immunoprecipitate was tested for DNA relaxation activity as described above, except that 7.5 mM MgCl2 was added to the reaction mixture. The presence of p53 was confirmed by Western blot analysis, as described above.
RESULTS
Topoisomerase I Does Not Activate the Sequence-Specific DNA Binding of p53.
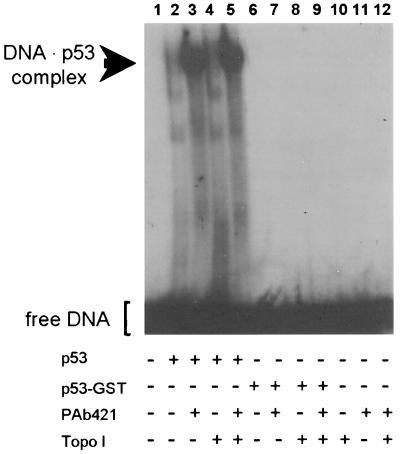
Wild-type recombinant p53 purified from bacteria exists in a latent form with weak sequence-specific DNA binding (19). However, the sequence-specific binding can be activated by physical interaction with certain other proteins such as PAb 421 monoclonal antibodies or high-mobility group protein-1 (HMG-1) (19, 30). An electrophoretic mobility-shift assay was used to determine whether the association with topoisomerase I activates p53. p53 alone showed weak binding to an oligonucleotide containing the GADD45 p53-binding site (Fig. 1, lane 2) whereas addition of monoclonal PAb 421 activated the sequence-specific DNA binding (Fig. 1, lane 3). The presence of topoisomerase I neither stimulated nor prevented the sequence-specific DNA-binding of p53 (Fig. 1, lanes 4 and 5). GST–p53 showed no sequence-specific DNA binding and could not be activated by PAb 421 or topoisomerase I (Fig. 1, lanes 6–9).
Figure 1.
Influence of topoisomerase I on the sequence-specific binding of p53 as measured by an electrophoretic mobility-shift assay. All lanes contained radiolabeled oligonucleotides carrying the p53-binding site of the GADD45 promoter. Lanes 2–5 and 6–9 contained 500 ng of p53 or 500 ng of p53–GST, respectively, in the presence of 1 μg of PAb 421 (lanes 3, 5, 7, and 9) or 500 ng of topoisomerase I (lanes 4, 5, 8, and 9). As control, oligonucleotides were incubated in the absence of p53 either alone (lane1), with topoisomerase I (lane 10), with PAb 421 (lane 11), or in the presence of both topoisomerase I and PAb 421 (lane12).
Latent and Activated Wild-Type p53 Stimulate the Catalytic Activity of Topoisomerase I Similarly.
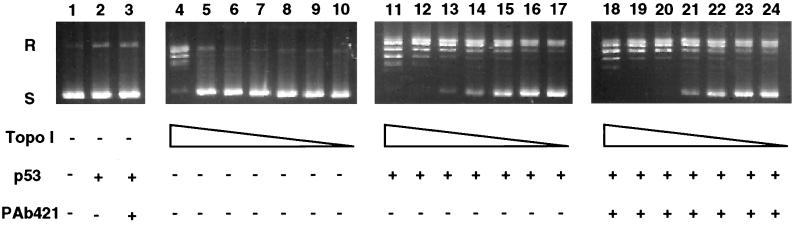
Purified recombinant human topoisomerase I and p53 were used to compare the ability of latent and activated wild-type p53 to stimulate the catalytic activity of topoisomerase I. The presence of p53 clearly stimulated the relaxation of supercoiled DNA by topoisomerase I (Fig. 2, compare lanes 4–10 with lanes 11–17). However, no differences were observed between latent and activated forms of p53 (Fig. 2, compare lanes 11–17 with lanes 18–24).
Figure 2.
