Abstract
Nucleotide binding site–leucine-rich repeat (NBS–LRR) proteins mediate pathogen recognition in both mammals and plants. The molecular mechanisms by which pathogen molecules activate NBS–LRR proteins are poorly understood. Here we show that RPS5, a NBS–LRR protein from Arabidopsis, is activated by AvrPphB, a bacterial protease, via an indirect mechanism. When transiently expressed in Nicotiana benthamiana leaves, full-length RPS5 protein triggered programmed cell death, but only when coexpressed with AvrPphB and a second Arabidopsis protein, PBS1, which is a specific substrate of AvrPphB. Using coimmunoprecipitation analysis, we found that PBS1 is in a complex with the N-terminal coiled coil (CC) domain of RPS5 before exposure to AvrPphB. Deletion of the RPS5 LRR domain caused RPS5 to constitutively activate programmed cell death, even in the absence of AvrPphB and PBS1, and this activation depended on both the CC and NBS domains. The LRR and CC domains both coimmunoprecipitate with the NBS domain but not with each other. Thus, the LRR domain appears to function in part to inhibit RPS5 signaling, and cleavage of PBS1 by AvrPphB appears to release RPS5 from this inhibition. An amino acid substitution in the NBS site of RPS5 that is known to inhibit ATP binding in other NBS–LRR proteins blocked activation of RPS5, whereas a substitution thought to inhibit ATP hydrolysis constitutively activated RPS5. Combined, these data suggest that ATP versus ADP binding functions as a molecular switch that is flipped by cleavage of PBS1.
Keywords: AvrPphB, disease resistance, NOD domain, Pseudomonas syringae, RPS5
Both plants and animals employ nucleotide binding site–leucine-rich repeat (NBS–LRR) proteins to mediate detection of pathogen molecules (1). There appear to be at least two distinct mechanisms by which NBS–LRR proteins detect pathogens: either by binding pathogen-derived molecules directly or by sensing the modification of host proteins by pathogen-derived molecules (2). It is presently unclear how either mechanism causes activation of signaling by NBS–LRR proteins. We have been investigating these processes by using the plant NBS–LRR protein RPS5, which mediates detection of the protease AvrPphB from the bacterial pathogen Pseudomonas syringae (3–6).
In plants, NBS–LRR proteins were first identified as the products of classically defined disease-resistance genes (R genes) (7–9), which are genes that confer resistance to infection by specific pathogen strains. R gene-mediated resistance is typically manifested by activation of a programmed cell death response referred to as the hypersensitive response (HR) that is localized to the site of pathogen ingress (10). In the last decade, R genes have been cloned from a large range of plant species, with the majority being found to encode NBS–LRR proteins (11). Plant NBS–LRR proteins can be subdivided into two broad categories defined by the presence of a Toll-interleukin receptor (TIR) domain or a non-TIR domain, most often a coiled-coil (CC) domain, at the amino terminus. The function of the CC and TIR domains in pathogen perception and signaling is unclear. The NBS domain is also known as the NOD, NACHT, CATERPILLAR, or NB-ARC domain and has been shown to bind and hydrolyze ATP in plants and animals (12–14) and to mediate ligand-induced homo-oligomerization of mammalian NBS proteins (1). ATP binding appears to be essential for NBS–LRR signaling, because mutations in the NBS domain that block ATP binding also block function (14). The LRR domain is a key determinant of protein–protein interactions (15) and was shown to determine recognition specificity for a number of plant NBS–LRR proteins (16–18) and thus represents a likely domain for binding pathogen proteins or modified host proteins.
In Arabidopsis, infection with P. syringae expressing AvrPphB induces an RPS5-dependent HR that depends on the Arabidopsis protein kinase, PBS1 (3, 5). AvrPphB protease activity is required for RPS5 activation, and PBS1 is a specific substrate of AvrPphB; thus, we proposed that RPS5 is indirectly activated by AvrPphB via proteolysis of PBS1 (3). Here we show that RPS5 forms a complex with PBS1 before pathogen exposure and that formation of this complex is mediated by the N-terminal CC domain of RPS5. We also show that deletion of the LRR domain constitutively activates RPS5, as does a mutation in the NBS domain thought to inhibit ATP hydrolysis. RPS5 activation is abolished by a mutation that blocks ATP binding, suggesting a model in which cleavage of PBS1 triggers an exchange of ADP for ATP, thereby activating RPS5.
