Abstract
We have constructed transgenic Drosophila melanogaster lines that express green fluorescent protein (GFP) exclusively in the nervous system. Expression is controlled with transcriptional regulatory elements present in the 5′ flanking DNA of the Drosophila Na+,K+-ATPase β-subunit gene Nervana2 (Nrv2). This regulatory DNA is fused to the yeast transcriptional activator GAL4, which binds specifically to a sequence motif termed the UAS (upstream activating sequence). Drosophila lines carrying Nrv2-GAL4 transgenes have been genetically recombined with UAS–GFP (S65T) transgenes (Nrv2-GAL4+UAS–GFP) inserted on the same chromosomes. We observe strong nervous system-specific fluorescence in embryos, larvae, pupae, and adults. The GFP fluorescence is sufficiently bright to allow dynamic imaging of the nervous system at all of these developmental stages directly through the cuticle of live Drosophila. These lines provide an unprecedented view of the nervous system in living animals and will be valuable tools for investigating a number of developmental, physiological, and genetic neurobiological problems.
Dynamic visualization of the nervous system in a living organism would provide new insights into neural development, maintenance, and function. In principle, the use of the green fluorescent protein (GFP) reporter gene would be an ideal tool to accomplish this goal because it is easily observed in transgenic model organisms and cultured cells (1–3). In several studies, discrete neuronal subsets of Drosophila or nematodes have been observed in living animals with GFP (4–6). A number of practical problems, however, must be overcome before GFP visualization can serve as a useful marker for dynamic-imaging studies in live animals. First, it is necessary to have a promoter or DNA-regulatory fragment driving expression exclusively in the nervous system. Second, the strength of expression must be sufficient to allow easy observation of GFP fluorescence in intact organisms. Third, expression must be robust at all developmental stages.
Recently, we isolated and characterized tissue-specific transcriptional regulatory elements for the two different Drosophila Na+,K+-ATPase (sodium pump) β-subunit genes (7). These two genes, termed Nervana 1 and 2, originally were identified and cloned (8, 9) on the basis of reactivity with anti-horseradish peroxidase (anti-HRP) antibodies, which are commonly used to identify Drosophila neurons (10). The Nervana 1 protein does not carry the carbohydrate epitope(s) recognized by anti-HRP and is expressed primarily in muscle cells. In contrast, Nervana 2 is decorated with the anti-HRP epitope. and its expression is restricted to nervous system (11). The tissue-specific expression of each isoform gene is controlled by separate cis-DNA regulatory elements located in the 5′ flanking DNA (7).
The sodium pump is a major component of the plasma membranes of all excitable cell types and serves to generate and maintain the asymmetric gradient of Na+ and K+ ions necessary to establish the membrane resting potential and drive a variety of secondary ionic exchanger and metabolite-uptake systems (12, 13). Because extremely high levels of pump subunits are expressed in neurons at all developmental stages, it seemed likely that the regulatory elements of the Nrv 2 gene would be an excellent candidate for a pan-neuronal promoter with appropriate strength, specificity, and temporal expression to use GFP fluorescence in dynamic-imaging studies of the nervous system.
In the present study, we fused the 5′ flanking DNA of the Nrv 2 gene to the coding sequence of the yeast transcriptional activator GAL4 and constructed transgenic fly lines. When these lines are genetically recombined with lines carrying cognate GAL4 recognition elements (UAS, upstream activating sequence) controlling expression of a GFP responder gene, nervous system-specific GFP fluorescence was observed. The levels of GFP fluorescence are particularly strong, allowing the nervous system to be dynamically observed in live animals at all developmental stages.
MATERIALS AND METHODS
Nrv2-GAL4 Construct.
We previously cloned the 7.7-kilobase Nrv2 5′ flanking DNA into the Drosophila P element transformation vector, pCaSpeR–AUG–β-galactosidase (14), in a modified NotI cloning site (7). The GAL4 gene from the pGaTN vector (15) was subcloned into that DNA construct to replace the lacZ reporter gene of pCaSpeR–AUG–β-galactosidase by using the NotI and XbaI cloning sites.
P Element Transformation.
Standard P element-mediated germ-line transformation of Drosophila melanogaster was performed (16). The Nrv2-GAL4 construct was injected into w− embryos at a concentration of 0.8 μg/μl along with pUChs Δ2–3 helper plasmid (0.1 μg/μl). A total of 14 independent lines (eye color selection) were generated with insertion of the transgene on the X, second, or third chromosomes. Linkage analysis of the inserted P elements was carried out by examining their segregation from second or third chromosomes marked with Cy or Sb, respectively. X-linked inserts were determined by standard crosses.