Latent and activated wild-type p53 stimulate the catalytic activity of topoisomerase I in a similar manner. Supercoiled plasmid DNA was incubated in the presence of decreasing concentrations of topoisomerase I (45, 22, 15, 11, 9.5, and 2 ng) either alone (lanes 4–10), with 300 ng of p53 (lanes 11–17), or in the presence of 300 ng of p53 and 1 μg of PAb 421 (lanes 18–24). As controls, plasmid DNA was incubated without topoisomerase I either alone (lane1), with p53 (lane 2), or with p53 and PAb 421 (lane 3). S, supercoiled plasmid DNA; R, relaxed DNA.
Latent and Activated Wild-Type p53 Stimulate the DNA Cleavage Activity of Topoisomerase I Similarly.
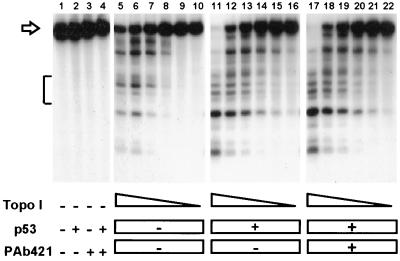
To determine the influence of p53 on topoisomerase I-mediated DNA cleavage, topoisomerase I was incubated with end-labeled plasmid DNA and camptothecin in the absence or presence of p53. The resulting covalent DNA–topoisomerase I complexes mask a single-stranded DNA break that can be revealed by proteolysis and DNA strand separation. The presence of topoisomerase I resulted in a dose-dependent increase in DNA cleavage, as revealed by a decrease in the amounts of full-length plasmid DNA (indicated by an arrow) and a concomitant increase in the appearance of cleavage products (Fig. 3, lanes 5 to 10). The presence of latent or activated wild-type p53 resulted in a comparable increase in topoisomerase I-mediated DNA cleavage for all topoisomerase I concentrations tested (Fig. 3, compare lanes 11–16 and 17–22). Interestingly, the presence of p53 partly changed the cleavage pattern of topoisomerase I, as indicated by the appearance of a new cleavage site and by differences in relative cleavage frequencies within a specific region (indicated with a bracket).
Figure 3.
Latent and activated wild-type p53 stimulate the DNA cleavage activity of topoisomerase I in a similar manner. End-labeled plasmid DNA was incubated in the presence of decreasing concentrations of topoisomerase I (400, 200, 100, 50, 25, and 12.5 ng) either alone (lanes 5–10), with 300 ng of p53 (lanes 11–16), or in the presence of 300 ng of p53 and 1 μg of PAb 421 (lanes 17–22). As controls, substrate DNA was incubated without topoisomerase I either alone (lane 1), with p53 (lane 2), with PAb 421 (lane 3), or with both p53 and PAb 421 (lane 4). The open arrow marks the migration of intact substrate DNA, whereas the bracket indicates a region with differential DNA cleavage in the presence and absence of p53.
Mutant and Truncated p53 Proteins Can Activate Topoisomerase I.
The ability of different forms of recombinant p53 to stimulate the catalytic activity of topoisomerase I was further studied. Full-length GST–p53 fusion protein, which lacks sequence-specific DNA binding (Fig. 1), is able to activate topoisomerase I, as is GST–p53 with an Arg175His mutation (Table 1). GST–p53 containing amino acids 1–362 was also able to activate topoisomerase I, whereas GST–p53 containing amino acids 320–393 was not. Because GST–p53 containing amino acids 299–390 from murine p53 (corresponding to amino acids 302–393 of human p53) has been reported to stimulate topoisomerase I (25), the interaction domain with topoisomerase I is most likely between amino acids 302–320. In agreement, a polypeptide containing amino acids 302–321 of human p53 also activated topoisomerase I, whereas control peptides had no effect or even inhibited the topoisomerase reaction under similar experimental conditions (results not shown).
Table 1.
