Abstract
By means of a computer search for upstream promoter elements (distal sequence element and proximal sequence element) typical of small nuclear RNA genes, we have identified in the human genome a number of previously unrecognized, putative transcription units whose predicted products are novel noncoding RNAs with homology to protein-coding genes. By elucidating the function of one of them, we provide evidence for the existence of a sense/antisense-based gene-regulation network where part of the polymerase III transcriptome could control its polymerase II counterpart.
Author Summary
After the sequence of the human genome was determined, it was immediately recognized that a large part of the regulation of the gene expression occurring in the cells under physiological, as well as under pathological conditions, is carried out by RNA molecules that do not code for proteins (the “noncoding portion” of the genome). Here, we focus on small RNA molecules transcribed by the RNA polymerase III and identify a novel set of approximately 30 noncoding (nc) RNA genes. We propose that these RNA transcripts play a key role in regulating the expression of specific protein-coding genes transcribed by the RNA polymerase II, thus constituting an unprecedented example of cogene/gene pairs. Furthermore, we provide evidence that the RNA polymerase III, in addition to the well-known task in the constitutive synthesis of small RNAs (such as 5S rRNA and tRNAs), also plays a key role in the area of gene-expression control. A detailed investigation of the function of one of the novel ncRNA genes, called 21A, revealed that its transcript plays a role in the control of the proliferation of some tumor cells. The above findings significantly expand our understanding of the ncRNA universe and open the way to further studies aimed at the elucidation of the molecular pathways involving this novel class of regulatory RNAs.
Introduction
Recent advances in mammalian genome studies are bringing to light the occurrence of a widespread transcription of noncoding (nc) regions devoted to the regulation of the protein coding genome expression [1–4]. The mechanisms of action of these transcripts are various and of different natures, although all of them are devoted to the regulation of fundamental genetic pathways involved in the determination of the cell phenotype. The concomitant evolution of noncoding regulatory transcripts and proteins that target different RNA:RNA or RNA:DNA complexes emphasizes the importance of studying the regulatory processes mediated by nucleic acid interactions. It has been demonstrated that in both prokaryotes and eukaryotes, cis-acting RNA regulatory regions (i.e., 3′ UTRs forming secondary structures with regulatory features or containing sequences recognized by proteins involved in RNA stability modulation) can be simultaneously regulated in trans by other noncoding RNAs (i.e., microRNAs) or by protein complexes. The simultaneous occurrence of cis- and trans-regulatory elements brings to light the complexity of this network where the coexistence of different ncRNAs plays a key role in the control of other target gene expression [5]. In this context a prominent role is played by the enlarging family of microRNAs (miRNAs) that act at a posttranscriptional level by inhibiting the translation of protein-coding genes [6]. The known miRNAs, similar to protein-coding mRNAs, are synthesized as polyadenylated precursor molecules by the RNA polymerase (pol) II transcription machinery [7]. The vast majority of the tools used in molecular biology are based on transcript collections obtained by oligo (dT) reverse transcriptase (RT)-PCR, thus encompassing only polyadenylated pol II products. However, a wide contribution of nonpolyadenylated transcripts to the human transcriptome has been revealed [8]. The role of such transcripts in pol II transcriptome–expression regulation remains largely unexplored.
Among the noncoding elements, one of the most investigated has been the Alu class of repetitive sequences that represents about one-tenth of the whole human genome. Although it is not yet possible to discern a peculiar role of Alu sequences, their short transcripts have been shown to be involved in several biological processes such as: RNA editing (where Alus are preferential sites for A to I RNA editing thus having profound implications both in gene-expression regulation and in mammalian genome evolution) [9], alternative splicing (internal exons that contain an Alu sequence are almost always alternatively spliced) [10], chromosomal recombination (the recombination between Alu elements is at the base of genomic deletions associated with many human genetic disorders) [11], gene-expression regulation (functioning as naturally occurring antisense RNAs) [12], cell-stress response (such as heat-shock response and/or translation inhibition) [13], and as putative miRNAs targets [14]. The physiological role of Alus and all the other 7SL-derived transcripts needs to be studied in more detail. In particular, the fact that their transcription is RNA pol III–dependent brings to light a previously unexpected role in gene-expression regulation of this enzyme that deserves investigation.
In this study, starting from the observation that pol III is specialized in transcription of ncRNA genes, we addressed the hypothesis that the human genome might contain pol III transcription units each specifically regulating one (or more) specific pol II genes, thus constituting functional “cogene”/gene pairs.
Results/Discussion
In Silico Identification of a Novel Set of Small Nuclear RNA Gene-Like Transcriptional Units in the Human Genome
To test our hypothesis we focused on pol III type 3–extragenic promoters, which are located upstream of the transcribed region. We screened the human genome for regions containing the consensus sequences characteristic of pol III type 3 promoters: the proximal sequence element (PSE) and the distal sequence element (DSE) [15,16]. First, we tested the PSE sequences of three well-characterized pol III type 3–ncRNA genes (U6, H1, and 7SK) as query sequences for the search of similar (if not equal) elements in the human genome by using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/BLAST; under “Nucleotide” subsection select “short, nearly exact matches” option, then pull down “Homo sapiens” organism database) (see Materials and Methods for sequences used as query). While the search with U6 and 7SK PSE sequences did not identify a significant number of homologous regions scattered throughout the genome, the H1 PSE element shared a high homology with 60 novel putative PSE sequences. Among these we selected (by a BLAST analysis) those that contained a DSE sequence element within an arbitrarily defined distance of 1,000 basepairs upstream the PSE. In addition to the expected H1, results evidenced 31 novel putative PSE/DSE-dependent promoters characterized by the concomitant occurrence of the PSE and the DSE sequences within that defined genomic distance. Moreover, a detailed sequence analysis showed that the vast majority of the distances between the PSE and a downstream TATA box or TATA-like element are within 18–22 basepairs, as expected for a canonical type 3 promoter [16]. Altogether these observations were taken as preliminary indication of a functional pol III type 3 structure of these novel promoters (Table S1).