Generation of Nrv2-GAL4+UAS–GFP Lines.
Fly lines carrying both a Nrv2-GAL4 driver and a UAS–GFP responder on the second or third chromosomes were generated by recombination (17). Flies with Nrv2-GAL4 insertion on the second or third chromosome were crossed with flies carrying a UAS–GFP (S65T) responder gene on the second or third chromosome (obtained from the Bloomington Drosophila Stock Center, B-1521, B-1522), respectively. The F1 female progeny were crossed with w− males, and the F2 larval progeny were observed by using a fluorescence microscope. The larvae with GFP expression in nervous system were selected and reared to make the Nrv2-GAL4+UAS–GFP homozygous lines. The experiments described in this paper all were performed by using these Nrv2-GAL4+UAS–GFP lines.
Double Labeling of Whole-Mount Embryos with Anti-GFP and Anti-HRP Antibodies.
Embryos were collected from Nrv2-GAL4+UAS–GFP flies and staged at 25°C. They were dechorionated, fixed, and processed for immunocytochemistry as described by Patel (18). After blocking in PBT+NGS (0.1% Triton X-100/0.1% BSA/5.0% normal goat serum in PBS), the embryos were incubated with a 1:5 dilution of rat anti-GFP mAb JFP-J1 (kindly provided by S. Fujita, Mitsubishi Kasei, Tokyo) and rabbit anti-HRP antibodies (1 μg/ml, Sigma). After overnight incubation at 4°C, the embryos were washed and probed with FITC-labeled goat anti-rat IgG (1:200 dilution; Pierce) and rhodamine-labeled goat anti-rabbit IgG (1:200 dilution; Pierce) for 2 h at room temperature (8). After washing, preparations were mounted in 70% (vol/vol) glycerol (in PBS) and viewed under a fluorescence microscope.
Visualization of GFP with Confocal Microscopy.
Dechorionated embryos or first-instar larvae were directly mounted in a 1:1 mixture of glycerol and PBS. Samples were observed directly by using a Zeiss LSM 410 confocal scanning microscope. Excitation was with the argon laser (488-nm excitation), and emission was at 527 nm.
Visualization of GFP by Using Wide Field Fluorescence Microscopy.
Mechanically dechorionated live embryos, larvae, or pupae were mounted in a 1:1 mixture of glycerol/PBS in a small viewing chamber constructed of a filter paper support for a standard coverslip and viewed directly. Adult flies were sealed in an imaging chamber (Molecular Probes) and observed directly. Fly parts (head, abdomen, leg, and wing) were collected from adults by manual dissection and mounted in 70% (vol/vol) glycerol (in PBS). Samples were observed with a Nikon Diavert or Olympus Vannox fluorescence microscope under Hg illumination by using standard FITC fluorescence filters. Digital images were recorded by using either a Cambridge Research (Cambridge, MA) Real-14 cooled CCD camera or a Kodak DC120 color CCD camera. Image files were processed by using scion image (Scion, Frederick, MD), image pro+ (Media Cybernetics, Silver Spring, MD), or photoshop (Adobe Systems, Mountain View, CA).
RESULTS
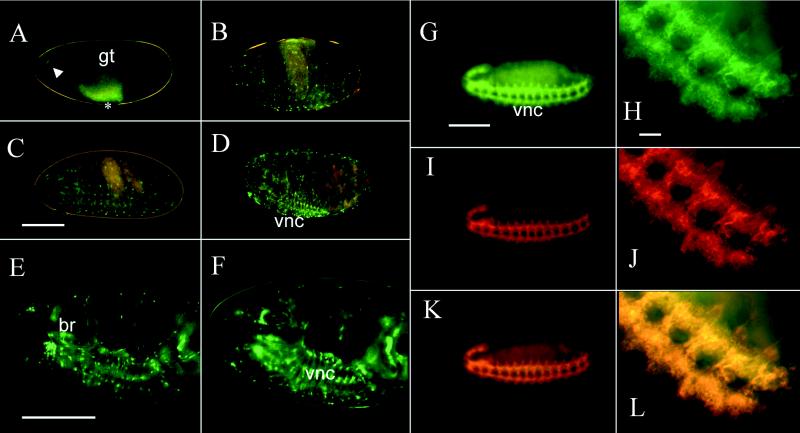
GFP Expression in Embryos. Exposure of embryos to the repeated periods of several minutes of UV light required for observation did not affect their viability. Most embryos (≈75%) completed development when they were observed repeatedly over a several-hour period and hatched into active larvae. GFP expression was easily monitored in the developing nervous system (both central and peripheral) at different embryonic stages (Fig. 1A–D). GFP fluorescence initially was detected in a group of cells at the anterior end of stage-12 embryos and a few cells in the ventral nerve cord, which are partially obscured by gut autofluorescence (Fig. 1A). As neural development proceeded, the fluorescence became visible in many cells in the brain and ventral nerve cord and was especially intense in some peripheral sensory neurons near the surface of the developing embryo (Fig. 1B–D). In stage-17 embryos, the pattern of fluorescence is similar to early larval stages (Fig. 1E and F).