Stimulation of topoisomerase I activity
| Proteins | Amino acids | Stimulation of topoisomerase I activity |
|---|---|---|
| GST–hp53 | 1–393 (wild type) | Yes |
| GST | — | No |
| GST-hp53 | Arg 175 His | Yes |
| GST-hp53 | 1–362 | Yes |
| GST-hp53 | 320–393 | No |
| GST-hp53* | 299–390 | Yes |
| Peptide | 302–321 | Yes |
After Albor et al., 1998 (25).
Transfection of p53 into p53-Null Cells Is Associated with Increased Topoisomerase I Activity.
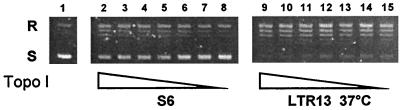
To determine whether p53 also stimulates topoisomerase I in living cells, the catalytic activities of topoisomerase I in nuclear extracts from different M1 cell lines were determined. S6 parental cells, which do not express p53 protein or mRNA, were transfected with a vector coding for a temperature-sensitive p53 protein containing the Ala135Val mutation. The transfected LTR13 cells constitutively express important levels of predominantly nuclear p53, which has mutant conformation at 37°C and wild-type conformation at 32°C (31, 32). Expression of either mutant (Fig. 4) and wild-type (results not shown) p53 was associated with an increase in the catalytic activity of topoisomerase I. Previous results have shown that p53 and topoisomerase I interact physically in the nucleus of the p53-transfected cells (24). Therefore, the interaction between topoisomerase I and both mutant and wild-type p53 is associated with a clear stimulation of topoisomerase I activities in transfected cells in the absence of DNA damage.
Figure 4.
Transfection of p53 into p53-null cells is accompanied by increased topoisomerase I activity. Nuclear extracts were prepared from M1 cells expressing no p53 (S6 cells) or mutant p53 (LTR13 cells at 37°C). The catalytic activity of topoisomerase I was determined by relaxation of supercoiled DNA in the presence of decreasing concentrations of nuclear extracts. S, supercoiled DNA; R, relaxed DNA.
The p53–Topoisomerase I Interaction in Nontransfected Cells Depends on p53 Status.
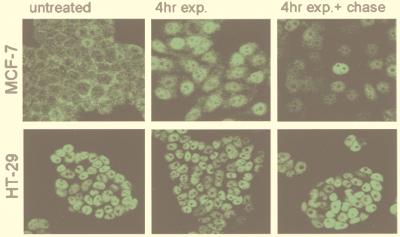
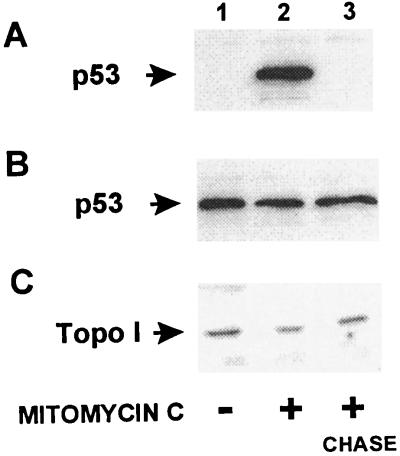
In order for topoisomerase I and p53 to interact in nontransfected cells, the two proteins have to be present in the same place at the same time. Topoisomerase I is predominantly a nuclear protein in both proliferating and growth-arrested cells (33, 34). In contrast, p53 may not always be present in the nucleus. Therefore, the cellular localization of p53 in untreated proliferating cells, in cells treated with the DNA-damaging agent mitomycin C, and in cells treated with mitomycin C followed by 20-hour chase in fresh growth medium was determined by immunohistochemistry (Fig. 5). Untreated MCF-7 cells, which have wild-type p53, exhibit weak cytoplasmic fluorescence. Mitomycin C treatment results in a strongly increased nuclear fluorescence, which, after a 20-hour chase in drug-free medium, is greatly diminished in most cells. The situation is different for HT-29 cells, which contain the Arg273His p53 mutation (35), because all HT-29 cells exhibit a strong nuclear fluorescence independent of whether the cells have been treated with mitomycin C or not. The nuclear localization of p53 was further confirmed by Western blot analysis of nuclear extracts (Fig. 6 A and B). In contrast, the nuclear levels of topoisomerase I in both MCF-7 and HT-29 cells were not affected by mitomycin C treatment (Fig. 6C and results not shown).