Since our search was based on pol III type 3 promoters, some additional features of these promoters needed to be considered: (i) the occurrence of a PSE consensus sequence does not identify per se a pol III type 3 promoter; that is, rather, the result of the simultaneous occurrence at an appropriate distance of the PSE and an A/T-rich (TATA-like) element. Indeed, it has been clearly shown that the occurrence of a PSE consensus that lacks a downstream A/T-rich element makes the promoter readable by RNA pol II, such as in the case of the U2 snRNA gene [17]. In this context, the transcription initiation region is not relevant for the choice of the RNA polymerase, at least in humans, although it seems to be of fundamental importance in Xenopus [18]. Therefore, the PSE/DSE-flanked, putative transcription units identified by our search might in theory be transcribed either by pol II or by pol III, depending on the occurrence of a functional A/T-rich region downstream of the PSE. (ii) Pol III transcription units are characterized by the presence of very simple and easily recognizable termination signals, consisting in a run of four or more consecutive T residues. We thus searched, within the collection of DSE/PSE-containing sequences, for the further occurrence of a TATA-like element downstream of the PSE and for the occurrence of a termination signal at a significant distance downstream of the hypothetical transcription start site, assumed to be located approximately 30 basepairs downstream of the TATA element. Such a search refinement revealed that most of the newly identified sequences have features compatible with a pol III type 3 promoter structure (Table S2).
Our in silico search was based on the H1 PSE sequence, which was used as a query only allowing for mismatches in the first and last positions. The search thus likely identified only those promoters whose structure is very similar to that of H1. This is supported by the fact that out of H1 no other previously known pol III type 3 promoters were found in our search. Given the divergence among the PSE sequences of U6, 7SK, and H1, it is to be expected that the use, as a query sequence, of a more degenerated PSE consensus, derived from the known pol III type 3 PSE sequences, would bring to light a considerably higher number of putative PSE-dependent transcription units in the human genome.
To further characterize in silico the novel transcription units, we arbitrarily assumed as transcribed the region starting from the 30th nucleotide downstream of the first nucleotide of the predicted TATA box. In addition, a run of at least four T residues was considered as a pol III transcription–termination signal, although events of “read-through” are possible at T4 sequences depending on sequence context features [19,20]. While it has to be emphasized that the transcribed region of each element of this collection needs to be experimentally determined, we selected 32 putative novel transcripts to be subjected to additional analysis on the basis of their in silico characterization.
To test if a common secondary structure could be a hallmark of the novel molecules, an in silico analysis of their secondary structure was performed by mfold algorithm (http://www.bioinfo.rpi.edu/applications/mfold/rna/form1.cgi) [21]. Results showed that although hairpins with short stems (5–7 basepairs) were frequent, no shared secondary structures were recurrent, indicating that a peculiar molecular organization is not the common hallmark of this set of noncoding molecules. Although their averaged free energy (ΔG) was extremely variable (−42.7 ± 41.2), a group of four transcripts (11A, 20A, 21A, and 29A) showed a ΔG value significantly lower than all the others (ΔG < −100). A statistical analysis of such ΔG differences was performed evidencing that the differences between this group of transcripts (11A, 20A, 21A, and 29A), and the rest of the pool is highly significant (Student's t-test, 33 degrees of freedom, α significance level = 0.1 corresponding to a p-value of 0.0001), thus keeping in line with their physiologically functional–secondary structure organization (Table S3).
In order to assess if the pool of transcription units was prevalently constituted by repeats such as retroposons, we analyzed their sequences by Repeat Masker algorithm (http://repeatmasker.org) evidencing that: (i) only two out of 32 (6.2%) are short interspersed nucleotide elements (21A and 29A, which were marked as AluJb elements); (ii) three of them (24A, 37A, and 38A) are part of long interspersed nucleotide elements; (iii) two sequences (17A and 40A) contained a mammalian interspersed repeat (MIR), and (iv) three sequences (30A, 32A, and 44A) contained different types of long terminal repeats (Table S4). Considering that Alus, long interspersed nucleotide elements, and MIRs constitute about 15%, 30%, and 1%–5% of the human genome, respectively, one would have expected a higher frequency of repeats in the pool of sequences. Altogether these observations provide evidence that the novel PSE-dependent transcripts are not associated to a specific class of repetitive sequences scattered throughout the human genome, but instead they constitute a novel heterogeneous set of type 3 promoter-driven elements.
When these noncoding sequences were used to challenge the human genome database (BLAST analysis), it was found that seven of them were internal to known or predicted protein-coding genes, four being in antisense and three in sense configuration. Most of the novel sequence elements not mapping in coding regions shared a high sequence homology (∼80%) to a pol II transcript/expressed sequence tag that maps in a different locus (Table S5). Such homologies reached even higher values (up to 90%), if only parts of the putative transcripts were considered. In fact, no expressed sequence tags entirely containing one of our transcription units were found, so that if a sense/antisense-based regulation would occur, it would likely be related to parts of the ncRNA sequences, while the other part could have structural properties that facilitate this regulatory action (perhaps by binding specific structural proteins). Based on these observations, a novel control mechanism of gene expression could be postulated where pol III (or pol III-like) elements act as trans-locus antisense of their homologous protein-coding RNAs. In this model, the pol III cogenes in antisense configuration with respect to one (or more) specific target gene(s) could regulate their expression either by interfering with its mRNA maturation (if the homologous region is internal to an intron) or by inhibiting protein translation (if the homology is associated to an exon).
21A as Cogene Experimental Model
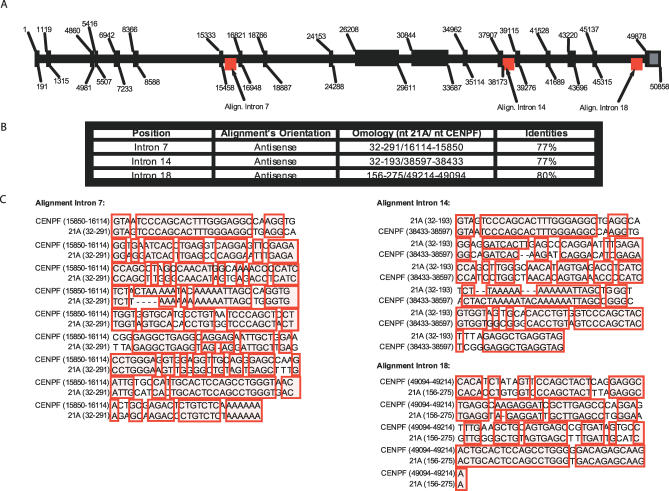
To test our hypothesis we selected one of the novel transcription units (here referred to as 21A) that maps in 8q24.13. If aligned to the human genome, it shows several homology hits among which the most highly significant were associated to multiple intronic regions of centromeric protein F gene (CENP-F; 1q32-q41) [22], thus constituting its putative natural trans-chromosomal antisense (Figure 1A–1C). Although, similarly to all of the 7SL/Alu-derived elements, 21A is expected to be primate-specific [23], an evolutionary conservation analysis was performed aligning its sequence with the mouse-predicted CENP-F gene. No significant similarities were found indicating that in rodents a putative CENP-F antisense regulatory role, if any, would be associated to a different class of noncoding elements. Despite its high sequence similarity with other human Alus, 21A lacks the Alu-specific intragenic consensus elements needed to promote its pol III transcription such as the blocks A and B [24]. This observation further pointed to a 21A transcription driven by an extragenic type 3 pol III promoter.