Figure 1.
GFP fluorescence and double-labeled antibody staining in embryos. A live stage-12 embryo (A) shows green fluorescent neurons in the anterior end (arrowhead) and in the ventral nerve cord (asterisk). The ventral nerve cord neurons are partially obscured by the strong yellow autofluorescence of the gut at this stage. In stage-15 embryos (B), some of the peripheral sensory neurons near the surface as well as many cells in the ventral nerve cord are visible. By stage 16 (C) and 17 (D) more cells in the ventral nerve cord and brain and a few peripheral sensory neurons show fluorescence. The same field of view is shown for a stage-17 embryo in E near the surface and F more internally. All embryos are oriented with anterior to the left and ventral to the bottom. A–D are color charge-coupled device images, E and F are false-colored black and white charge-coupled device images. Fixed, stage-12 embryos were double stained with anti-GFP (G and H) and anti-HRP (I and J) antibodies. The hazy green area in G is yellow autofluorescence of the gut. The complete nervous system is stained with anti-GFP antibody (G) and is shown at higher power (H) or is stained with anti-HRP antibody (I) and is shown at higher power (J). Images in G and I as well as H and J were merged to produce K and L, respectively. There is a complete correspondence of the anti-GFP and anti-HRP staining. Note also the difference between GFP fluorescence in a live stage-12 embryo (A) and anti-GFP antibody staining of a similarly aged fixed embryo (G and H). The images in G–L are false-colored. br, brain; gt, developing gut; vnc, ventral nerve cord. (Bar = 100 μm for whole embryos, 10 μm for higher power views in H, J, and L.)
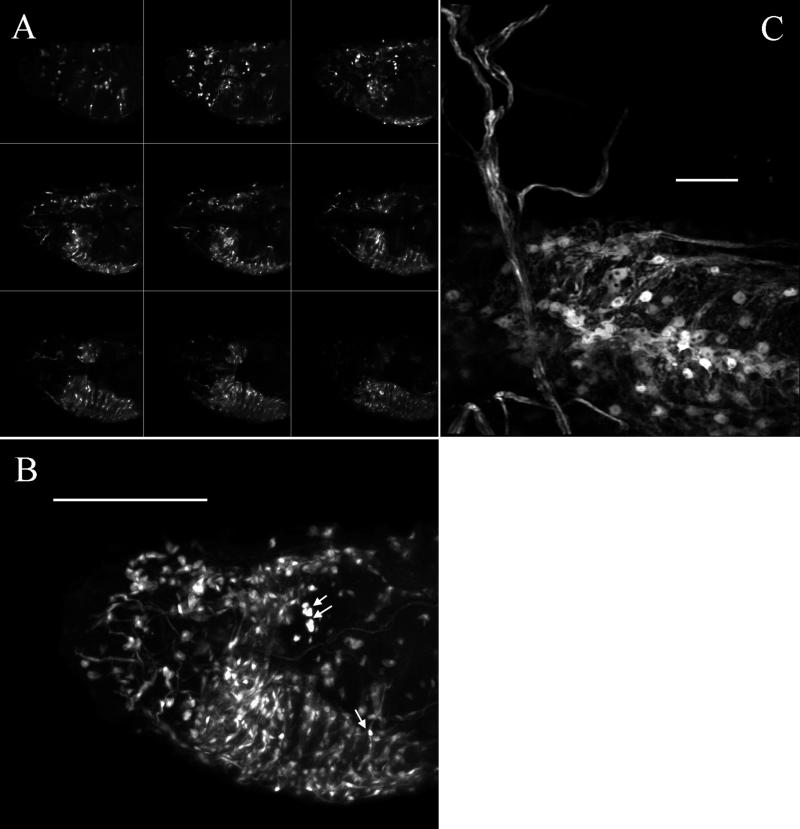
We also have observed embryos at different developmental stages by using confocal microscopy. In a stage-17 embryo, the GFP signal is easily observed in neurons and neural processes of both the central (brain, ventral nerve cord) and peripheral (Fig. 2 A and B) nervous system. A three-dimensional projection of confocal optical sections clearly reveals the extent and relationship of the GFP fluorescent cells and processes (Fig. 2B). An animated three-dimensional projection of confocal sections can be viewed as supplemental data on the PNAS web site (www.pnas.org). A higher-power three-dimensional projection of optical sections through an early first-instar larval ventral nerve cord shows the cellular morphology and fluorescent nerve fibers (Fig. 2C).
