Abstract
The aconitase protein of Bacillus subtilis was able to bind specifically to sequences resembling the iron response elements (IREs) found in eukaryotic mRNAs. The sequences bound include the rabbit ferritin IRE and IRE-like sequences in the B. subtilis operons that encode the major cytochrome oxidase and an iron uptake system. IRE binding activity was affected by the availability of iron both in vivo and in vitro. In eukaryotic cells, aconitase-like proteins regulate translation and stability of iron metabolism mRNAs in response to iron availability. A mutant strain of B. subtilis that produces an enzymatically inactive aconitase that was still able to bind RNA sporulated 40× more efficiently than did an aconitase null mutant, suggesting that a nonenzymatic activity of aconitase is important for sporulation. The results support the idea that bacterial aconitases, like their eukaryotic homologs, are bifunctional proteins, showing aconitase activity in the presence of iron and RNA binding activity when cells are iron-deprived.
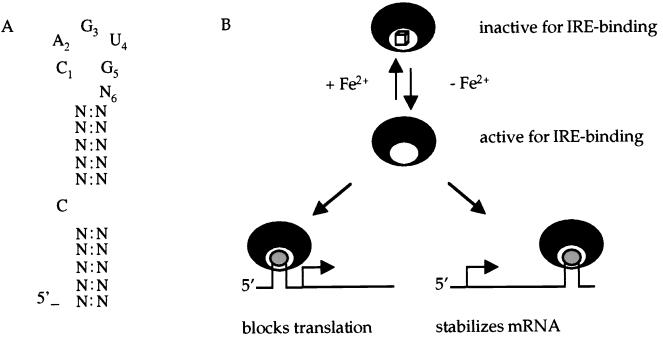
An important component of the response to iron deprivation and oxidative stress in eukaryotic cells depends on a nonenzymatic activity of cytosolic aconitase (ACN) and other iron regulatory proteins (IRPs) (1, 2). ACN enzymes interconvert citrate and isocitrate. IRPs, whether they have enzymatic activity or not, bind to iron-response elements (IREs), i.e., conserved stem-loop structures, in various mRNAs encoding proteins involved in iron sequestration and either protect the mRNAs against degradation or inhibit their translation (Fig. 1). For instance, the IRE of rabbit ferritin mRNA is located in the 5′ untranslated region (UTR); when this region is bound by a cytosolic ACN (also known as IRP1), translation of ferritin mRNA is inhibited (3). On the other hand, the IRE of transferrin receptor mRNA is located in the 3′ UTR; binding of IRPs to this region increases the half-life of the mRNA and thereby increases accumulation of transferrin receptor (4, 5). In either case, binding only occurs when cells are depleted of iron or, in vitro, when iron is absent (6). ACNs are iron-requiring enzymes, containing a 4Fe-4S cluster (2). Thus, when iron is limiting, enzymatic activity of cytosolic aconitase (IRP1) is greatly reduced whereas RNA binding is increased.
Figure 1.
Iron responsive elements and iron regulatory proteins. (A) Consensus sequence of the eukaryotic IRE (1). (B) IREs are stem-loop structures located in 5′ or 3′ UTRs (shown in gray) of mRNAs encoding proteins involved in iron metabolism. If the IRE is located in the 5′ UTR, binding of IRP (black crescents) inhibits initiation of translation and the level of the protein product decreases. If the IRE is located in the 3′ UTR, binding of IRP protects the mRNA against degradation and the level of the protein increases. In the presence of iron, IRPs are converted to a form that is inactive for RNA binding but may have aconitase enzyme activity. The cube signifies a 4Fe-4S cluster that is essential for aconitase enzyme activity.
Cytosolic ACNs belong to a family of highly conserved proteins among prokaryotes and eukaryotes (7). Experiments in prokaryotes have suggested a role for aconitase in iron metabolism and virulence (8, 9). Because Bacillus subtilis aconitase is required for sporulation, an iron-dependent process (10–13), we were led to test whether B. subtilis ACN is an iron-responsive RNA binding protein.