Results and Discussion
AvrPphB-Mediated Activation of RPS5 Can Be Reconstituted in Nicotiana benthamiana.
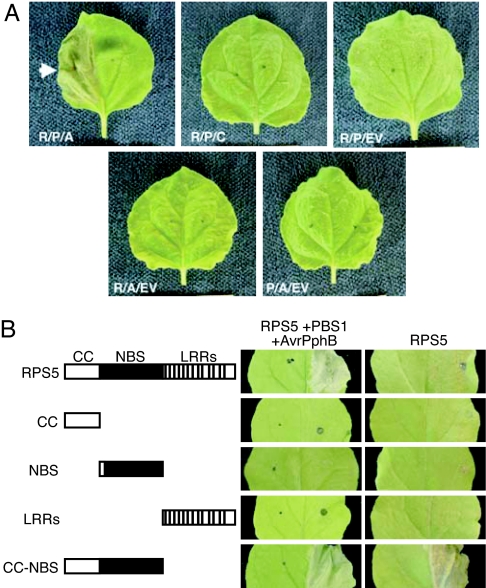
To investigate the molecular mechanism of RPS5 activation, we developed a transient expression system in N. benthamiana that reconstitutes an HR-like response that depends on coexpression of RPS5, PBS1, and functional AvrPphB (Fig. 1A). Coexpression of all three proteins resulted in a rapid collapse of leaf tissue in the infiltrated region, which was visible within 12 h of inducing protein expression. This collapse was not observed when any of the three proteins were left out or when a protease-inactive form of AvrPphB (C98S) was used. Thus, RPS5 can activate a classic HR-like response in N. benthamiana that depends on both the active pathogen effector protein, AvrPphB, and on the target of the effector, PBS1, indicating that RPS5-dependent signaling can be assayed using a transient expression system. These data also indicate that whatever lies downstream of RPS5 in the activation of programmed cell death must be conserved between N. benthamiana and Arabidopsis.
Fig. 1.
Reconstitution of the RPS5- mediated HR in N. benthamiana. (A) Coexpression of RPS5, PBS1, and AvrPphB induces an HR-like response in N. benthamiana. Leaves were photographed 20 h after induction of transgene expression. The arrowhead indicates the HR. R, RPS5; P, PBS1; A, AvrPphB; C, AvrPphB (C98S); EV, empty pTA7002 vector control. (B) Assessment of the RPS5 domains required for the HR. The indicated RPS5 domains were coexpressed in the presence of PBS1 and AvrPphB or alone in N. benthamiana. Leaves were photographed 20 h after induction of transgene expression.
Although RPS5, PBS1, and AvrPphB are being highly overexpressed in this transient expression system, several lines of evidence support the validity of our conclusions. First, RPS5-induced cell death is observed only when RPS5, PBS1, and AvrPphB are coexpressed; thus, overexpression of wild-type RPS5 does not trigger cell death on its own. Second, PBS1 and RPS5 mutants that fail to function in Arabidopsis also fail to function in this transient system (data not shown). Third, several PBS1 mutations that block RPS5 signaling in Arabidopsis also block RPS5–PBS1 coimmunoprecipitation (co-IP) in N. benthamiana (see below). This observation argues that the RPS5–PBS1 interaction is quite specific and correlates with active signaling.
The LRR Domain of RPS5 Inhibits HR Activation.
Using this transient system, we tested which of the CC, NBS, and LRR domains of RPS5 are required for HR activation. To rule out problems with protein stability, we first verified that all constructs generated products clearly detectable on an immunoblot [see supporting information (SI) Fig. 6]. When expressed with PBS1 and AvrPphB, none of the individual RPS5 domains were sufficient to induce an HR. However, a construct that contained both the CC and NBS domains induced an HR-like leaf collapse (Fig. 1B). This result suggested two possibilities: either the LRR domain of RPS5 is dispensable for both recognition and downstream signaling, or the LRR domain inhibits an otherwise constitutive downstream signaling. To distinguish between these two possibilities, we repeated the experiment in the absence of PBS1 and AvrPphB. Unlike the full-length RPS5, the CC–NBS construct was able to activate an HR on its own (Fig. 1B), indicating that the LRR domain inhibits the constitutive activation of defense signaling by RPS5. Because the independent expression of the CC and NBS domains showed no such phenotype, we conclude that the combination of both domains is necessary and sufficient for RPS5 downstream signaling.