Figure 2.
Confocal microscopy. (A) The nine small images are individual optical sections (spaced 6 μm apart) taken from the anterior of a stage-17 embryo. (B) Twenty-eight optical sections from the same embryo (spaced 2 μm apart) are projected to reconstruct the complete three-dimensional distribution of fluorescence. The single and double arrows indicate especially bright peripheral neurons. An animation of this projection series is available as supplemental data on the PNAS web site (www.pnas.org). (C) A higher power view of fluorescence in the central part of the ventral nerve cord of an early first-instar larvae. Twenty optical sections (spaced 2.5 μm apart) are projected to reconstruct the three-dimensional view. The fluorescence is clearly present in most central nervous system cells and processes. All images are oriented with anterior to the left and ventral down. [Bar = 100 μm (B) and 10 μm (C).]
To establish the cellular specificity of GFP expression in Nrv2-GAL4+UAS–GFP embryos, we compared the immunocytochemical staining of stage-12 embryos by using an anti-GFP antibody to the staining pattern obtained with anti-HRP antibodies, a known pan-neuronal marker (10). Double labeling of whole-mount embryos with anti-GFP and anti-HRP antibodies showed an indistinguishable pattern of immunocytochemical staining, indicating that GFP expression is likely to be pan-neuronal at early embryonic stages (Fig. 1 G–L). GFP expression is detected a few hours earlier by antibody staining than by fluorescence. This is most likely because of the time required for posttranslational modification (oxidation) required to produce GFP fluorescence (19, 20). One difference between the GFP expression pattern in later- stage embryos and anti-HRP staining is the absence of fluorescence in many of the v, v′, l, and d peripheral sensory neurons (21).
GFP Expression in Larvae.
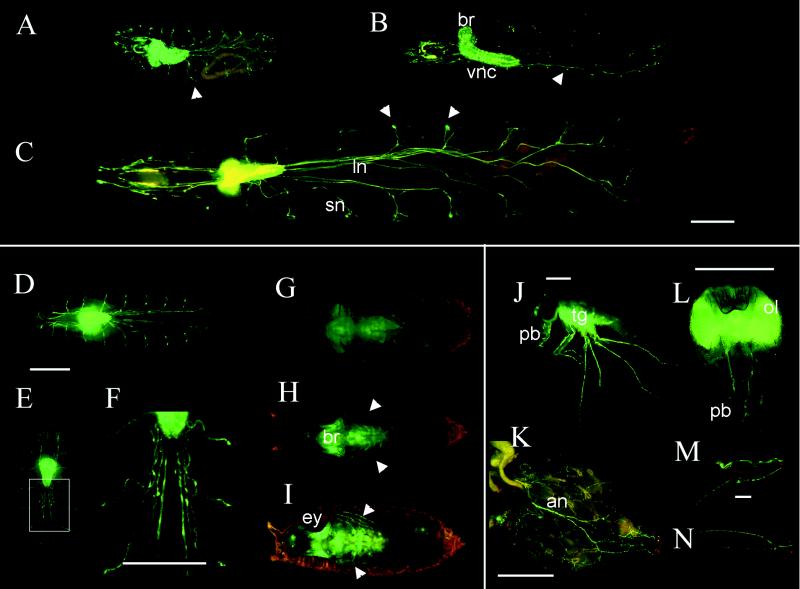
The exceptionally strong GFP signal in larvae allows direct visualization of the nervous system in fully active living animals. Especially striking when viewing fully active larvae is an appreciation of the dynamic mechanical forces that the brain and nerves are subjected to during movement. The brain and ventral ganglion as well as the nerve fibers are rapidly stretched and compressed as the animals move about. A quicktime movie of a fluorescent larvae is available for viewing as supplemental data on the PNAS web site (www.pnas.org). Extremely bright GFP fluorescence is specifically observed in the brain and ventral ganglion of all three larval instars. Because of the number of fluorescent cells, it is difficult to resolve individual cell types by using conventional fluorescence microscopy. The long longitudinal nerve fibers, segmental branches, and fine fibers at the distal ends of the segmental nerves are also clearly seen, as are the nerve fibers in the anterior part of the larvae (Fig. 3 A–C). The bright fluorescent spots in the lateral regions of the larvae may be peripheral glial cells based on their position. Nervous system structures that do not fluoresce in larvae include most of the peripheral sensory neurons and the differentiating photoreceptor neurons of the third-instar larval eye disk.