MATERIALS AND METHODS
Strains, Plasmids, and Growth Conditions.
The bacterial strains and plasmids used are listed in Table 1. B. subtilis strains were grown in liquid DS medium (0.8% nutrient broth/0.1% KCl/0.025% MgSO4-7H2O/1 mM Ca(NO3)2/10 μM MnCl2/1 μM FeSO4). For plates, agar was added to 17 g per liter. In some cases, FeSO4 was omitted, as indicated. Spore formation was quantitated by measuring the fraction of cells in an overnight culture that survived treatment at 80°C for 10 min. The oligonucleotides used are listed in Table 2.
Table 1.
Strains and plasmids used in this study
| Name | Genotype | Reference or source |
|---|---|---|
| B. subtilis strains | ||
| SMY | Wild-type B. subtilis strain | Lab stock |
| JH642 | trpC2 pheA1 | J. A. Hoch (Scripps Research Institute, La Jolla, CA) |
| MAB160 | trpC2 pheA1 Ω citB∷spc | Ref. 10 |
| CA4 | pheA1 citB517 | This work |
| Plasmids | ||
| pTZM1 | Rabbit ferritin IRE under T7 promoter | Ref. 14 |
| pBluescript SK(−) | Stratagene | |
| pCA60 | OCA1 and OCA2 were annealed and ligated to pBluescript cut with XbaI-HindIII | This work |
| pCA61 | OCA3 and OCA4 were annealed and ligated to pBluescript cut with XbaI-HindIII | This work |
Table 2.
Oligonucleotides used in this study
| Name | Sequence | Description |
|---|---|---|
| OCA1 | AGCTTAAAAAACCCTCTTCAGTGGAAGAGGGTTTTTT | qoxD sense strand (HindIII overhang) |
| OCA2 | CTAGAAAAAACCCTCTTCCACTGAAGAGGGTTTTTTA | qoxD antisense strand (XbaI overhang) |
| OCA3 | CTAGACAAAACTAATTCAGCGTAGGTTTTTGTA | feuAB antisense strand (XbaI overhang) |
| OCA4 | AGCTTACAAAAACCTACTCTGAATTAGTTTTGT | feuAB sense strand (HindIII overhang) |
| OCA5 | GTAGTAGATCTGGCTTCACTGCGT | citB sequence (5′ to 3′) from nucleotides 310–333 |
| OCA6 | AAGCGGACCTTAGTTCCCGATAGCTGTTGTACAGCCGTACCCAACGAG | citB sequence (3′ to 5′) from nucleotides 1578–1531 |
| OCA7 | CTCGTTGGGTACGGCTGTACAACAGCTATCGGGAACTCAGGTCCGCTT | citB sequence (5′ to 3′) from nucleotides 1531–1578 |
| OCA8 | TCAGGACTGCTTCATTTTTTCACG | citB sequence (3′ to 5′) from nucleotides 2707–2730 |
The bases changed in order to mutate aconitase residue cysteine 517 to alanine are shown in bold.
In Vitro Transcription.
pTZM1, pCA60, and pCA61 were linearized and used as templates for the transcription of rabbit ferritin IRE and of IRE-like sequences located downstream of the qoxD and feuA genes, respectively. Ten-microliter reactions contained 100 ng of DNA template, 20 μCi of α32P-UTP (600 Ci/mmol), 0.5 mM ATP, CTP, and GTP, 50 μM unlabeled UTP, 20–25 units of RNasin (Promega), 10 mM DTT, and 25 units of T7 RNA polymerase. The reactions were incubated at 37°C for 1 hour before adding 10 μg of yeast tRNA and 25 units of RNasin. One unit of RNase-free DNase I was added and the reaction was further incubated for 15 min at 37°C. After extraction with phenol, RNA was precipitated with 0.5 M ammonium acetate and one volume of isopropanol at −20°C. The labeled RNA was dissolved in water. To allow proper folding, the RNA was heated to 85°C and was cooled slowly to 4°C.