Figure 5.
Immunolocalization of p53 in cells with wild-type or mutant p53. MCF-7 breast carcinoma cells (wt p53) and HT-29 colon carcinoma cells (mutant p53) were treated with the DNA-damaging agent mitomycin C (10 μg/ml) for 4 hours followed by a chase in drug-free media for an additional 20 hours. The cellular localization of p53 was determined by immunohistochemistry with the PAb 1801 monoclonal anti-p53 antibody followed by FITC-conjugated anti-mouse secondary antibodies.
Figure 6.
Western blot analysis of nuclear extracts from mitomycin C-treated cells. MCF-7 breast carcinoma cells (wt p53) and HT-29 colon carcinoma cells (mutant p53) were treated with the DNA-damaging agent mitomycin C (10 μg/ml) for 4 hours followed by a chase in drug-free media for an additional 20 hours. Nuclear extracts from MCF-7 cells (A and C) and HT-29 cells (B) were subjected to Western blot analysis with antibodies directed toward p53 (A and B) or topoisomerase I (C).
Both Wild-Type and Mutant p53 Proteins Are Associated with Topoisomerase I.
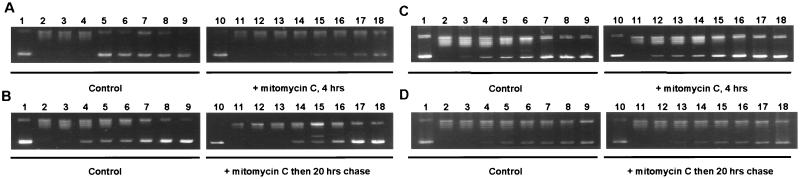
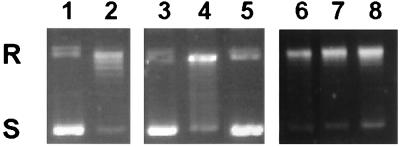
The influence of nuclear p53 on the catalytic activity of topoisomerase I was determined by the capacity of partially purified nuclear extracts to relax supercoiled DNA. The results show that the nuclear translocation of p53 in mitomycin-treated MCF-7 cells is associated with a strong increase in topoisomerase I activity (Fig. 7 A and B). After a 20-hour incubation in drug-free medium, the catalytic activity of topoisomerase I had diminished and was only slightly above what is observed for untreated control cells. In contrast, mitomycin C treatment had no detectable effect on the catalytic activity of topoisomerase I in HT-29 cells (Fig. 7 C and D). To establish whether the activation of topoisomerase I in MCF-7 cells was due to the formation of topoisomerase–p53 complexes, nuclear p53 was immunoprecipitated, and the coimmunoprecipitation of topoisomerase I was determined by relaxation of supercoiled DNA. Immunoprecipitates from mitomycin C-treated MCF-7 cells contained topoisomerase I activity (Fig. 8, lane 4). This cannot be because of cross-reactivity of the p53 antibody, because neither untreated control cells nor treated cells subjected to a 20-hour chase showed any detectable DNA-relaxation activity (Fig. 8, lanes 3 and 5). Similar experiments with HT-29 cells showed that all p53 immunoprecipitates contain topoisomerase I activities, strongly suggesting that the interaction between p53 and topoisomerase I is independent of DNA damage in these cells (Fig. 8, lanes 6–8). The disappearance of the supercoiled substrate is unlikely to be a result of a nonspecific coprecipitating nuclease, as indicated by the presence of different DNA isomers typical of topoisomerase reactions (Fig. 8, lanes 4 and 8). Furthermore, no DNA relaxation was observed in the presence of the specific topoisomerase I inhibitor, camptothecin, strongly suggesting that the relaxation is mediated by topoisomerase I and not by another topoisomerase (results not shown).