Figure 3.
GFP fluorescence in larvae, pupae, and adults. First- (A), second- (B), and third- (C) instar larvae show intense fluorescence in the brain and ventral ganglion as well as the longitudinal and segmental nerves and nerve branches. The large bright structures at the ends of segmental nerves may be peripheral glia (indicated by arrowheads). The specimens in A and C are viewed from the ventral surface whereas B is a lateral view. The image in C has been constructed from images of three smaller fields. In early pupae (D), the fluorescence is similar to late third-instar larvae. As metamorphosis proceeds, the nerves begin to appear wavy and frayed (E, and at higher power in F). Later stages of pupation show decreased fluorescence (G) and a shape more characteristic of adult central nervous system. Leg nerves are visible in late-stage pupae (H) or a few hours before hatching (I) and are indicated with arrowheads. Whole adults show bright fluorescence in the optic lobes, brain, thoracic ganglion, and many kinds of nerves. In J, the thoracic ganglion, leg, and probosal nerves are evident, while the optic lobe and brain staining is obscured by the red eye. An adult female abdomen shows major nerves with many finer branches at higher power (K). A dissected head viewed from the posterior shows the intense fluorescence in the brain, optic lobes, and proboscal nerves (L). The leg nerve (M) and wing nerve (N) are also clearly visible in dissected parts from adults. an, abdominal nerve; br, brain; ey, eye; ln, longitudinal nerve; ol, optic lobe; pb, proboscis; sn, segmental nerve; tg, thoracic ganglion; vnc, ventral nerve cord. (Bars = 100 μm.)
GFP Expression in Pupae.
The intensity of GFP fluorescence is strong enough that we can monitor the morphological changes of the nervous system in developing pupae directly through the pupal case during metamorphosis (Fig. 3 D–I). The nervous system in early pupae is similar to that of third-instar larvae (Fig. 3D). As metamorphosis proceeds, the nerve-fiber bundles become wavy and begin to appear defasiculated (Fig. 3 E and F). Later, the fluorescence intensity of nerve fibers and cell bodies decreases, and eventually the fibers disappear in 1-day-old pupae (Fig. 3G). The central nervous system also undergoes morphological changes in overall shape, replacing the larval central nervous system shape with an adult shape. The GFP fluorescence is seen in brain, optic lobes, and thoracico-abdominal ganglion in the 1-day-old pupae (Fig. 3G). As metamorphosis continues, the intensity of GFP fluorescence increases along with the development of new nervous system structures (Fig. 3 H and I). GFP fluorescence can clearly be seen in peripheral nervous system structures, such as antennae, labial palps, maxillary palps, and even leg nerves, at later stages of pupation. The optic lobes are extremely bright in pupae close to eclosion (Fig. 3I).
GFP Expression in Adults.
Remarkably, we can even observe GFP fluorescence directly through the pigmented cuticle of intact living adults (Fig. 3J). Fluorescence is localized in brain, optic lobes, and thoracic ganglion, as well as in parts of the peripheral nervous system. For example, strong fluorescence is seen in antennae and the ocellus (photoreceptor organ located at the top of the head). In dissected adult fly parts, it is easier to recognize finer details of the various nerve-fiber tracts. In adult heads, in addition to the intense fluorescence in the brain and optic lobes, the maxillary and labial nerves in the proboscis are visible (Fig. 3L). GFP fluorescence is also clearly observed in the nerves of the abdomen, leg, and wing (Fig. 3 K, M, and N).