PBS1 Complexes with the CC Domain of RPS5 Before AvrPphB Exposure.
How does AvrPphB elicitation release RPS5 activation from its inhibition by the LRR domain? Because PBS1 is required for RPS5 activation and is a substrate of AvrPphB (3), we have proposed two plausible models of RPS5 activation. AvrPphB may cleave free molecules of PBS1, with only the cleavage products capable of binding to and activating RPS5. Alternatively, RPS5 might preassociate with PBS1, with cleavage of PBS1 in the complex inducing a conformational change in RPS5. In the latter case, the PBS1-induced conformation change in RPS5 might release the inhibition by the LRR domain.
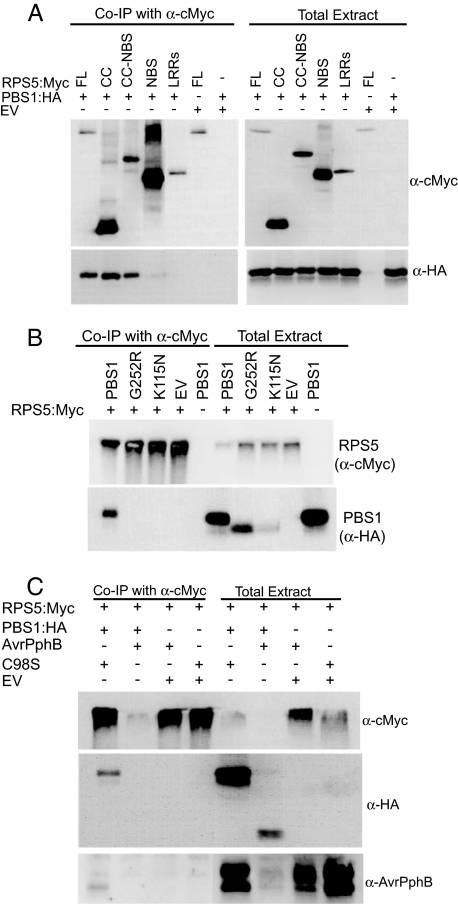
To distinguish between these two models, we coexpressed epitope-tagged forms of RPS5 and PBS1 in N. benthamiana and assayed for physical associations by using co-IP analysis on total protein extracted from leaves. PBS1 immunoprecipitated with RPS5 in the absence of AvrPphB (Fig. 2A), demonstrating that PBS1 and RPS5 are in the same complex before AvrPphB exposure. To determine which domains of RPS5 are required for complexing with PBS1, we performed co-IP experiments with the RPS5 deletion derivatives shown in Fig. 1B. These analyses revealed that the N-terminal CC domain of RPS5 is both necessary and sufficient for association with PBS1 (Fig. 2A).
Fig. 2.
RPS5 and PBS1 coimmunoprecipitate. (A) Coimmunoprecipitation of RPS5 and PBS1 requires the RPS5 CC domain and occurs independent of AvrPphB. FL, full-length RPS5; EV, empty pTA7002 vector control. (B) Coimmunoprecipitation of RPS5 and PBS1 requires PBS1 kinase activity. (C) AvrPphB coimmunoprecipitates with RPS5 only in the presence of PBS1. The indicated combinations of constructs were transiently expressed in N. benthamiana. Proteins were immunoprecipitated with anti-cMyc, and immunoblots were performed with the antibodies indicated on the right. G252R and K115N indicate amino acid substitutions in PBS1 that eliminate kinase activity of PBS1 in vivo. C98S, the protease-inactive form of AvrPphB.
The co-IP analyses shown in Fig. 2A revealed that PBS1 usually runs as a doublet on SDS/PAGE gels and that only the slower migrating form immunoprecipitates with RPS5. This observation suggested that PBS1 may autophosphorylate in vivo and that only phosphorylated PBS1 associates with RPS5. To test this hypothesis directly, we assessed whether kinase-inactive forms of PBS1 could immunoprecipitate with RPS5. The PBS1 mutations G252R and K115N, which abolish kinase activity (3, 5), disrupted the association of PBS1 with RPS5 (Fig. 2B), indicating that PBS1 autophosphorylation is required for forming this complex. Because these amino acid substitutions also prevent AvrPphB-mediated activation of RPS5 (3, 5), but do not affect the cleavage of PBS1 by AvrPphB (3), these data also suggest that PBS1/RPS5 complex formation is required for activation of RPS5 by AvrPphB.