Figure 1. CENP-F as 21A-Specific Molecular Target.
(A) Human CENP-F gene structure as resulting from GI:89161185 (region 212843155 - 212904537).
(B) The positions of the 21A antisense homologous regions are reported together with their percentage of identity.
(C) Sequence alignment of 21A/CENP-F homologous regions.
To check for 21A expression in cultured cells, we performed Northern blot analysis on total HeLa cell RNA and skin fibroblast RNA using a 21A dsDNA probe. Two positive bands were detected: one corresponding in size to the expected 21A transcript (∼300 nucleotides), and the other one corresponding to a high molecular mass transcript (as expected for CENP-F mRNA) (Figure 2A). However, considering that the 21A double-strand cDNA probe would detect transcription of 21A-similar Alus from multiple loci, we also amplified a 21A-specific cDNA from total RNA samples, extracted from skin fibroblasts and four tumor cell lines (293T, LAN5, HCT, and HeLa), by random hexamer-based RT-PCR in order to better identify a 21A-specific transcription product (Figure 2B). The DNA band obtained was then purified and sequenced, evidencing that the amplification product was the expected 21A. In addition, to better assess 21A transcription, we fused its promoter region to a luciferase silencer hairpin and cotransfected this construct with a plasmid-expressing luciferase. Results showed a halved luciferase activity 48 h after transfection, thus demonstrating an efficient transcription directed by 21A promoter. In the same experiment, a set of five novel promoters from our collection was tested, demonstrating an active transcription of the hairpin promoted by four of them (Figure 2C). These data support the conclusion that the majority of the novel putative transcription units is under the control of active extragenic PSE/TATA-containing promoters.
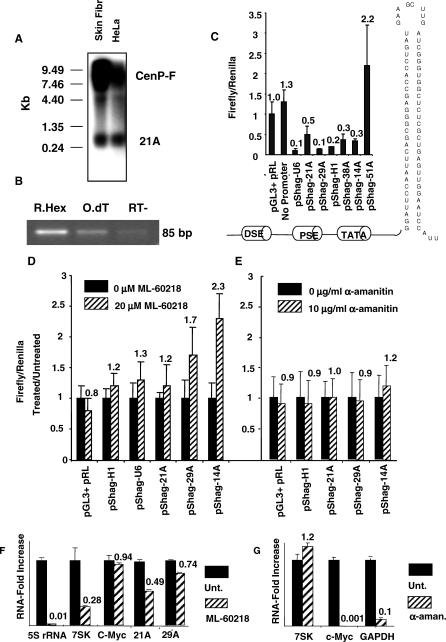
Figure 2. Pol III-Dependent Synthesis of Novel Transcription Units.
(A) Northern blot analysis of human skin fibroblasts and HeLa cells. Results show two bands: the first (detected at about 300 nucleotides) being the 21A endogenous product and the second (of a very high molecular mass) representing CENP-F mRNA.
(B) 21A-specific RT-PCR amplification. As expected for nonpolyadenylated transcripts, an efficient amplification product was obtained only in the random hexamers-primed reactions.
(C) Promoter activity transfection assay. A specific luciferase-silencing hairpin is transcribed by six PSE/DSE-dependent promoter elements (11A [H1 RNA], 14A, 21A, 29A, 38A, 51A). A view of the silencing constructs including the hairpin nucleotide sequence is enclosed. The promoter region encompasses the putative pol III type 3 regulatory regions (TATA box, PSE, and DSE). pGL3 + pRL, negative control; pSHAG-U6, canonical pol III promoter; No promoter, hairpin without PSE/DSE-dependent promoter thus resulting transcriptionally inactive.
(D and E) Promoter-activity transfection assay in presence/absence of 20 μM ML-60218 cell-permeable pol III inhibitor or 10 μg/ml α-amanitin pol II–specific inhibitor. Results are reported as luciferase emission of treated versus untreated samples.
(F) Real-time RT-PCR analysis of transcript levels for 21A and 29A transcription units, two known pol III–transcribed genes (7SK and 5S rRNA), and one pol II–transcribed gene (c-Myc) in ML-60218 treated/untreated cells, as resulting after normalization of each treated sample with its untreated counterpart. All the measures were referred to a ML-60218 unaffected pol II housekeeping gene (GAPDH).
(G) Real-time RT-PCR analysis of the RNA level of two known pol II–transcribed (c-Myc and GAPDH) and one pol III–transcribed (7SK) genes in α-amanitin treated/untreated cells as resulting after normalization of each treated sample with its untreated counterpart. All the measures were referred to an α-amanitin unaffected pol III gene (5S rRNA).
Pol III–Dependence of the Novel Transcription Units
The same experiment as above was repeated after 24 h of cell treatment with ML-60218, a cell-permeable indazolo-sulfonamide compound that displays broad-spectrum inhibitory activity against pol III [25]. Results showed an efficient luciferase-silencing activity in the absence of the pol III inhibitor (as evidenced by a decreased luciferase emission), while after treatment with ML-60218 the luciferase signal was increased (Figure 2D). As a control, a similar experiment was performed by treating the cells for 12 h with a pol II inhibitor (α-amanitin). In this case, no major transcription variations were observed either for the novel transcript units or for the well-known pol III-dependent H1 gene (Figure 2E).
To better assess pol III dependence of 21A transcription, we directly measured the endogenous 21A (and 29A) RNA amount in ML-60218 treated cells versus untreated control samples. Results evidenced a significant transcription downregulation of the two novel pol III units (50% and 30% inhibition in the case of 21A and 29A, respectively), thus keeping in line with the occurrence of a specific effect of ML-60218 on their promoters.
To further assess the specificity of action of the two inhibitors, we analyzed by real-time RT-PCR the transcription activity of two pol III (5S rRNA and 7SK) and two pol II-dependent genes (c-Myc and glyceraldehyde 3 phosphate dehydrogenase [GAPDH]) both in ML-60218 and in α-amanitin-treated cells. Results showed a significant inhibition of the pol III–dependent genes after ML-60218 treatment and a stable transcription level of the pol II–dependent genes in the same samples (Figure 2F). On the contrary, pol III transcription activity was stable in α-amanitin–treated cells, while in the same samples the pol II–transcribed genes were downregulated, thus demonstrating the specificity of action of the two inhibitors in these experimental conditions (Figure 2G). Altogether, these results provide evidence that the novel PSE/TATA-containing transcription units are transcribed by pol III.