DISCUSSION
The Nrv2-GAL4+UAS–GFP lines we describe here provide a view of the nervous system in live Drosophila at all developmental stages. The advantages of using Nrv2-specific transcriptional regulatory elements are the extremely robust expression and the apparently complete restriction of expression to the nervous system. Part of the robust expression is derived from the use of the strong yeast transcriptional activator GAL4, but part is also because of the high-level activity of the Nrv2 regulatory DNA. The sodium pump is an important homeostatic regulator with exceptionally high levels of expression in excitable cells, such as neurons. We also have observed even more robust muscle-specific GFP fluorescence in transgenic lines constructed by using the Nrv1 5′ flanking DNA (unpublished observations).
A number of laboratories have observed GAL4-mediated nervous system-specific expression of GFP in live Drosophila or in Caenorhabditis elegans; (1, 4, 22). Many of the Drosophila GAL4 lines that have been described show expression in subsets of glial and/or neurons (for recent review, see ref. 23). These subset-specific lines provide excellent tools for dynamic visualization of particular types of neurons or glia. In most cases, the GFP fluorescence patterns have been analyzed only during early embryogenesis and have not been described at later developmental stages. Most subset-specific expression of GFP relies on the use of rather specialized types of promoters or enhancer trap lines to direct GFP expression either directly or with the GAL4 activation system (23, 24). Many neuronal and glial genes, especially those with very specialized functions, may not be expressed at high enough levels to be useful for GFP fluorescence in living animals.
Other homeostatic or housekeeping genes with potentially strong promoters, such as cellular actin or microtubule protein genes, also could be used in principle to drive high-level expression of GFP fluorescence. Unfortunately, most are expressed too broadly to target individual tissue types such as nervous system (25, 26). Fortunately, Drosophila has a single Nrv2 sodium pump β-subunit gene whose expression is restricted to nervous system (9, 11). The expression of Nrv2 is independently regulated by separate transcriptional elements relative to the other Drosophila sodium pump β-subunit gene (Nrv1). These separate regulatory elements are located in the 5′ flanking DNA (7), and this independent regulation is essential to achieve the specificity of tissue-specific expression we report here. The Nrv2 gene is alternatively spliced to produce two different proteins (Nrv 2.1 and 2.2) with differential expression in head and thoracic ganglia (11), but apparently this further specificity occurs posttranscriptionally. In other species, individual α- and β-subunit genes of Na+,K+-ATPase are more numerous than in Drosophila. Several specific isoforms also are expressed at high levels in neurons in particular combinations (27), but the patterns are more complex than the case for Drosophila β-subunit expression.
Only a few genes have been described that are expressed specifically in most or all neurons throughout development. In addition to Nrv2 expression, the Drosophila elav gene product has been detected immunocytochemically in neurons at all developmental stages (28–30). We have compared the Nrv2-GAL4+UAS–GFP lines described here with two different elav-GAL4 lines (31). The patterns of elav-GAL4- and Nrv2-GAL4-directed GFP fluorescence are similar, with a few notable differences. First, the GFP fluorescence is much more intense in our Nrv2-GAL4+UAS–GFP lines. Typical integration times for the charge-coupled device images presented in this paper are less than 0.1 sec for Nrv2-GAL4 lines, whereas the equivalent stage elav-GAL4 lines need integration times of 30 sec–>1 min. Second, we do not see fluorescence in many peripheral sensory neurons in the Nrv2-GAL4 lines in late embryo and larval stages that are observed in elav-GAL4 lines. Finally, we do not see fluorescence in developing photoreceptor cell neurons in third-instar larval eye disks in the Nrv2-GAL4 lines that are seen in elav-GAL4 lines. The elav-GAL4 lines thus seem to express GFP fluorescence in more types of neurons than the Nrv2-GAL4 lines. Our failure to detect fluorescence in the latter stage peripheral nervous system and photoreceptor neurons could indicate that the Nrv2 construct is missing some regulatory elements or that the signal in these types of neurons may be below the detection level of fluorescence microscopy.
The ability to view the nervous system in living embryos makes our lines an excellent preparation to construct time lapse records of early nervous system development. One important aspect of Na+,K+-ATPase expression is that it is required to generate and maintain the ionic gradients essential for functionally active neurons and glia. Certain cells therefore may express the pump at different times in their developmental life histories, and such differences could indicate when the neurons become functionally active. It will be interesting to combine observations on our Nrv2-GAL4+UAS–GFP lines with other markers of specific neuronal and glial cell types that may be recognized before the particular cells become functional.