AvrPphB, PBS1, and RPS5 Form a Single Complex.
If this conclusion is correct, then a complex of AvrPphB, PBS1, and RPS5 should form at least transiently during RPS5 activation. In our previous work, we demonstrated that a protease inactive form of AvrPphB(C98S) can immunoprecipitate with PBS1 (3). Here we show that AvrPphB(C98S) can also immunoprecipitate with RPS5, but only if PBS1 is present (Fig. 2C). This finding shows that PBS1 acts as a bridge between AvrPphB and RPS5 and demonstrates the formation of a complex between a pathogen effector, its target, and its cognate resistance protein (R protein). The fact that the protease inactive form of AvrPphB associates with the PBS1–RPS5 complex but fails to trigger an HR also indicates that cleavage of PBS1 is essential for RPS5 activation.
We also attempted to immunoprecipitate the RPS5–PBS1–AvrPphB complex by using wild-type AvrPphB but were unable to detect RPS5, likely because it is rapidly degraded as a consequence of HR activation. Such rapid degradation upon activation has previously been reported for the NBS–LRR protein RPM1, which mediates recognition of the AvrB and AvrRpm1 proteins in Arabidopsis (19). To determine whether this degradation was a consequence of specific degradation of RPS5 after activation or a more general consequence of HR-activation, we coexpressed RPS5 with AvrB, a P. syringae protein that induces an RPS5-independent HR when transiently expressed in N. benthamiana. The AvrB-triggered HR did not reduce RPS5 accumulation, indicating that RPS5 degradation triggered by AvrPphB is likely a specific consequence of its activation (see SI Fig. 7).
The CC, NBS, and LRR Domains Can Mediate Self-Association of RPS5.
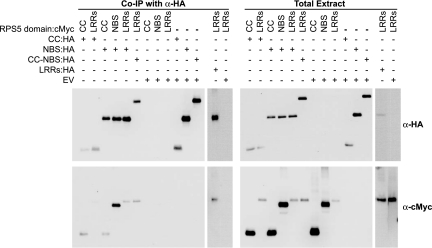
In animal NBS-containing proteins such as Apaf-1, activation by ligands causes self-association mediated by the NBS domains, which in turn brings together downstream signaling proteins bound to the N-terminal domains of the NBS proteins (1, 20). This “induced proximity” of downstream effector proteins is thought to be a key step in the activation of Apaf-1 signaling. In addition, it has recently been reported that the N protein from tobacco, which belongs to the TIR–NBS–LRR subfamily, self-associates upon activation by the p50 fragment of the replicase protein of tobacco mosaic virus and, furthermore, that the TIR domains by themselves can oligomerize (21). In light of these results, we tested for interactions among RPS5 domains. We coexpressed epitope-tagged versions of the RPS5 truncation series with epitope-tagged full-length RPS5 and performed co-IP analyses. Full-length RPS5-HA coimmunoprecipitated with full-length RPS5-Myc, indicating that RPS5 can form dimers or possibly oligomers (see SI Fig. 8). In addition, each of the RPS5 domains, individually or in combination, coimmunoprecipitated with full-length RPS5 (see SI Fig. 8). The reciprocal experiments provided identical results. To get a more detailed view of RPS5 domain interactions, pair-wise combinations of RPS5 domains were coexpressed and assayed by co-IP (Fig. 3). The N-terminal domain was found to immunoprecipitate with itself and with the NBS domain, but not with the LRR domain. The NBS domain was found to immunoprecipitate with the N-terminal domain, with the LRR domain, and with itself. Likewise, the LRR domain also immunoprecipitated with itself. Reciprocal co-IPs produced identical results (see SI Fig. 9).
Fig. 3.
RPS5 domains show homo- and heterotypic interactions. The indicated combinations of constructs were transiently expressed in N. benthamiana. Proteins were immunoprecipitated with anti-HA antibodies, and immunoblots were performed with the indicated antibodies. The two lanes on the far right of each blot were run on a separate gel from the other lanes.