Figure 7.
Catalytic activity of topoisomerase I in nuclear extracts from mitomycin C-treated cells. MCF-7 breast carcinoma cells (wt p53, A and B) and HT-29 colon carcinoma cells (mutant p53, C and D) were treated with the DNA-damaging agent mitomycin C (10 μg/ml) for 4 hours followed by a chase in drug-free media for an additional 20 hours. Different dilutions of partially purified nuclear extracts were tested for topoisomerase I activities as measured by relaxation of supercoiled DNA. The catalytic activity of untreated control cells was compared with either treated cells (A and C) or with treated cells postincubated in drug-free medium for 20 hours (B and D).
Figure 8.
Topoisomerase I and p53 are physically associated in mitomycin C-treated cells. MCF-7 breast carcinoma cells (wt p53) and HT-29 colon carcinoma cells (mutant p53) were treated with the DNA-damaging agent mitomycin C (10 μg/ml) for 4 hours followed by a chase in drug-free media for an additional 20 hours. Nuclear extracts were treated with PAb 1801 monoclonal anti-p53 antibodies, and the topoisomerase I activity associated with the p53 immunoprecipitates was determined by relaxation of supercoiled DNA. Lanes 3–8 show the catalytic activity of p53 immunoprecipitates from untreated control cells (lanes 3 and 6), cells treated with mitomycin C for 4 hours (lanes 4 and 7), or cells treated with mitomycin C for 4 hours followed by a 20-hour chase in drug-free media (lanes 5 and 8) obtained from either MCF-7 cells (lanes 3–5) or HT-29 cells (lanes 6–8). In comparison, the first two lanes show the migration of the DNA substrate alone (lane 1) or in the presence of purified topoisomerase I (lane 2). S, supercoiled DNA; R, relaxed DNA.
DISCUSSION
Topoisomerase I and p53 are part of the DNA damage response at several different stages. Topoisomerase I is able to recognize different types of DNA lesions, including UV-induced cyclobutane pyrimidine dimers (14–17). This is an early response to DNA damage, because covalent DNA–topoisomerase I complexes can be detected within minutes in UV-irradiated HeLa cells (17). Topoisomerase I is known to interact with both isolated and TFIID-associated TATA box-binding protein (4, 5). It has recently has been shown that TATA box-binding protein/TFIID binds selectively to, and is sequestered by, cisplatin- or UV-damaged DNA in the context of a multiprotein complex including TFIIH, which contains the XPB and XPD (Xeroderma pigmentosum complementation groups B and D) helicases (36). Thus, the mobilization of topoisomerase I is likely part of a complex response to DNA damage resulting in the assembly of multiprotein damage-recognition complexes.
In contrast to topoisomerase I, the mobilization of p53 appears to be a delayed response. Elevated levels of wild-type, nuclear p53 are rarely, if ever, present immediately after the occurrence of DNA damage. In MCF-7 cells, nuclear p53 is first observed after at least 1 hour of treatment with mitomycin C or cisplatin to reach a plateau after 4 to 6 hours of treatment. Even in cell types where p53 is not cytoplasmic, the base levels of nuclear p53 are very low and depend on new protein synthesis for up-regulation. Relatively little is known about the upstream events that lead to translocation and up-regulation of p53 after genotoxic lesions, although formation of DNA strand breaks seems to be an important factor (37). These lesions can be generated either directly by DNA damage inflicted by ionizing radiation or by repair intermediates such as those present in nucleotide excision repair, the major type of repair to UV damage. It has previously been shown that topoisomerase I-concealed DNA strand breaks can be converted into frank DNA strand breaks by the helicase-driven separation of duplex DNA that occurs during DNA replication and DNA repair (38–40). Thus, it is likely that topoisomerase I/helicase-induced DNA strand breaks might contribute to the initial activation of p53 after genotoxic stress.