Gel Shift Assays.
The radiolabeled RNA fragment (≈1 pmol per reaction) obtained by in vitro transcription, as described above, was incubated with protein samples in 20 μl reactions containing 10 mM Tris⋅HCl (pH 8.0), 50 mM KCl, 1 μg of total yeast RNA, and 10% glycerol. The reactions were incubated for 15 min at room temperature and then were loaded on a running 4% nondenaturing polyacrylamide gel.
Determination of ACN Activity and Activation of the Enzyme.
Enzymatic activity was determined by incubation of 5–20 μl of protein preparation in a 1-ml reaction containing 90 mM Tris⋅HCl (pH 8) and isocitrate (20 mM) as substrate. One unit is defined as the amount of enzyme necessary to increase the OD at 240 nm by 0.0003 (because of accumulation of cis-aconitate). Aconitase-specific activity is expressed in units of activity per minute per milligram of protein. In some cases, aconitase was activated by treatment with 8 mM DTT and 0.8 mM Fe(NH4)2(SO4)3 in Tris⋅HCl buffer (50 mM, pH 8) for 10 min at 25°C.
Mutagenesis of citB.
To introduce a change in codon 517 of the aconitase gene of B. subtilis, a two-step PCR strategy was used. In the first PCR, the N-terminal segment (codons 104 to 526) and a C-terminal segment (codons 511 to 910) of citB were amplified by using oligonucleotides OCA5, OCA6, OCA7, and OCA8, respectively (Table 2). OCA6 and OCA7 are complementary and both carry the desired mutation [TGT (cysteine) to GCT (alanine)]. The resulting DNA was used as a template for a second PCR using the external oligonucleotides (OCA5 and OCA8). The second PCR product then was directly introduced into strain JH642 (phe trp) by cotransformation with the chromosomal DNA of a tryptophan prototroph. Transformants were selected on minimal medium containing phenylalanine but lacking tryptophan. To find mutants lacking aconitase enzyme activity, the resulting colonies were screened for glutamine auxotrophy. The lack of aconitase activity was further confirmed by enzymatic assay (data not shown).
Searching for IRE-Like Sequences.
Two approaches were used to search for IRE-like sequences from the sequenced B. subtilis genome (15). First, the B. subtilis genome was scanned by using the rnabob program (http://www.genetics.wustl.edu/eddy/software/#rnabob) for a stem-loop structure with the sequence N′ N′ N′ N′ C N′ N′ N′ N′ C A G U G N N" N" N" N" N" N" N" N" (where N is any base and N′ and N" are complementary but otherwise not specified). Next, the genome was scanned for the sequence C N N N N N C A G U G located within 100 base pairs upstream of a start codon or within 100 base pairs downstream of a stop codon by using the pattern search program of subtilist (http://www.pasteur.fr/Bio/SubtiList). Sequences to be tested were synthesized as complementary oligonucleotides and were annealed and ligated to pBluescript SK(−).
Preparation of Crude Extracts and Purification of ACN and ACNC517A.
Crude extracts and purified aconitase were prepared as described by Dingman and Sonenshein (11). In brief, to obtain crude extracts, wild-type and citB mutant strains were grown in 200 ml of DS medium without FeSO4 supplementation at 37°C with vigorous aeration. The cells were harvested by centrifugation (12,000 × g, 4°C) and were broken by two passes through a French pressure cell [15,000 psi (1 psi = 6.89 kPa)]. The crude extracts were separated from the cell debris by centrifugation. To obtain purified aconitase, the crude extracts were fractionated by ammonium sulfate precipitation (aconitase precipitated between 60 and 85% of saturation with ammonium sulfate). The pellet was dissolved in 20 mM Tris⋅citrate (pH 7.35) and was desalted and concentrated by using a Centricon 30 Microconcentrator (Amicon). The partially purified aconitase was then sequentially chromatographed on columns of DEAE-Sephacel and Sephadex G-100. Fractions containing aconitase were pooled and were stored at −20°C. Purified rabbit IRP1 was a gift from W. Walden (University of Illinois, Chicago).