These results highlight the following points. First, the LRR domain likely interacts with the NBS domain, consistent with our hypothesis that the LRR domain may inhibit constitutive RPS5 signaling by binding and inhibiting the function of the NBS domain. In addition, the RPS5 NBS domain appears to self-associate, which is similar to NBS domains of animal proteins (1, 20) but is not yet reported for plant NBS-containing proteins. Likewise, the N-terminal CC domain and the LRR domain of RPS5 both appeared to be capable of homotypic association, suggesting that they may play a role in RPS5–RPS5 intermolecular interactions. Finally, interactions between the N-terminal and NBS domains and the NBS and LRR domains suggest that these domains participate in intramolecular and intermolecular interactions.
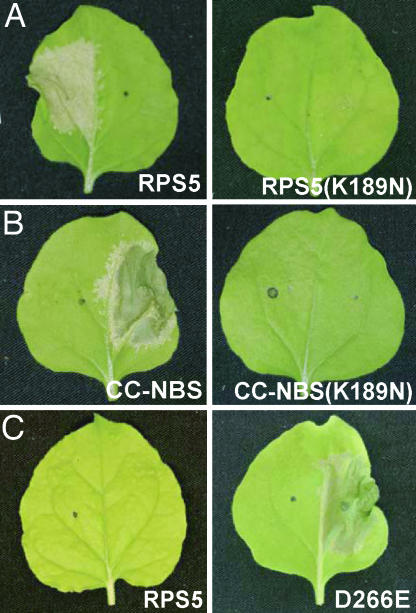
Given that PBS1 complexes with the CC domain, which is itself associated with the NBS domain, how might cleavage of PBS1 activate RPS5? Work on animal NBS-containing proteins and recent work on the I-2 and N proteins of tomato and tobacco has indicated that ATP binding state may play a key role in NBS–LRR protein activation (13, 14, 22, 23). In the I-2 protein, amino acid substitutions that reduce ATP hydrolysis, but not ATP binding, cause constitutive activation of I-2, whereas substitutions that inhibit ATP binding eliminate signaling (14). These observations suggest that the ATP-bound form of I-2 is active and that the ADP-bound form and unbound forms are inactive. To test whether this hypothesis might also be true for RPS5, we made an aspartate-to-glutamate substitution at residue 266 (D266E), which is located in the Walker B box of RPS5 (VLLLDD). In I-2 and other ATPases, this residue is thought to function as a catalytic base in ATP hydrolysis (24, 25). Transient expression of full-length RPS5 (D266E) by itself induced a clear HR-like response in N. benthamiana, showing that this substitution also makes RPS5 autoactive (Fig. 4). Conversely, a lysine-to-asparagine substitution in the P-loop of RPS5 (K189N), which is expected to block ATP binding (13, 14), eliminated the activity of wild-type RPS5 when coexpressed with PBS1 and AvrPphB and also eliminated the autoactivity of the RPS5 CC–NBS construct (Fig. 4).
Fig. 4.
Mutations in the ATP binding site affect RPS5 signaling. (A) A K189N substitution in the P-loop blocks the RPS5-mediated HR. Wild-type RPS5 or the K189N mutant was coexpressed with PBS1 and AvrPphB in N. benthamiana leaves. (B) The K189N mutation also blocks the HR induced by the autoactive RPS5 CC–NBS construct. (C) A D266E substitution in the Walker B motif autoactivates RPS5. (B and C) The indicated constructs were expressed by themselves. All photos were taken 24 h after induction.
Conclusions
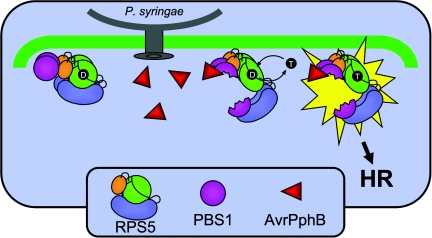
We propose the following model to explain the above results (Fig. 5). In the absence of PBS1, the N-terminal and LRR domains of RPS5 interact with the NBS domain, forming an inactive structure that is bound to ADP. Although RPS5 is capable of oligomerizing in this state, such self-association is not sufficient to cause activation of programmed cell death, thus additional conformational changes must be required to activate RPS5. Under normal conditions, RPS5 is bound to PBS1, priming the cell for AvrPphB detection. In this state, we hypothesize that the LRR domain is still bound to the NBS domain, inhibiting RPS5 activation. Cleavage of PBS1 by AvrPphB results in a structural change of RPS5, possibly due to engagement of the LRR domain by the cleaved PBS1 protein. This structural change then removes the autoinhibition of the LRR domain of RPS5, allowing the NBS domain to exchange ADP for ATP. This nucleotide exchange then causes further conformational changes that reveal new binding sites for additional RPS5-interacting proteins. Such binding sites may be located on the CC domain, which would explain why it is required for downstream signaling.