21A Acts as CENP-F Regulatory Cogene Modulating Its Expression at Posttranscriptional Level
To test whether the 21A transcript acts as an antisense inhibitor of CENP-F expression, we measured by Western analysis CENP-F protein level in HeLa cells transiently transfected with four different 21A constructs carrying: (i) the whole 21A region containing both DSE and PSE elements (p21A); (ii) its upstream moiety, which contains the DSE and a MIR element, but not the CENP-F homology region (p21A-1); (iii) the novel pol III type 3–transcription region (which includes an Alu Jb module) (p21A-2); and (iv) an empty vector as mock control (pMock). As shown in Figure 3, starting at 24 h from transfection of the whole 21A region, inhibition of CENP-F accumulation (followed by a rapid degradation) was observed. Such inhibition was specifically associated to constructs expressing the 21A RNA (p21A, p21A-2), while the MIR element in the upstream moiety of the fragment (p21A-1 construct) was ineffective (Figure 3A–3D).
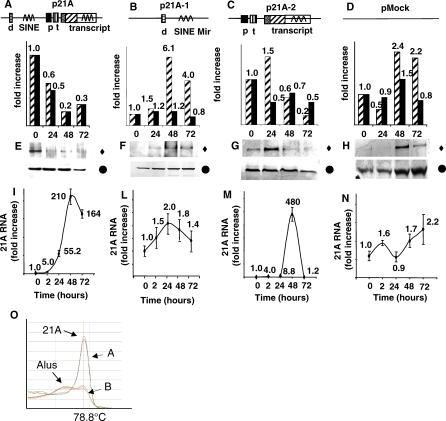
Figure 3. 21A-Driven CENP-F Expression Regulation.
(A–D) 21A constructs. d, DSE Element; p, PSE Element; p21A, whole transcription unit; p21A-1, upstream, DSE-containing region; p21A-2, transcription region; pMock, empty vector; t, TATA box.
(E–H) CENP-F protein expression level after 0, 24, 48, and 72 h of constructs transfection. Full triangle, anti-CENP-F antibody; Full circle, anti-tubulin antibody (for protein loading normalization); Striped columns, quantitative determination of CENP-F expression modulation as determined by Western blot analysis; Full columns, quantitative determination of CENP-F mRNA expression modulation as determined real-time RT-PCR analysis.
(I–N) 21A RNA level in transfected samples indicating that the exogenous 21A expression inversely correlates with CENP-F protein expression.
(O) Dissociation curve of 21A amplification products. A, 21A-transfected HeLa cells; B, untransfected HeLa cells showing the very low basal 21A transcription level.
In this context, it has to be noted that a slight delay occurred in 21A-2 inhibitory action (and in the expression of 21A-specific RNA), compared to what was observed with the complete 21A construct (more rapid increase in 21A RNA expression and decrease of CENP-F protein levels), suggesting a positive transcriptional role of the DSE element. The actual occurrence of 21A transcription in transfected cells was analyzed by real-time quantitative RT-PCR. As expected, a very high amount of 21A transcript was detected in p21A and p21A-2–transfected cells (210- and 480-fold, respectively, at 48 h from transfection), while the 21A RNA content of samples transfected with pMock control plasmid and/or with a construct containing the promoter lacking the transcribed region (p21A-1 construct) were essentially stable, showing a very low basal level of 21A expression in untransfected HeLa cells (Figure 3I–3N). All the PCR products were analyzed in their dissociation curve, showing a single characteristic pick (at 78–79 °C) in p21A/p21A2-transfected samples significantly reduced in pMOCK/p21A-1. On the contrary, the cells transfected with the two control plasmids (pMock/p21A-1) showed a dissociation pattern characteristic of a heterogeneous population of molecules (Figure 3O). Again these results confirmed an active synthesis of the exogenous 21A ncRNA transcript in p21A/p21A-2–transfected samples that was strongly reduced at a very low endogenous–basal level in the samples lacking the transcript region (pMOCK/p21A-1). As a consequence of 21A very active transcription, the level of CENP-F mRNA (as determined by real-time RT-PCR) was significantly decreased in p21A/p21A-2–transfected cells, while no major CENP-F mRNA variations were observed in pMOCK/p21A-1–transfected cells (Figure 3E–3H). Altogether these results demonstrate an inverse correlation between 21A transcription and CENP-F expression. Therefore, considering the high sequence homology between 21A transcript and three CENP-F hnRNA intronic portions, we suggest a mechanism of antisense inhibition of CENP-F mRNA maturation by the 21A transcript.
21A Overexpression Specifically Inhibits Cell Proliferation in Humans
Given the central role of CENP-F in mitosis, we tested the effect of ectopic 21A expression on cell proliferation. By measuring [3H]-thymidine incorporation, we observed a dramatic arrest of cell proliferation after 48 h in 21A-transfected cells. Again, the effect was specifically associated to the downstream 21A transcribed region (p21A/p21A-2 constructs), while transfection of the MIR-containing upstream moiety (p21A-1 construct) did not alter cell proliferation (Figure 4A). Although at the present state we cannot exclude a contribution to this effect by Alus from other loci, this experiment demonstrates an inverse correlation of 21A transcription and cell proliferation that is in accord with the inhibition of CENP-F synthesis demonstrated above.
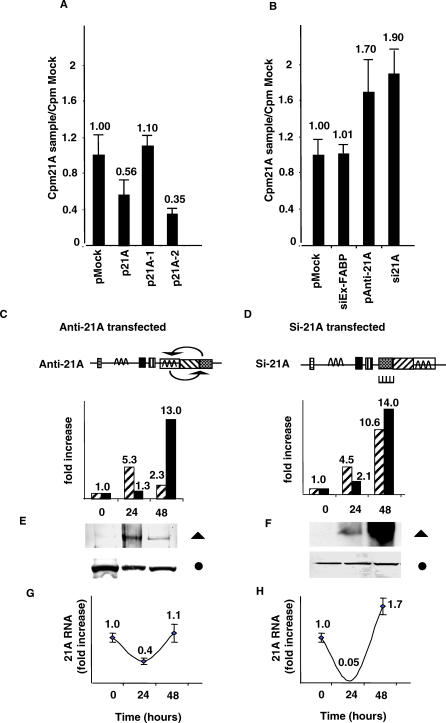
Figure 4. Modification of Cell Proliferation Rate in 21A-Overexpressing HeLa Cells.