RESULTS
B. subtilis Aconitase Is an RNA Binding Protein. As a first test of the ability of B. subtilis aconitase to bind RNA, we assayed the interaction of the enzyme with a 90-nucleotide RNA containing the IRE normally found in the 5′ UTR of rabbit ferritin mRNA (14). The RNA probe was synthesized in vitro and was allowed to fold by heating and slow cooling. Extracts of wild-type cells proved to contain an IRE-interacting component(s), but only when prepared from cells harvested after the end of exponential growth phase (Fig. 2B). This corresponds to the time at which ACN synthesis is induced in B. subtilis (Fig. 2A). Extracts of an aconitase null mutant (MAB160; ref. 10) had no IRE-binding component (Fig. 2B, rightmost lane) or aconitase enzyme activity (10). B. subtilis has a single aconitase gene, citB (10), whose product is similar to AcnA of Escherichia coli (7, 16).
Figure 2.
Ferritin IRE binding activity of B. subtilis cell extracts. (A) Crude extracts of B. subtilis wild-type and MAB160 (citB∷spc) strains were prepared from cells grown in DS medium without iron supplementation and harvested at the four time points indicated (1–4 hours after the end of exponential growth phase). Aconitase activity of the wild-type crude extracts, after activation with iron, is shown for each time point. The MAB160 extracts had no detectable aconitase activity. (B) A rabbit ferritin IRE-containing, radiolabeled 90-nt RNA (≈20 cps per reaction) was mixed with extracts of wild-type or MAB160 (50 μg of total protein per reaction), was incubated at room temperature for 15 min, and then was subjected to polyacrylamide gel electrophoresis.
When a purified preparation of aconitase was used in RNA binding assays as described above, an IRE/aconitase complex also was obtained. The mobility of the complex was the same as that obtained by using cell extracts (Fig. 3A). Moreover, the mobility of the B. subtilis ACN/IRE complex was very similar to that of an IRP1/IRE complex (Fig. 3A). The apparent affinity of B. subtilis aconitase for the ferritin IRE was lower than the affinity of rabbit IRP1 for the same substrate.
Figure 3.
Ferritin IRE binding activity of purified aconitase. (A) Binding reactions were performed as described in Fig. 2 using successive 2-fold dilutions of purified B. subtilis aconitase (see Materials and Methods) starting from 5 μg/ml (lanes 2–5). Lanes 7 and 8 show similar binding reactions using wild-type crude extracts. Lanes 9 and 10 show the complex formed by rabbit IRP1 (5 and 2.5 μg/ml respectively) with the same RNA probe. Unbound RNA was run in lanes 1 and 6. (B) The sequence complementary to the ferritin IRE (asIRE) was synthesized in vitro and was used as a target for binding (lanes 1 and 2). In lanes 3 and 4, the IRE probe was tested with or without B. subtilis ACN (5 μg/ml). (C) Ferritin IRE binding activity of purified ACNC517A. Binding reactions were set up with radiolabeled ferritin IRE and either wild-type or ACNC517A protein preparations (see Materials and Methods). The respective protein concentrations are indicated.
The RNA Binding Activity of B. subtilis Aconitase Is Sequence-Specific.
All of the binding assays described above were performed in the presence of at least 100-fold excess of yeast tRNA. To prove that the binding was sequence-specific, a radioactive RNA corresponding to the complement of the IRE sequence was synthesized and tested in a binding assay. Even though the sequence complementary to the IRE is likely to form a stem-loop structure similar to that of the IRE, there was no formation of a defined complex (Fig. 3B).