Fig. 5.
Model for RPS5 activation. Free RPS5 is unable to activate defense responses because of the negative regulatory action of the LRR domain. In the uninfected cell, most, if not all RPS5 is bound to PBS1 and ADP (D) and primed for a response to pathogen attack. PBS1 is cleaved through the cysteine protease action of AvrPphB after injection by P. syringae. PBS1 cleavage is detected by RPS5, resulting in a conformational change that enables exchange of ATP (T) for ADP. The ATP-bound form of RPS5 then engages downstream signaling molecules, activating the defense response.
The above model is consistent with recently published data for the human Nod2 protein, which mediates detection of bacterial peptidoglycan and activates the NF-κB transcription factor (26). Deletion of the Nod2 LRR domain constitutively activates Nod2 signaling, and this signaling depends on the N-terminal caspase recruitment domain of Nod2 (26). A role for the LRR domain in repressing signaling has also been suggested by work on the Rx protein from potato, which mediates recognition of potato virus X. Deletion of the LRR domain from Rx enhances an HR-like response that is observed when Rx is transiently overexpressed in tobacco leaves, and, in addition, specific missense mutations in the LRR domain induce a cell death response in tobacco even when expressed from its own promoter (27). Similarly, truncation of the LRR domains of the Arabidopsis CC–NBS–LRR protein RPS2 and the TIR–NBS–LRR protein RPP1A also causes autoactivation, at least when transiently overexpressed (28, 29).
Although the above data indicate that the LRR domain functions at least in part to inhibit activation of NBS protein signaling in the absence of pathogens, other data indicate that the LRR domain also plays a positive role in signaling. For example, the rps5–2 mutation, which causes a complete loss of RPS5 function (as assayed by resistance to P. syringae strains expressing AvrPphB), is located in the LRR region (6). Consistent with this observation, we noted that the HR-like response induced by the truncated RPS5 protein lacking the LRR domain was not as strong as that observed when full-length RPS5 was coexpressed with AvrPphB and PBS1, despite the fact that truncated RPS5 protein accumulated to higher levels than full-length protein (see SI Fig. 6). Similarly, activation of an HR-like response by autoactivating mutant forms of the potato Rx protein also requires the LRR domain (30). These data suggest that the LRR domain of NBS–LRR proteins may also contribute to engagement of downstream signaling proteins or, alternatively, to converting the NBS domain from an inactive conformation to an active conformation.
The latter hypothesis is supported by recent work on I-2, in which autoactive mutant forms require the LRR domain to induce cell death (24). Furthermore, the I-2 protein lacking the LRR domain displays a low dissociation rate for ADP in vitro. It is tempting to speculate that the LRR domain contributes to the exchange of ADP for ATP upon detection of the pathogen signal.
A key question is what role the LRR domain plays in detection of the pathogen signal. Our data indicate that it is the CC domain, not the LRR domain, that associates with the target of the pathogen effector protein and thus associates indirectly with the pathogen-derived signal. Similarly, yeast two-hybrid data have shown that the CC domain of the RPM1 protein interacts with RIN4, a target of three different pathogen effectors: AvrB, AvrRpm1, and AvrRpt2 (31, 32). Most recently, the N-terminal domain of the tomato NBS–LRR protein Prf has been shown to coimmunoprecipitate with Pto, a target of the effector AvrPto (33). These findings are unexpected, because experiments involving swapping of LRR domains between closely related NBS–LRR proteins have indicated that the LRR domain is an important determinant of pathogen specificity (16–18, 34). This conclusion is supported by sequence comparisons between closely related NBS–LRR proteins, which indicate that specific residues within the LRR domain are under diversifying selection (35). Furthermore, the specificity of the potato Rx protein, which confers resistance to potato virus X, can be broadened to include additional viruses by altering specific amino acids in the Rx LRR domain (36). We did not observe any interactions between AvrPphB and the LRR domain, nor between PBS1 and the LRR domain. It is plausible that cleavage of PBS1 that is already bound to RPS5 causes PBS1 to interact with the LRR domain, which then enables signaling to occur. According to this scenario, specificity for pathogen recognition occurs at the level of the LRR domain interacting with the modified target of the pathogen effector that is first bound to the N-terminal domain.