(A) Proliferation inhibition of HeLa cells after 48 h of 21A constructs transfection. Results emphasize the specificity of the Alu Jb-containing region as a source of proliferation inhibitory transcripts.
(B) Proliferation increase of HeLa cells after 48 h of pAnti-21A and si21A transfection. siEx-FABP, unrelated chicken-specific siRNA (negative control).
(C and D) Constructs structures. Anti-21A, the transcript region is inverted and the construct maintains 21A promoter as well as its termination site. si21A, siRNA 21A-specific.
(E and F) CENP-F protein expression level after 0, 24, and 48 h of constructs transfection. Full triangle, anti-CENP-F antibody; full circle, anti-tubulin antibody; striped columns, quantitative determination of CENP-F expression modulation as determined by Western blot analysis; full columns, quantitative determination of CENP-F mRNA expression modulation as determined real-time RT-PCR analysis.
(G and H) 21A RNA level in transfected samples indicating that the endogenous 21A expression is inhibited after 24 h of anti-21A/si21A transfection.
To further support the antisense role of 21A, we transfected HeLa cells with a construct expressing the transcript in antisense configuration (here referred to as pAnti-21A), thus quenching the activity of the endogenous 21A molecules. Results showed an increased cell proliferation 24–48 h after transfection. Similar results were obtained when a 21A-specific small interfering RNA (siRNA)–expressing construct was transfected in HeLa cells, while the negative control sample (cells transfected with an unrelated chicken-specific siRNA) maintained a cell-proliferation rate similar to that of pMock-transfected cells (Figure 4B). In both of the experiments an increased CENP-F expression was detected both at the protein and mRNA levels (Figure 4E and 4F). As evidenced by real-time RT-PCR in the same experiment, a concomitant 21A-RNA decrease was observed after 24 h of transfection in anti/si21A treated cells, although a complete recovery of 21A RNA synthesis occurred after 48 h of transfection (Figure 4G–4H). As shown in these experiments CENP-F modulation and 21A RNA decrease were analyzed only at 0, 24, and 48 h after transfection, rather than at 0, 24, 48, and 72 h as in the previous experiments. In fact, at 72 h after transfection the CENP-F synthesis determinations would be strongly biased by an early cell culture overconfluence caused by the proliferation increase that follows 21A downregulation.
These data suggest that the decreased amount of 21A transcript consequent to its siRNA-mediated silencing, as well as its suppression by antisense technology, specifically increases CENP-F synthesis, thus keeping in line with the proposed role of 21A as CENP-F regulatory cogene. In addition, it has to be considered that the increased proliferation rate observed here supports the idea of a widespread regulatory action of 21A that may control at the posttranscriptional level the expression of several target genes similarly to what has been proposed for miRNAs [26].
The 21A Regulatory Effect Is Human Specific
Considering that a 21A-driven cell-proliferation inhibition is expected to be primate specific (Alu sequences were not found in other mammalian orders), we tested for its possible occurrence in mouse. We found that, after transfection of p21A, p21A-1, p21A-2, and pMock, the murine fibroblast NIH 3T3 cells did not show any proliferation decrease as assessed by [3H]-thymidine incorporation (Figure 5). The species-specificity of 21A action, together with its inability to cause a nonspecific cell reaction that leads to a proliferative blockade in mice, further strengthens a 21A-specific (perhaps multilocus) regulatory role.
Figure 5. Mouse NIH-3T3 Cell Proliferation Rate after Transfection of 21A Constructs.
No proliferation decrease was observed.
In fact, considering these data we rule out a nonspecific effect of 21A on cell proliferation, perhaps due to the activation of a more general biological process, such as the interferon response (an antiviral cell reaction shared by all mammals), rather than a specific multilocus 21A regulatory action.
21A Endogenous ncRNA Is Downregulated in Tumor Cell Lines
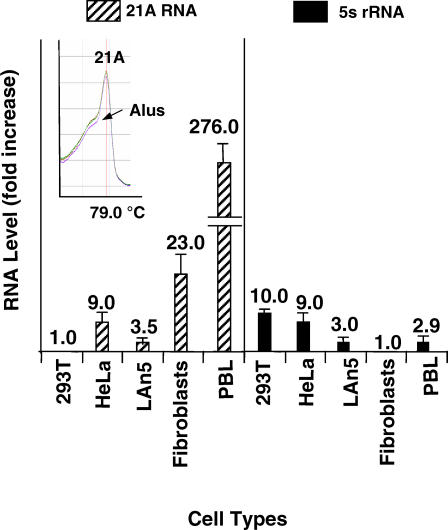
As demonstrated by transfection experiments, 21A overexpression is inversely correlated to cell proliferation. In accordance with this finding, its basal expression level is very low in fully proliferating HeLa cells. To better investigate the inverse correlation between the endogenous 21A expression and cell proliferation, we analyzed by quantitative real-time RT-PCR the 21A expression levels in cell types characterized by different proliferation potentials. Results showed that in three immortalized, fully proliferating cell lines analyzed here (HeLa as cervical adenocarcinoma; 293T as renal epithelial adenovirus transformed cells; LAN5 as neuroblastoma), the level of 21A transcription was very low if compared to the unproliferating/resting PBL (peripheral blood lymphocyte) cells, in which a 276-fold-increased–21A transcription was evidenced. In the same experiment, according to an inverse correlation between endogenous 21A transcription and the cell proliferation rate, the 21A RNA level in primary skin fibroblasts (of which the proliferation rate is significantly lower than that of the tumor cell lines analyzed here) showed a 23-fold increase compared to 393 T cells, and a very low expression level if compared to the resting/unproliferating PBL (Figure 6). Again the dissociation curve analysis of 21A amplification product showed in PBL a peak at 78–79 °C, characteristic of a single specific molecular species that resembled the one obtained in 21A/21A-2 transfected cells (where the amount of 21A transcripts was strongly increased), although a slight shoulder, most likely due to a cross-amplification of other very similar transcripts, revealed a detectable endogenous Alu transcription background (Figure 6). Altogether these results evidence a very active 21A transcription in PBL/resting cells that further strengthens the idea of 21A as a novel key factor of cell-proliferation control.
Figure 6. Real-Time RT-PCR Analysis of 21A Endogenous RNA in Different Cell Types with Respect to the Less Abundant Samples (293T for 21A RNA and Fibroblasts for 5S rRNA).
Striped columns, 21A RNA; full columns, 5S rRNA. The dissociation curve of 21A amplification product in PBL is reported in the inset.