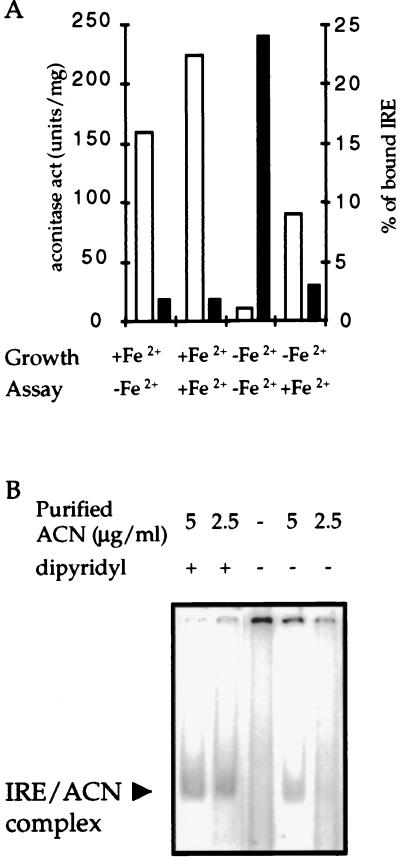
The RNA Binding Activity of B. subtilis Aconitase Is Iron Responsive. IRP1 RNA-binding and aconitase activities are mutually exclusive in an iron-dependent fashion. Incubation of purified IRP1 with iron salts in the presence of a reducing agent decreases binding to RNA while increasing aconitase activity. On the other hand, incubation of the protein with an iron-specific chelator has the opposite effect (6). These characteristics are crucial to the role of IRP1 in iron homeostasis (1, 2). To test whether B. subtilis aconitase also shows an interconversion between enzymatic and RNA-binding forms, crude extracts were obtained from wild-type cultures grown in medium with or without iron supplementation. Neither the level of expression of the citB gene nor the amount of aconitase protein in the extracts was changed as a result of altering the iron concentration in the medium (data not shown). The crude extracts were tested for both aconitase and IRE-binding activities (Fig. 4A) before and after treatment in vitro with iron under reducing conditions. A complex was formed with the extract obtained from the iron-deprived culture but not with the extract from the iron-supplemented culture. On the other hand, high enzymatic activity was only seen when iron was added in vivo or in vitro. Addition of iron to an extract prepared from iron-deprived cells inhibited IRE-binding activity (Fig. 4A).
Figure 4.
Iron-responsiveness of ferritin IRE binding activity. (A) A wild-type strain was grown either in DS medium or in the same medium without addition of FeSO4. Cells were harvested at the start of the stationary phase, and crude extracts were prepared. All samples were assayed for aconitase activity (open bars) and IRE binding activity (filled bars) before and after activation in vitro with Fe2+ and DTT (see Materials and Methods). The resulting binding gels were scanned, and both free and bound IRE were quantified by using the program image quant 1.2 (Molecular Dynamics). (B) Binding reactions of purified aconitase in binding buffer with or without addition of dipyridyl (0.5 mM). In this experiment, the unbound probe ran off the bottom of the gel.
The RNA-binding activity of B. subtilis aconitase was increased by treatment with dipyridyl, a chelator of divalent cations, particularly Fe2+ (Fig. 4B). Iron and dipyridyl had opposing effects on aconitase activity and RNA binding; a concentration of iron that stimulated enzymatic activity inhibited binding (Fig. 4A) whereas a concentration of dipyridyl that inhibited enzyme activity stimulated binding (Fig. 4B). Thus, B. subtilis ACN has both enzymatic and RNA-binding activities that are interconvertible in response to the availability of iron.
ACNC517A, an Enzymatically Inactive B. subtilis ACN, Still Binds IRE.
An enzymatically inactive form of B. subtilis ACN (ACNC517A) was constructed (see Materials and Methods) by changing to alanine one of the three cysteines implicated in iron binding (17). The strain (CA4) that produces ACNC517A is a glutamate auxotroph resulting from the lack of any detectable aconitase activity (data not shown). MAB160, a null citB mutant, and CA4 are both defective in sporulation, but the defect in sporulation was less severe in CA4. Approximately 0.04% of the cells of a CA4 strain culture in DS medium formed heat-resistant spores whereas only 0.001% of MAB160 cells did so. ACNC517A was purified by using the same protocol used for the wild-type protein. The protein was able to bind ferritin IRE, although the mutant had a lower affinity for the IRE than did wild-type aconitase (Fig. 3C). ACNC517A formed a complex with the IRE even when obtained from a culture grown with iron supplementation (Fig. 3C).