During larval stages, our lines can be used to observe the dynamics of nervous system movements accompanying the movement of larvae. They also can be useful for tracing the main axonal pathways. The ability to see GFP fluorescence through the pupal case also will make it possible to record time-lapse movies of the remodeling of the nervous system during metamorphosis. Our lines provide excellent visualization of particular neurons and nerve fibers that may be accessible for electrophysiological recording or lesioning studies.
The strength of Drosophila as a model organism is in its genetic definition. We foresee many uses for our lines in genetic experiments. For example, the Nrv2-GAL4+UAS–GFP could serve as a marker to view the effects of particular mutations on nervous system structure and development. Existing axonal pathfinding or neurodegenerative mutants would be good candidate genes to examine in this regard (32–34). Our lines also could serve as a genetic background for generating new mutations that effect axonal pathfinding or neurodegeneration because they could easily be screened for phenotypes by simple observation with a fluorescence microscope.
We have not attempted to increase the utility of our lines by modifying the GFP reporter gene. Others have produced GFP fusion proteins, for example, that incorporate protein motifs that direct the reporter to a particular intracellular compartment such as the plasma membrane (35), microtubules (36), neuronal processes (37), or nucleus (38). The flexible design of the GAL4 activation system will make it easy to incorporate these types of GFP-fusion reporters in future work. It also will be possible to direct a variety of other functional or mutant genes to the nervous system by using the Nrv2-GAL4 driver. One limitation of using GFP fluorescence is the time required for posttranslational oxidation to generate a fluorescent protein (19). We have used the S65T mutated form of GFP, which has been reported to speed up the acquisition of fluorescence (39), but generation of the fluorochrome still lags expression of GFP protein, as assessed by anti-GFP antibody staining. Future improvements in GFP design may lessen this time lag and thus make our lines more useful for studying early neurogenesis in living animals.
Supplementary Material
Acknowledgments
We thank our colleagues Drs. T. Kitamoto, K. Ikeda, E. Roberts, L. Iverson, S. Novak, and R. Williamson for their helpful advice and criticism. This work was supported by a grant from the American Heart Association.
ABBREVIATIONS
- GFP
green fluorescent protein
- HRP
horseradish peroxidase
- UAS
upstream activating sequence
References
- 1.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 2.Prasher D C. Trends Genet. 1995;11:320–323. doi: 10.1016/s0168-9525(00)89090-3. [DOI] [PubMed] [Google Scholar]
- 3.Gerdes H H, Kaether C. FEBS Lett. 1996;389:44–47. doi: 10.1016/0014-5793(96)00586-8. [DOI] [PubMed] [Google Scholar]
- 4.Brand A. Trends Genet. 1995;11:324–325. doi: 10.1016/s0168-9525(00)89091-5. [DOI] [PubMed] [Google Scholar]
- 5.Fleming J T, Squire M D, Barnes T M, Tornoe C, Matsuda K, Ahnn J, Fire A, Sulston J E, Barnard E A, Sattelle D B, Lewis J A. J Neurosci. 1997;17:5843–5857. doi: 10.1523/JNEUROSCI.17-15-05843.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Labrousse A M, Shurland D L, van der Bliek A M. Mol Biol Cell. 1998;9:3227–3239. doi: 10.1091/mbc.9.11.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu, P., Sun, B. & Salvaterra, P. M. (1999) Gene, in press. [DOI] [PubMed]
- 8.Sun B, Salvaterra P M. J Neurochem. 1995;65:434–443. doi: 10.1046/j.1471-4159.1995.65010434.x. [DOI] [PubMed] [Google Scholar]