In order to check if the endogenous 21A overexpression in unproliferating cells was related to a widespread increased RNA polymerase III activity rather than a 21A-specific activation, we measured by real-time RT-PCR the 5S rRNA expression level in the same samples. The results showed no direct correlation between 5S rRNA expression and the cell-proliferation rate variations, evidencing that the 21A overexpression in resting cells was the consequence of a 21A-specific transcription activation rather than a wider, nonspecific increase of pol III activity (Figure 6). These data thus suggest the existence of an unexpectedly specific expression regulation of 21A promoter (related to the cell proliferation state) that needs to be investigated in detail.
Conclusions
We propose that the noncoding fraction of the human genome includes a larger than expected number of ncRNA genes controlled by DSE and PSE promoter elements. Due to their promoter structure, a number of these genes is likely to be transcribed by pol III. We refer to them as cogenes since they could specifically coact with a protein-coding pol II gene. Given the very high sequence homology between pol III and pol II transcript pairs, and in light of the results we have obtained investigating the regulatory activity of the 21A transcription unit, we propose that a large part of these novel elements may act as antisense inhibitors of protein translation and/or mRNA maturation, although some of them (those whose homology with the pol II target gene is in sense configuration) could play a role in gene-expression regulation with different mechanisms. Altogether these findings provide evidence for the existence of an ncRNA gene set associated to PSE/DSE-containing promoters, whose products coact with a corresponding set of protein-coding targets.
In conclusion, this study provides: (i) a collection of novel noncoding transcripts to be investigated for their potential regulatory action with respect to pol II target genes; (ii) a novel source of PSE-dependent promoters useful for the identification of common regulatory regions specific for this type of promoters; (iii) a novel class of molecules involved in the RNA-dependent gene expression regulation; and (iv) a novel transcript (21A), whose role in tumor cell proliferation deserves further investigation in the context of cancer studies.
Materials and Methods
Databases and searches.
All of the sequence searches and alignments were carried out by means of BLAST at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST). The sequences used to query were the following: H1 PSE, nCACCATAAAnGTGAAAn or nTTTCACnTTTATGGTGn; U6 PSE, CTTACCGTAACTTGAAAGT; 7SK PSE (as reported in PMID: 2011518), TTGACC-TAAGTG; DSE (Octamer-Binding Transcription Factor 1 [Oct1] consensus sequence), ATTTGCAT or ATGCAAAT with or without a single base of mismatch.
Cell culture, transfection, and luciferase assay.
For transient transfections, HeLa cells (grown in DMEM supplemented with 10% FCS), were grown in multiwell petri dishes 16 h before transfection. The expression (21A, 21A[1], 21A[2], 21A[3]) constructs containing the regions of interest cloned in the pTopo vectors (Invitrogen, http://www.invitrogen.com) were introduced into the cells using the Fugene 6 transfection reagent (Roche, http://www.roche.com/home.html) according to the manufacturer's instructions. A plasmid expressing luciferase was used as a control of transfection efficiency (to which all the results were normalized). Cells were harvested 24, 48, and 72 h after transfection, and firefly luciferase activity was measured by dual-luciferase reporter–assay system (Promega, http://www.promega.com) according to the manufacturer's protocol. To specifically inhibit RNA polymerase III and/or RNA polymerase II, a cell-permeable chlorobenzenesulfonamide (ML-60218) (Calbiochem, http://splash.emdbiosciences.com) and/or α-amanitin (Roche, http://www.roche.com) were used at the concentration of 20 μM and 10 μg/ml, respectively, in the medium for 25 h (ML-60218) and 12 h (α-amanitin) before the luciferase activity detection.
RNA interference–silencing assay.
To test the promoter activity of the novel transcription units, we prepared six plasmid constructs expressing a firefly luciferase-silencing hairpin (Gregory Hannon, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, United States); transcription was driven by the 11A, 14A, 21A, 29A, 38A, 51A promoters, respectively. The hairpin sequence (targeting a firefly luciferase mRNA from a cotransfected expression plasmid [Promega]) is: 5′-GGAUUCCAUUCAGCGGAGCCACCUGAUGAAGCUUGAUCGGGUCUCGCUGAGUUGGAAUCCAUU-3′. Oligos used to subclone the novel pol III type III promoters within Not I/HinD III restriction sites (uppercase letters) were the following:
11AFprom Not I: 5′-atgcGCGGCCGCatttgcatgtcgctatgtg-3′
11ARprom HinD III: 5′-gatcAAGCTTcatcaggtggctcccgctgaattggaatccacgcactcagctcgtg-3′
14AFprom Not I: 5′-atgcGCGGCCGCaactgatgtatgattatatctt-3′
14ARprom HinD III: 5′-gatcAAGCTTcatcaggtggctcccgctgaattggaatccattattatctcctttgttctgt-3′
21AFprom Not I: 5′-atgcGCGGCCGCacagctgtagcagatgct-3′
21ARprom HinD III: 5′-gatcAAGCTTcatcaggtggctcccgctgaattggaatccaccacacttggtcaactat-3′
29AFprom Not I: 5′-atgcGCGGCCGCttctcacctaaaggagtc-3′
29ARprom HinD III: 5′-gatcAAGCTTcatcaggtggctcccgctgaattggaatccttctaatcctcctaagatca-3′
38AFprom Not I: 5′-atgcGCGGCCGCttcactaagatccagtgc-3′
38Arprom HinD III: 5′-gatcAAGCTTcatcaggtggctcccgctgaattggaatccgattcatgaacacagaatatt3′
51AFprom Not I: 5′-atgcGCGGCCGCgttgaacatttaactctgtat-3′
51Arprom HinD III: 5′-gatcAAGCTTcatcaggtggctcccgctgaattggaatccctcatggcacttggagat-3′
In this analysis, the above constructs were cotransfected with a pGL3 plasmid-expressing (Promega) firefly luciferase as a target to be silenced and with a pRL plasmid expressing (Promega) renilla luciferase to which all the determinations were normalized. Cells were harvested 24, 48, and 72 h after transfection, and firefly/renilla luciferase activities were measured by dual-luciferase reporter–assay system (Promega) according to the manufacturer's protocol.
Plasmid constructs generation and sequencing.