It has been shown that wild-type p53 isolated from mammalian cells exists in both latent and activated forms (41). Therefore, it is tempting to speculate that the interaction with topoisomerase I and other components of the TFIID and TFIIH complexes might serve to target p53 toward the DNA lesions, where the sequence-specific binding of p53 is activated. Although topoisomerase I by itself is not able to activate the sequence-specific DNA binding of p53, it may influence the activation of p53. At least two proteins known to associate with topoisomerase I, casein kinase II and HMG-1, are able to activate the sequence-specific DNA binding of p53 (19, 30, 42, 43). Furthermore, the amino acids 302–321 of p53, which contain the putative topoisomerase I-binding domain, are also part of the casein kinase II binding region (amino acids 287–340) (44), suggesting that topoisomerase I may influence the phoshorylation of p53 by casein kinase II. Taken together, these results suggest that wild-type p53 may be activated in the vicinity of DNA lesions and fine-tuned by interaction between the different components of the damage-recognition complex.
p53 contains an acidic transactivation domain that stimulates transcription of genes with a p53-binding site in their promoter region. The transcription is further stimulated if the consensus site is proximal to TATA sequences (45). In addition, p53 can repress genes that lack the p53-binding site but contain TATA elements (45). It is interesting that the topoisomerase I-mediated stimulation of transcription depends on the presence of both acidic activators and TFIID, whereas repression of basal transcription can be mediated be either TFIID or TATA box-binding protein (4, 5). Therefore, it is possible that topoisomerase I might cooperate with p53 in both stimulation and repression of gene transcription. Another interesting aspect is that the putative topoisomerase I binding site on p53 contains Ser-315. This residue is the phosphorylation site for cdc2 kinase and has been reported to activate the sequence-specific binding of p53 in a promoter-dependent manner (28).
In contrast to the highly regulated, transient interaction between topoisomerase I and wild-type p53, topoisomerase I is constitutively associated with mutant p53. This is particularly important because many tumor cells, like HT-29, contain elevated levels of nuclear, mutant p53. A nonregulated, continuous activation of topoisomerase I is likely to increase the level of genetic instability, considering the potent recombinase activities of topoisomerase I (8, 11).
It is generally accepted that the prevalence of mutant p53 in human tumors suggests that mutant p53 somehow contributes to the oncogenic process. Given the considerable efforts in this area, it now seems unlikely that the effects of mutant p53 are mediated by transcription of new gene products. In contrast, many forms of mutant p53 preserve the ability to associate with other cellular proteins, as documented here for the interaction with topoisomerase I. Whereas the interaction between wild-type p53 and topoisomerase I is highly regulated and only takes place during brief periods of cellular stress, the interaction between mutant p53 and topoisomerase I is deregulated and results in constitutive stimulation of topoisomerase I. This is likely to increase the cellular levels of illegitimate recombination, thereby leading to increased genetic instability and oncogenic progression. Thus, we propose a model in which the ability of mutant p53 to associate with other nuclear proteins in a nonregulated manner leads to constant activation of certain proteins such as topoisomerase I and probably to constitutive inhibition of others. This feature of mutant p53 might be the crucial factor leading to selection of tumor cells with mutant p53 protein.
Acknowledgments
This work was supported in part by Fondation de France and by the French-Polish Scientific and Technological Cooperation project of the Ministère des Affaires Etrangères, France and the Committee for Scientific Research (KBN) Poland (98192). Céline Gobert is a fellow of La Ligue Nationale Contre le Cancer. We thank Madame Ginette Chyzak for her expert assistance with the DNA relaxation assays.