- 9.Sun B, Salvaterra P M. Proc Natl Acad Sci USA. 1995;92:5396–5400. doi: 10.1073/pnas.92.12.5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jan L Y, Jan Y N. Proc Natl Acad Sci USA. 1982;79:2700–2704. doi: 10.1073/pnas.79.8.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun B, Wang W, Salvaterra P M. J Neurochem. 1998;71:142–151. doi: 10.1046/j.1471-4159.1998.71010142.x. [DOI] [PubMed] [Google Scholar]
- 12.Sweadner K J. Biochim Biophys Acta. 1989;988:185–220. doi: 10.1016/0304-4157(89)90019-1. [DOI] [PubMed] [Google Scholar]
- 13.Lingrel J B, Orlowski J, Shull M M, Price E M. Prog Nucleic Acid Res Mol Biol. 1990;38:37–89. doi: 10.1016/s0079-6603(08)60708-4. [DOI] [PubMed] [Google Scholar]
- 14.Thummel C S, Boulet A M, Lipshitz H D. Gene. 1988;74:445–456. doi: 10.1016/0378-1119(88)90177-1. [DOI] [PubMed] [Google Scholar]
- 15.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 16.Rubin G M, Spradling A C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 17.Greenspan R J. Fly Pushing: The Theory and Practice of Drosophila Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. [Google Scholar]
- 18.Patel N H. Methods Cell Biol. 1994;44:445–487. doi: 10.1016/s0091-679x(08)60927-9. [DOI] [PubMed] [Google Scholar]
- 19.Heim R, Prasher D C, Tsien R Y. Proc Natl Acad Sci USA. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inouye S, Tsuji F I. FEBS Lett. 1994;341:277–280. doi: 10.1016/0014-5793(94)80472-9. [DOI] [PubMed] [Google Scholar]
- 21.Ghysen A, Dambly-Chaudiere C, Aceves D, Jan L Y, Jan Y N. Roux’s Arch Dev Biol. 1986;195:281–289. doi: 10.1007/BF00376060. [DOI] [PubMed] [Google Scholar]
- 22.Yeh E, Gustafson K, Boulianne G L. Proc Natl Acad Sci USA. 1995;92:7036–7040. doi: 10.1073/pnas.92.15.7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brand A. Methods Cell Biol. 1999;58:165–181. doi: 10.1016/s0091-679x(08)61955-x. [DOI] [PubMed] [Google Scholar]
- 24.Itoh K, Urban J, Technau G M. Roux’s Arch Dev Biol. 1995;204:284–307. doi: 10.1007/BF02179499. [DOI] [PubMed] [Google Scholar]
- 25.Endow S A. Methods Cell Biol. 1999;58:153–163. doi: 10.1016/s0091-679x(08)61954-8. [DOI] [PubMed] [Google Scholar]
- 26.Edwards K A, Demsky M, Montague R A, Weymouth N, Kiehart D P. Dev Biol. 1997;191:103–117. doi: 10.1006/dbio.1997.8707. [DOI] [PubMed] [Google Scholar]
- 27.Blanco G, Mercer R W. Am J Physiol. 1998;275:F633–F650. doi: 10.1152/ajprenal.1998.275.5.F633. [DOI] [PubMed] [Google Scholar]
- 28.Robinow S, White K. Dev Biol. 1988;126:294–303. doi: 10.1016/0012-1606(88)90139-x. [DOI] [PubMed] [Google Scholar]
- 29.Robinow S, White K. J Neurobiol. 1991;22:443–461. doi: 10.1002/neu.480220503. [DOI] [PubMed] [Google Scholar]
- 30.Yao K M, White K. J Neurochem. 1994;63:41–51. doi: 10.1046/j.1471-4159.1994.63010041.x. [DOI] [PubMed] [Google Scholar]
- 31.Luo L, Liao Y J, Jan L Y, Jan N J. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- 32.Baird D H, Koto M, Wyman R J. J Neurobiol. 1993;24:971–984. doi: 10.1002/neu.480240710. [DOI] [PubMed] [Google Scholar]
- 33.Hoang B, Chiba A. J Neurosci. 1998;18:7847–7855. doi: 10.1523/JNEUROSCI.18-19-07847.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holmes A L, Raper R N, Heilig J S. Genetics. 1998;148:1189–1201. doi: 10.1093/genetics/148.3.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee T, Luo L. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 36.Theurkauf W E, Hazelrigg T I. Development (Cambridge, UK) 1998;125:3655–3666. doi: 10.1242/dev.125.18.3655. [DOI] [PubMed] [Google Scholar]
- 37.Murray M J, Merritt D J, Brand A H, Whitington P M. J Neurobiol. 1998;37:607–621. [PubMed] [Google Scholar]
- 38.Davis I, Girdham C H, O’Farrell P H. Dev Biol. 1995;170:726–729. doi: 10.1006/dbio.1995.1251. [DOI] [PubMed] [Google Scholar]
- 39.Heim R, Cubitt A B, Tsien R Y. Nature (London) 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.