The plasmid constructs p21A, p21A(1), and p21A(2) were generated amplifying from a genomic DNA preparation of the regions of interest; the PCR products were then subcloned into the pNEB193 vector. The oligos used to generate p21A PCR fragments were the following:
21A forward: 5′-GGAAATCTTACCTTCCTGCC-3′
21A reverse: 5′-TGGCTAGGTCATGTGACCAT-3′
21A(1) forward: 5′-GGAAATCTTACCTTCCTGCC-3′
21A(1) reverse: 5′-TTCATTCATTCATTCATTGATTCAC-3′
21A(2) forward: 5′–CAGCTGCAGCAGATGCTAGCAGGGC-3′
21A(2) reverse: 5′–TGGCTAGGTCATGTGACCATTC-3′
The plasmid construct pAnti-21A was generated amplifying the transcribed region from p21A plasmid using the following oligos:
Anti-21A Terminator-containing forward: 5′-CTGAAAAAGTAGTCCCAGCACTTTG-3′
Anti-21A Bam HI-containing reverse: 5′-ATGCGGATCCGAGACAGGGTCTTGCTC-3′
Thus the transcribed region was generated in antisense configuration. The pAnti-21A promoter was obtained by amplifying p21A promoter with the following oligos:
21A Forward: 5′-GGAAATCTTACCTTCCTGCC-3′ p21A Bam HI-containing reverse: 5′-ATGCGGATCCGAGCCACCACACTTGGTC-3′. The PCR products were digested with the restriction enzyme Bam HI, purified by gel electrophoresis, and ligated by T4 ligase (Invitrogen). The insert obtained was then subcloned in pTOPO vector (Invitrogen) following the manufacturer's instructions. Prior to transfection all of the plasmids were sequenced by DNA sequencing kit (Applied Biosystems, www.appliedbiosystems.com) following the manufacturer's instructions.
RT-PCR reactions.
To isolate and sequence a partial 21A cDNA, we performed different RT-PCR reactions. Starting from about 5 μg of total RNA, cDNA was synthesized by using an oligo (dT)12–18 primer or a random hexamers mix and a superscript first-strand synthesis system for RT-PCR (Invitrogen). cDNAs were diluted 10–50 times, then subjected to PCR reactions. The oligos used to isolate 21A RT-PCR product were: oligo forward 21AF 5′-gctcacgtagtcccagcacttt-3′ and oligo reverse 21AR 5′-actatgttgcccaagctggtct-3′. PCR products were separated on 1.5%–2% agarose gel. The DNA bands were cut, purified by the Millipore DNA gel extraction kit (http://www.millipore.com), and sequenced.
Real-time quantitative RT-PCR.
The RNA for 21A was measured by real-time quantitative RT-PCR using the PE ABI PRISM@ 7700 sequence detection system (PerkinElmer, http://www.perkinelmer.com) and Sybr Green (Applied Biosytems). The sequences of 21A forward and reverse primers as designed by the Primer Express 1.5 software (Applied Biosystems) were 5′-GCTGAGGCAGGAGGATCACT-3′ and 5′-GCACTACCACACCCAGCTAATTTT-3′. The sequences of CENP-F forward and reverse primers were 5′-CTGCAGAAAGAACTCTCTCAACTTC-3′ and 5′-TCAACAATTAAGTAGCTGGAACCA-3′. For endogenous control, the expression of GAPDH gene was examined. The sequences for human GAPDH primers were 5′-GAAGGTGAAGGTCGGAGTC-3′ and 5′- GAAGATGGTGATGGGATTTC-3′. The sequences for human 5S rRNA primers were 5′-TACGGCCATACCACCCTGAA-3′ and 5′-GCGGTCTCCCATCCAAGTAC-3′. The sequences for human 7SK RNA primers were 5′-AGGACCGGTCTTCGGTCAA-3′ and 5′-TCATTTGGATGTGTCTGCAGTCT-3′. The sequences for human c-Myc primers were 5′-CGTCTCCACACATCAGCATAA-3′ and 5′-GACACTGTCCAACTTGACCCTCTT-3′. Relative transcript levels were determined from the relative standard curve constructed from stock cDNA dilutions and divided by the target quantity of the calibrator following manufacturer's instructions.
Anti-21A siRNA synthesis.
The Anti-21A siRNA was synthesized against a region of the 21A transcript of no homology with CENP-F so that the silencing effect was specific for the pol III regulatory RNA and did not interfere with CENP-F RNA stability. The siRNA synthesis was carried out taking advantage of the siRNA construction kit (Ambion, http://www.ambion.com) according to the manufacturer's protocol. The sense/antisense oligos used were: 5′-aaGTGTGGTGGCTCACcctgtctc-3′ and 5′-aaGTGAGCCACCACACcctgtctc-3′.
Proliferation assay.
We tested proliferation of HeLa cells transfected with 21A, 21A-1, 21A-2, 21A-3, and Anti-21A constructs plating 5 × 105 cells per well in round-bottomed 96-well plates, incubated for 24, 48, and 72 h after transfection, and pulsed with [3H]-thymidine (1.0 μCi/10 μl/well) (Amersham Biosciences, http://www5.amershambiosciences.com) for the last 18 h. We harvested the cells and evaluated cell proliferation by counting the thymidine uptake. We calculated the averaged proliferation rate, measured as counts per minute, and standard deviation for the triplicate wells of each sample.
RNA isolation and Northern blot analysis.
Based on a single-step acid-phenol–guanidium method, total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Total RNAs, from HeLa cells, were electrophoresed through 1.5% agarose gels in the presence of formaldehyde and blotted onto Hybond N membranes (Amersham). The blot was hybridized with an 85-bp–long probe contained in the region from nucleotide 1,194 to nucleotide 1,278 of the 21A reported sequence (Table S2), spanning a region internal to the transcript. The probe was obtained by PCR (using the 21A plasmid construct as template) using the following oligos: 21AF 5′- GCTCACGTAGTCCCAGCACTTT-3′ and 21AR 5′-AGACCAGCTTGGGCAACATAGT-3′. Blot prehybridization was performed at 65 °C for 2 h in 333 mM NaH2PO4 (pH 7.2), 6.66% sodium dodecyl sulphate, and 250 mg/ml denatured salmon sperm DNA. Blot hybridization was performed at 65 °C for 18 h in the same solution containing 106 counts per minute/ml of denatured and labeled probes. After hybridization the blots were washed twice at 65 °C for 30 min in 0.2% sodium dodecyl sulphate, 2× SSPE and once at 65 °C for 30 min in 0.2% sodium dodecyl sulphate, and 0.2× SSPE. Membranes were exposed to autoradiographic films for 24–48 h and then developed.
Western blot analysis.