ABBREVIATIONS
- TFIID
transcription factor IID
- GST
glutathione S-transferase
References
- 1.Wang J C. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 2.Fleischmann G, Pflugfelder G, Steiner E K, Javaherian K, Howard G C, Wang J C, Elgin S C. Proc Natl Acad Sci USA. 1984;81:6958–6962. doi: 10.1073/pnas.81.22.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egyhazi E, Durban E. Mol Cell Biol. 1987;7:4308–4316. doi: 10.1128/mcb.7.12.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merino A, Madden K R, Lane W S, Champoux J J, Reinberg D. Nature (London) 1993;363:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 5.Kretzschmar M, Meisternst M, Roeder R G. Proc Natl Acad Sci USA. 1993;90:11508–11512. doi: 10.1073/pnas.90.24.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossi F, Labourier E, Fomé T, Divita G, Derancourt J, Riou J-F, Antoine E, Cathala G, Brunel C, Tazi J. Nature (London) 1996;381:80–82. doi: 10.1038/381080a0. [DOI] [PubMed] [Google Scholar]
- 7.Tazi J, Rossi F, Labourier E, Gallouzi I-E, Brunel C, Antoine E. J Mol Med. 1997;75:786–800. doi: 10.1007/s001090050168. [DOI] [PubMed] [Google Scholar]
- 8.Cheng C, Kussie P, Pavletich N, Shuman S. Cell. 1998;92:841–850. doi: 10.1016/s0092-8674(00)81411-7. [DOI] [PubMed] [Google Scholar]
- 9.Bullock P, Champoux J J, Botchman M. Science. 1985;230:954–958. doi: 10.1126/science.2997924. [DOI] [PubMed] [Google Scholar]
- 10.Christiansen K, Westergaard O. J Biol Chem. 1994;269:721–729. , 1994. [PubMed] [Google Scholar]
- 11.Zhu J, Schiestl R H. Mol Cell Biol. 1996;16:1805–1812. doi: 10.1128/mcb.16.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson R D, Berger N A. Mutat Res. 1994;309:109–142. doi: 10.1016/0027-5107(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 13.Larsen A K, Skladanowski A. Biochem Biophys Acta. 1998;1400:257–274. doi: 10.1016/s0167-4781(98)00140-7. [DOI] [PubMed] [Google Scholar]
- 14.Yeh Y C, Liu L F, Ellis C A, Lu A L. J Biol Chem. 1994;269:15498–15504. [PubMed] [Google Scholar]
- 15.Pourquier P, Ueng L M, Kohlhagen G, Mazzumder A, Gupta M, Kohn K W, Pommier Y. J Biol Chem. 1997;272:7792–7796. doi: 10.1074/jbc.272.12.7792. [DOI] [PubMed] [Google Scholar]
- 16.Lanza L, Tornaletti S, Rodolfo C, Scanavini M C, Pedrini A M. J Biol Chem. 1996;271:6978–6986. doi: 10.1074/jbc.271.12.6978. [DOI] [PubMed] [Google Scholar]
- 17.Subramanian D, Rosenstein B S, Muller M T. Cancer Res. 1998;58:976–984. [PubMed] [Google Scholar]
- 18.Kern S E, Kinzler K W, Bruskin A, Jarosz D, Friedman P, Prives C, Vogelstein B. Science. 1991;252:1708–1711. doi: 10.1126/science.2047879. [DOI] [PubMed] [Google Scholar]
- 19.Hupp T R, Meek D W, Midgley C A, Lane D P. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 20.Ko L J, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 21.Hollstein M, Sidransky D, Vogelstein B, Harris C C. Science. 1991;253:49–52. doi: 10.1126/science.1905840. , 1991. [DOI] [PubMed] [Google Scholar]
- 22.Levine A J, Zambetti G P. FASEB J. 1993;7:855–865. doi: 10.1096/fasebj.7.10.8344485. [DOI] [PubMed] [Google Scholar]
- 23.Gobert C, Bracco L, Rossi F, Olivier M, Tazi J, Lavelle F, Larsen A K, Riou J-F. Biochemistry. 1996;35:5778–5786. doi: 10.1021/bi952327w. [DOI] [PubMed] [Google Scholar]