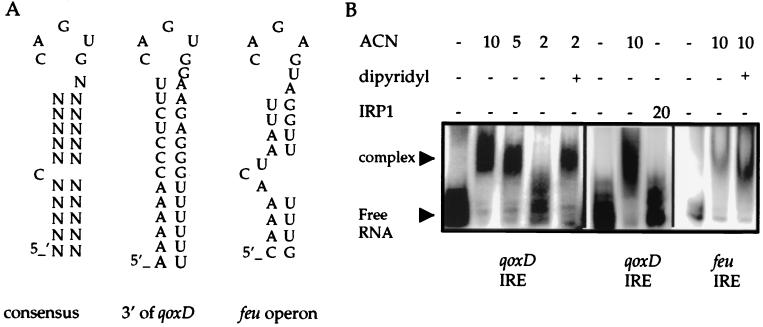
IRE-Like Structures in the B. subtilis Genome. The sequence-specific, iron-responsive RNA-binding activity of B. subtilisACN would be consistent with a role in posttranscriptional regulation of iron metabolism genes. As a first approach to determining which genes are targets for such regulation, we searched the genome for IRE-like structures located in UTRs. Many candidates were found. For our initial analysis, we chose two putative IRE-like sequences, one located in the UTR downstream of the qoxD gene and another located between the feuA and feuB genes (Fig. 5A). The former locus was chosen because it encodes a major iron-containing protein involved in electron transport [the fourth subunit of the cytochrome aa3 oxidase (18)]. The latter was chosen because it is involved in iron uptake (19).
Figure 5.
B. subtilis IRE-like structures. (A) Sequences of the consensus eukaryotic IRE and putative IREs located 3′ of the qoxD coding sequence and in-between the feuA and feuB genes of B. subtilis. (B) The sequences shown in A were cloned and transcribed in vitro and were tested for binding to purified B. subtilis aconitase preparations. Similar binding reactions were performed by using a purified preparation of rabbit IRP1. Dipyridyl, to a final concentration of 0.5 mM, was added to the binding reactions as indicated.
Both IRE-like sequences were cloned downstream of a T7 promoter and were transcribed in vitro to obtain radiolabeled IRE-like structures. Aconitase was able to bind to both putative B. subtilis IREs (Fig. 5B). The affinity of the binding was increased in the presence of dipyridyl, suggesting that the binding to the IRE-like sequences is affected by the availability of iron. Rabbit IRP1 was not able to bind to the putative IRE located downstream of the qoxD gene at the concentration tested (Fig. 5B).
DISCUSSION
Iron metabolism is tightly regulated in both eukaryotes and prokaryotes (1, 20) because iron is both vital and potentially toxic for the cell. Although iron-containing enzymes fulfill essential functions such as electron transport and deoxynucleotide synthesis, high levels of iron catalyze the formation of highly reactive free radicals leading to oxidative damage to DNA and other cellular components (21–23). Posttranscriptional regulation by IRPs has been shown to be an important part of such regulation in eukaryotes (1). Although prokaryotic ACNs are highly similar to IRPs (7), they had not previously been shown to have such regulatory activity. In this work we present direct evidence that B. subtilis ACN is an RNA-binding protein and is potentially involved in regulation of iron metabolism in B. subtilis. If so, such regulation would be in addition to transcriptional regulation of iron metabolism genes mediated by the Fur protein (24).