Our finding that proper regulation of RPS5 signaling requires both the pathogen effector protein and the target of this effector has important implications for transfer of disease-resistance specificities between plant species. Transfer of R genes across plant families typically fail, either because they simply do not function or because they induce constitutive defense responses (37, 38). On the basis of our work and that of Day et al. (38), it appears that the solution to both of these problems is to identify the target of the pathogen effector molecule in the species from which the R gene is originating and then to coexpress the target with the cognate NBS–LRR protein in the recipient. This approach may provide a vast gene pool for mining disease-resistance genes that are effective against economically important pathogens.
Materials and Methods
Plasmid Constructs.
All PBS1 and AvrPphB constructs have been described in refs. 3–5. RPS5:Myc, RPS5:HA, and their derivative constructs were generated in two steps. First, the RPS5 ORF and deletion derivatives were PCR-amplified from a cDNA template and cloned into the SalI/NotI sites of the pBluescript plasmid (Stratagene, La Jolla, CA). The RPS5 deletion derivatives encoded amino acids 1–183 (CC domain), 157–512 (NBS domain), 513–899 (LRR domain), and 1–512 (CC–NBS domains). A five-copy cMyc tag or three-copy HA tag was then PCR-amplified from the Myc-containing plasmid pGem7Z f(+) (kindly provided by the Arabidopsis Biological Resource Center, Ohio State University, Columbus, OH) and an HA-containing pBluescript plasmid using forward and reverse primers containing NotI and NheI/NotI restriction sites, respectively. The PCR products were digested with NotI and cloned into the NotI site of pBluescript::RPS5. These recombinant plasmids were digested with SalI and NheI to release the tagged RPS5 fragments, which were then inserted into the XhoI/SpeI sites of the dexamethasone-inducible plasmid pTA7002 (39).
Plant Material.
N. benthamiana plants were grown under a 9-h photoperiod at 24°C in Metro-Mix 360 potting mixture (Sun Gro Horticulture, Bellevue, WA).
Agrobacterium Transient Expression Assays.
Agrobacterium tumefaciens GV3101(pMP90) strains carrying the various dexamethasone-inducible constructs were grown and prepared for transient expression as described in ref. 40. Agrobacterium cultures were resuspended in water at an OD600 = 0.8 for experiments involving coinjection of two or more strains and an OD600 = 0.4 when injected individually. For experiments requiring coexpression of AvrPphB, PBS1, RPS5, or empty vector, suspensions were mixed in a 1:1 ratio when two constructs were coexpressed or in a 1:1:1 ratio when three constructs were coexpressed. Bacterial suspensions were infiltrated into expanding leaves of 4-week-old N. benthamiana. Plants were sprayed with 50 μM dexamethasone 40 h after injection to induce expression. Samples were collected for protein extractions 4 h after dexamethasone application and were flash-frozen in liquid nitrogen. For HR tests, leaves were scored for hypersensitive phenotypes at 6 and 20 h after hormone application.
Immunoprecipitations and Immunoblots.
Immunoprecipitations were performed as described in ref. 3 by using anti-cMyc Ab-agarose beads (BD Biosciences, San Jose, CA) or anti-HA affinity matrix (Roche, Indianapolis, IN). The immunocomplexes were resuspended in 50 μl of 1× SDS loading buffer and boiled for 5 min, and 15 μl was separated in a SDS/10% PAGE gel. As a control, 50 μg of total protein was loaded on the same gels. Proteins were transferred to a nitrocellulose membrane and probed with anti-cMyc peroxidase or anti-HA peroxidase (Roche). The blot was stripped and reprobed with anti-AvrPphB polyclonal antibody, anti-HA peroxidase (Roche) or anti-cMyc peroxidase (Roche). For some experiments, dupli-cate gels and filters were prepared rather than stripping and reprobing a single membrane.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resources Center for providing the cMyc plasmid. This work was supported by National Institutes of Health Grant R01 GM046451 from the National Institute of General Medical Sciences (to R.W.I.).
Abbreviations
- co-IP
coimmunoprecipitation
- HR
hypersensitive response
- LRR
leucine-rich repeat
- NBS
nucleotide binding site
- R gene
resistance gene
- R protein
resistance protein
- TIR
Toll-interleukin receptor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0608779104/DC1.