- 24.Larsen A K, Gobert C, Gilbert C, Markovits J, Bojanowski K, Skladanowski A. Acta Biochim Pol. 1998;45:535–544. [PubMed] [Google Scholar]
- 25.Albor A, Kaku S, Kulesz-Martin M. Cancer Res. 1998;58:2091–2094. [PubMed] [Google Scholar]
- 26.Zieve G W, Penman S. J Mol Biol. 1981;145:501–523. doi: 10.1016/0022-2836(81)90542-8. [DOI] [PubMed] [Google Scholar]
- 27.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Prives C. Nature (London) 1995;376:88–91. doi: 10.1038/376088a0. [DOI] [PubMed] [Google Scholar]
- 29.Larsen A K, Grondard L, Couprie J, Desoize B, Comoe L, Jardillier J-C, Riou J-F. Biochem Pharmacol. 1993;46:1403–1412. doi: 10.1016/0006-2952(93)90105-6. [DOI] [PubMed] [Google Scholar]
- 30.Jayaraman L, Moorthy N C, Murrthy K G, Manley J L, Bustin M, Prives C. Genes Dev. 1998;15:462–472. doi: 10.1101/gad.12.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yonish-Rouach E, Resnitzky D, Lotem J, Sachs L, Kimchi A, Oren M. Nature (London) 1991;352:345–347. doi: 10.1038/352345a0. [DOI] [PubMed] [Google Scholar]
- 32.Skladanowski A, Larsen A K. Cancer Res. 1997;57:818–823. [PubMed] [Google Scholar]
- 33.Heck M M, Hittelman W N, Earnshaw W S. Proc Natl Acad Sci USA. 1988;85:1086–1090. doi: 10.1073/pnas.85.4.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker S D, Wadkins R M, Stewart C F, Beck W T, Danks M K. Cytometry. 1995;19:134–145. doi: 10.1002/cyto.990190208. [DOI] [PubMed] [Google Scholar]
- 35.Rodrigues N R, Rowan A, Smith M E F, Kerr I B, Bodmer W F, Gannon J V, Lane D P. Proc Natl Acad Sci USA. 1990;87:7555–7559. doi: 10.1073/pnas.87.19.7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vichi P, Coin F, Renaud J-P, Vermeulen W, Hoeijmakers J H J, Moras D, Egly J-M. EMBO J. 1997;16:7444–7456. doi: 10.1093/emboj/16.24.7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson W G, Kastan M B. Mol Cell Biol. 1994;14:1815–1823. doi: 10.1128/mcb.14.3.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holm C, Covey J M, Kerrigan D, Pommier Y. Cancer Res. 1989;49:6365–6368. [PubMed] [Google Scholar]
- 39.Matson S W, Bean D W, George J W. BioEssays. 1994;16:13–22. doi: 10.1002/bies.950160103. [DOI] [PubMed] [Google Scholar]
- 40.Hsiang Y H, Lihou M G, Liu L F. Cancer Res. 1989;49:5077–5082. [PubMed] [Google Scholar]
- 41.Hupp T R, Lane D P. Curr Biol. 1994;4:865–875. doi: 10.1016/s0960-9822(00)00195-0. [DOI] [PubMed] [Google Scholar]
- 42.Kordiyak G J, Jakes S, Ingebritsen T S, Benbow R M. Biochemistry. 1994;33:13484–13491. doi: 10.1021/bi00249a037. [DOI] [PubMed] [Google Scholar]
- 43.Javaherian K, Liu L F. Nucleic Acids Res. 1983;11:461–472. doi: 10.1093/nar/11.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Appel K, Wagner P, Boldyreff B, Issinger O-G, Montenarh M. Oncogene. 1995;11:1971–1978. [PubMed] [Google Scholar]
- 45.Stengler J E, Tegtmeyer P, Mayr G A, Reed M, Wang Y, Wang P, Hough P V C, Mastrangelo I A. EMBO J. 1994;13:6011–6020. doi: 10.1002/j.1460-2075.1994.tb06947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mack D H, Vartikar J, Pipas J M, Laimins L A. Nature (London) 1993;363:281–283. doi: 10.1038/363281a0. [DOI] [PubMed] [Google Scholar]