Although we have shown that B. subtilis ACN is able to bind specifically to IRE-like sequences and that the presence of iron mediates the conversion between the enzymatic and RNA binding activities of the protein, the mechanistic link between aconitase and iron metabolism is still unclear. Further research is necessary to reveal the ways in which the reported RNA binding activity influences B. subtilis physiology. Indirect evidence for such a regulatory role comes from the sporulation phenotypes of aconitase mutant strains. A citB null mutant is severely impaired in sporulation, being blocked at stage 0 (10). A similar block is seen if wild-type cells are grown under conditions of iron limitation (C.A., R. Pagliarini, and A.L.S., unpublished work). In this work, we show that a strain that produces a mutant form of ACN that lacks enzymatic activity but is still able to bind RNA sporulates 40× more efficiently than does the null mutant, even though neither mutant produces any detectable ACN activity. This result suggests a nonenzymatic role for the B. subtilis ACN, at least with respect to sporulation. As seen for eukaryotic IRP1 (17, 25), mutation of one of the cysteines implicated in the formation of the iron-sulfur cluster fixes the B. subtilis protein in the RNA-binding form independently of the availability of iron. Another aconitase mutant deleted only for the C-terminal 50% of ACN also sporulates more efficiently than does the null mutant (J. E. Craig and A.L.S., unpublished work). Thus, it is conceivable that it is within the N-terminal domain of B. subtilis ACN that the nonenzymatic regulatory role resides. In fact, the N-terminal region of B. subtilis ACN includes a 10-amino acid sequence nearly identical to that of a peptide in IRP1 shown to be closely associated with bound RNA (26).
Indirect evidence for a role of ACN in iron metabolism in prokaryotes comes from studies in Xanthomonas campestris and Pseudomonas aeruginosa. A null mutation in the ACN gene decreases the production of a X. campestris toxin involved in pathogenicity. The decreased level of the toxin is associated with a decreased concentration of intracellular iron (9). In P. aeruginosa, a low level of iron inactivates aconitase activity and increases the synthesis of exotoxin A (8). However, posttranscriptional regulation by ACN mediated through RNA binding is not the only possible explanation for these observations. When the gene encoding ACN is mutated, the level of citrate in the cell increases because the Krebs cycle is blocked. It has been suggested that it is the high level of citrate that blocks sporulation in B. subtilis citB null mutants by chelating iron and manganese and perhaps other divalent cations (10). Similar phenomena also could explain the altered expression of pathogenicity factors in ACN mutants of other bacteria.
We have shown that ACN is able to bind IRE-like sequences encoded in the B. subtilis chromosome. One such structure was found between the first two genes of the feu operon, which encodes an iron transport system. Computer analysis has identified similar sequences in the E. coli chromosome (27). Interestingly, one of the identified sequences in the E. coli genome is located upstream of fepB, which is also involved in the transport of iron and is a homolog of feuA. It is not clear whether the relationship seen in eukaryotes between the location of the IRE (whether 3′ or 5′ of the coding sequence) and the level of the proteins encoded in the mRNA is conserved in prokaryotes. There are important differences to take into account, such as the polycistronic nature of bacterial operons. Given the high conservation of the ACN sequence in the prokaryotic world, it is likely that RNA-binding activity similar to that of the B. subtilis enzyme would be found in other bacteria as well.
Acknowledgments
We thank W. Walden for much helpful advice and gifts of strains and IRP1 and B. Belitsky, N. Mani, C. Squires, and A. Wright for helpful discussions and comments on the manuscript. This work was supported by a research grant from the U.S. Public Health Service (GM 42219).