Equal amounts of proteins (10 μg/sample) from each sample were loaded on standard 4%–12% NU-PAGE gradient gels (Invitrogen). Blotting onto Protran nitrocellulose membranes (Schleicher & Schuell, www.schleicher-schuell.com) was performed in the X-Cell Sure Lock Electrophoresis Cell (Invitrogen), according to the manufacturer's instructions. The membranes were saturated overnight in 3% nonfat milk in TTBS buffer (500 nM NaCl; 20 mM Tris/Cl [pH 7.5]; 0.05% Tween-20) and incubated for 4 h at room temperature with the human anti-mitosin/CENP-F ab90 (ABCAM, http://abcam.com) and/or anti-alpha tubulin (OMIM 191110) (Sigma-Aldrich, www.sigmaaldrich.com) mouse monoclonal antibodies. The anti-mitosin antibody recognized a weak signal at a very high apparent molecular mass (350–400 kDa), while the anti-alpha tubulin showed a clear signal at 45 kDa. The immunoreactive band was revealed by an alkaline phosphate-conjugated affinity-purified monoclonal anti-rabbit–mouse IgG (Sigma-Aldrich), and (in the experiment indicated in Figure 1C) the enzymatic chemiluminescence (ECL) detection system (Amersham), or (in the experiment indicated in Figure 1E) the alkaline phosphatase substrate BCIP/NBT (ICN Biomedicals, http://www.mpbio.com).
Supporting Information
(52 KB PPT)
(104 KB DOC)
(429 KB PPT)
(45 KB RTF)
(74 KB DOC)
Accession Numbers
The National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) accession numbers for the genes and gene products discussed in this paper are: U6 (M14486), 7SK (001445), H1 (002312), U2 (002716), CENP-F (016343), 5S rRNA (V00589), c-Myc (002467), and GAPDH (002046).
Acknowledgments
We thank Silvio Garofalo and Lucio Luzzatto for helpful discussions and suggestions. pSHAG vectors were obtained from Gregory Hannon (Cold Spring Harbor Laboratory).
Abbreviations
- CENP-F
centromeric protein F
- DSE
distal sequence element
- GAPDH
glyceraldehyde 3 phosphate dehydrogenase
- MIR
mammalian interspersed repeat
- miRNAs
microRNAs
- nc
noncoding
- pol
polymerase
- PSE
proximal sequence element
- siRNA
small interfering RNA
- snRNA
small nuclear RNA
- RT-PCR
reverse transcriptase PCR
Footnotes
Competing interests. The authors have declared that no competing interests exist.
A previous version of this article appeared as an Early Online Release on November 20, 2006 (doi:10.1371/journal.pgen.0030001.eor).
Author contributions. AP conceived and designed the experiments. AP, MC, FT, and RF performed the experiments. AP, MC, FT, RF, GD, and RC analyzed the data. AP, MC, FT, and RC contributed reagents/materials/analysis tools. AP wrote the paper.
Funding. GD was supported by grants from the Human Frontier Science Program (grant RGY0011/2002-C) and by the Italian Ministry of Education, University and Research (MIUR, 2005 PRIN, and FIRB Programs).
References
- Reis EM, Nakaya HI, Louro R, Canavez FC, Flatschart AV, et al. Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer. Oncogene. 2004;23:6684–6692. doi: 10.1038/sj.onc.1207880. [DOI] [PubMed] [Google Scholar]
- Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, et al. Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- Dahary D, Elroy-Stein O, Sorek R. Naturally occurring antisense: Transcriptional leakage or real overlap? Genome Res. 2005;15:364–368. doi: 10.1101/gr.3308405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. RNA regulation: A new genetics? Nat Rev Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Non-coding RNA [review] Hum Mol Genet. 2006;15:17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Levanon K, Eisenberg E, Rechavi G, Levanon EY. Letter from the editor: Adenosine-to-inosine RNA editing in Alu repeats in the human genome [review] EMBO Rep. 2005;6:831–835. doi: 10.1038/sj.embor.7400507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen SK, Han K, Wang J, Lee J, Wang H, et al. Human genomic deletions mediated by recombination between Alu elements. Am J Hum Genet. 2006;79:41–53. doi: 10.1086/504600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JJ, Egry LA, Wong GH, Kaspar RL. The 3′ UTR of human MnSOD mRNA hybridizes to a small cytoplasmic RNA and inhibits gene expression. Biochem Biophys Res Commun. 2000;27:641–648. doi: 10.1006/bbrc.2000.3189. [DOI] [PubMed] [Google Scholar]
- Liu WM, Chu WM, Choudary PV, Schmid CW. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995;23:1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Torvik VI. Alu elements within human mRNAs are probable microRNA targets. Trends Genet. 2006;22:532–536. doi: 10.1016/j.tig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- Myslinski E, Ame JC, Krol A, Carbon P. An unusually compact external promoter for RNA polymerase III transcription of the human H1RNA gene. Nucleic Acids Res. 2001;29:2502–2509. doi: 10.1093/nar/29.12.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo SM, Ifill S, Hernandez N. Cis-acting elements required for RNA polymerase II and III transcription in the human U2 and U6 snRNA promoters. Nucleic Acids Res. 1990;18:2891–2899. doi: 10.1093/nar/18.10.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj IW, Dathan NA, Parry HD, Carbon P, Krol A. Changing the RNA polymerase specificity of U snRNA gene promoters. Cell. 1988;55:435–442. doi: 10.1016/0092-8674(88)90029-3. [DOI] [PubMed] [Google Scholar]
- Cozzarelli NR, Gerrard SP, Schlissel M, Brown DD, Bogenhagen DF. Purified RNA polymerase III accurately and efficiently terminates transcription of 5S RNA genes. Cell. 1983;34:829–835. doi: 10.1016/0092-8674(83)90540-8. [DOI] [PubMed] [Google Scholar]
- Braglia P, Percudani R, Dieci G. Sequence context effects on oligo (dT) termination signal recognition by Saccharomyces cerevisiae RNA polymerase III. J Biol Chem. 2005;280:19551–19562. doi: 10.1074/jbc.M412238200. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Liao H, Winkfein RJ, Mack G, Rattner JB, Yen TJ. CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J Cell Biol. 1995;130:507–518. doi: 10.1083/jcb.130.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Labuda D. CORE-SINEs: Eukaryotic short interspersed retroposing elements with common sequence motifs. Proc Natl Acad Sci U S A. 1999;96:2869–2874. doi: 10.1073/pnas.96.6.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolella G, Lucero MA, Murphy MH, Baralle FE. The Alu family repeat promoter has a tRNA-like bipartite structure. EMBO J. 1983;2:691–696. doi: 10.1002/j.1460-2075.1983.tb01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Pan J, Thoroddsen V, Wysong DR, Blackman RK, et al. Novel small-molecule inhibitors of RNA polymerase III. Eukaryot Cell. 2003;2:256–264. doi: 10.1128/EC.2.2.256-264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(52 KB PPT)
(104 KB DOC)
(429 KB PPT)
(45 KB RTF)
(74 KB DOC)