ABBREVIATIONS
- ACN
aconitase
- IRE
iron responsive element
- IRP
iron regulatory protein
- UTR
untranslated region
References
- 1.Hentze M W, Kuhn L C. Proc Natl Acad Sci USA. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rouault T A, Klausner R D. Trends Biochem Sci. 1996;21:174–178. [PubMed] [Google Scholar]
- 3.Aziz N, Munro H N. Proc Natl Acad Sci USA. 1987;84:8478–8482. doi: 10.1073/pnas.84.23.8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casey J L, Di Jeso B, Rao K, Klausner R D, Harford J B. Proc Natl Acad Sci USA. 1988;85:1787–1791. doi: 10.1073/pnas.85.6.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Owen D, Kühn L C. EMBO J. 1987;6:1287–1293. doi: 10.1002/j.1460-2075.1987.tb02366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Constable A, Quick S, Gray N K, Hentze M W. Proc Natl Acad Sci USA. 1992;89:4554–4558. doi: 10.1073/pnas.89.10.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gruer M J, Artymiuk P J, Guest J R. Trends Biochem Sci. 1997;22:3–6. doi: 10.1016/s0968-0004(96)10069-4. [DOI] [PubMed] [Google Scholar]
- 8.Somerville G, Mikoryak C A, Reitzer L J. J Bacteriol. 1999;181:1072–1078. doi: 10.1128/jb.181.4.1072-1078.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson T J, Bertrand N, Tang J L, Feng J X, Pau M Q, Barber C E, Dow J M, Daniels M J. Mol Microbiol. 1998;28:961–970. doi: 10.1046/j.1365-2958.1998.00852.x. [DOI] [PubMed] [Google Scholar]
- 10.Craig J E, Ford M J, Blaydon D C, Sonenshein A L. J Bacteriol. 1997;179:7351–7359. doi: 10.1128/jb.179.23.7351-7359.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dingman D W, Rosenkrantz M S, Sonenshein A L. J Bacteriol. 1987;169:3068–3075. doi: 10.1128/jb.169.7.3068-3075.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fortnagel P, Freese E. J Biol Chem. 1968;243:5289–5295. [PubMed] [Google Scholar]
- 13.Yousten A A, Hanson R S. J Bacteriol. 1972;109:886–894. doi: 10.1128/jb.109.2.886-894.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown P H, Daniels-McQueen S, Walden W E, Patino M M, Gaffield L, Bielser D, Thach R E. J Biol Chem. 1989;264:13383–13386. [PubMed] [Google Scholar]
- 15.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, et al. Nature (London) 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 16.Rose M, Entian K D. Microbiology. 1996;142:3097–3101. doi: 10.1099/13500872-142-11-3097. [DOI] [PubMed] [Google Scholar]
- 17.Hirling H, Henderson B R, Kühn L C. EMBO J. 1994;13:453–461. doi: 10.1002/j.1460-2075.1994.tb06280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santana M, Kunst F, Hullo M F, Rapoport G, Danchin A, Glaser P. J Biol Chem. 1992;267:10225–10231. [PubMed] [Google Scholar]
- 19.Liu H, Haga K, Yasumoto K, Ohashi Y, Yoshikawa H, Takahashi H. Microbiology. 1997;143:2763–2767. doi: 10.1099/00221287-143-8-2763. [DOI] [PubMed] [Google Scholar]
- 20.Crosa J H. Microbiol Mol Biol Rev. 1997;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keyer K, Gort A S, Imlay J A. J Bacteriol. 1995;177:6782–6790. doi: 10.1128/jb.177.23.6782-6790.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reif D W. Free Radical Biol Med. 1992;12:417–427. doi: 10.1016/0891-5849(92)90091-t. [DOI] [PubMed] [Google Scholar]
- 23.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. J Bacteriol. 1995;177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann J D. Mol Microbiol. 1998;29:189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- 25.DeRusso P A, Philpott C C, Iwai K, Mostowski H S, Klausner R D, Rouault T A. J Biol Chem. 1995;270:15451–15454. doi: 10.1074/jbc.270.26.15451. [DOI] [PubMed] [Google Scholar]
- 26.Basilion J P, Rouault T A, Massinople C M, Klausner R D, Burgess W H. Proc Natl Acad Sci USA. 1994;91:574–578. doi: 10.1073/pnas.91.2.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dandekar T, Beyer K, Bork P, Kenealy M R, Pantapoulos K M, Hentze M, Sonntag-Buck V, Flouriot G, Gannon F, Keller W, Schreiber S. Bioinformatics. 1998;14:271–278. doi: 10.1093/bioinformatics/14.3.271. [DOI] [PubMed] [Google Scholar]