References
- 1.Inohara N, Chamaillard M, McDonald C, Nunez G. Annu Rev Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- 2.Jones JD, Dangl JL. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 3.Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW. Science. 2003;301:1230–1233. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- 4.Shao F, Merritt PM, Bao Z, Innes RW, Dixon JE. Cell. 2002;109:575–588. doi: 10.1016/s0092-8674(02)00766-3. [DOI] [PubMed] [Google Scholar]
- 5.Swiderski MR, Innes RW. Plant J. 2001;26:101–112. doi: 10.1046/j.1365-313x.2001.01014.x. [DOI] [PubMed] [Google Scholar]
- 6.Warren RF, Henk A, Mowery P, Holub E, Innes RW. Plant Cell. 1998;10:1439–1452. doi: 10.1105/tpc.10.9.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitham S, Dinesh-Kumar SP, Choi D, Hehl R, Corr C, Baker B. Cell. 1994;78:1101–1115. doi: 10.1016/0092-8674(94)90283-6. [DOI] [PubMed] [Google Scholar]
- 8.Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- 9.Mindrinos M, Katagiri F, Yu GL, Ausubel FM. Cell. 1994;78:1089–1099. doi: 10.1016/0092-8674(94)90282-8. [DOI] [PubMed] [Google Scholar]
- 10.Goodman RN, Novacky AJ. The Hypersensitive Reaction in Plants to Pathogens. St. Paul, MN: Am Phytopathol Soc; 1994. [Google Scholar]
- 11.Martin GB, Bogdanove AJ, Sessa G. Annu Rev Plant Biol. 2003;54:23–61. doi: 10.1146/annurev.arplant.54.031902.135035. [DOI] [PubMed] [Google Scholar]
- 12.Kim HE, Du F, Fang M, Wang X. Proc Natl Acad Sci USA. 2005;102:17545–17550. doi: 10.1073/pnas.0507900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tameling WI, Elzinga SD, Darmin PS, Vossen JH, Takken FL, Haring MA, Cornelissen BJ. Plant Cell. 2002;14:2929–2939. doi: 10.1105/tpc.005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tameling WI, Vossen JH, Albrecht M, Lengauer T, Berden JA, Haring MA, Cornelissen BJ, Takken FL. Plant Physiol. 2006;140:1233–1245. doi: 10.1104/pp.105.073510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobe B, Deisenhofer J. Trends Biochem Sci. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 16.Dodds PN, Lawrence GJ, Ellis JG. Plant Cell. 2001;13:163–178. doi: 10.1105/tpc.13.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellis JG, Lawrence GJ, Luck JE, Dodds PN. Plant Cell. 1999;11:495–506. doi: 10.1105/tpc.11.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen QH, Zhou F, Bieri S, Haizel T, Shirasu K, Schulze-Lefert P. Plant Cell. 2003;15:732–744. doi: 10.1105/tpc.009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyes DC, Nam J, Dangl JL. Proc Natl Acad Sci USA. 1998;95:15849–15854. doi: 10.1073/pnas.95.26.15849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inohara N, Nunez G. Oncogene. 2001;20:6473–6481. doi: 10.1038/sj.onc.1204787. [DOI] [PubMed] [Google Scholar]
- 21.Mestre P, Baulcombe DC. Plant Cell. 2006;18:491–501. doi: 10.1105/tpc.105.037234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ueda H, Yamaguchi Y, Sano H. Plant Mol Biol. 2006;61:31–45. doi: 10.1007/s11103-005-5817-8. [DOI] [PubMed] [Google Scholar]
- 23.DeYoung BJ, Innes RW. Nat Immunol. 2006;7:1243–1249. doi: 10.1038/ni1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takken FL, Albrecht M, Tameling WI. Curr Opin Plant Biol. 2006;9:383–390. doi: 10.1016/j.pbi.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Hanson PI, Whiteheart SW. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 26.Tanabe T, Chamaillard M, Ogura Y, Zhu L, Qiu S, Masumoto J, Ghosh P, Moran A, Predergast MM, Tromp G, et al. EMBO J. 2004;23:1587–1597. doi: 10.1038/sj.emboj.7600175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bendahmane A, Farnham G, Moffett P, Baulcombe DC. Plant J. 2002;32:195–204. doi: 10.1046/j.1365-313x.2002.01413.x. [DOI] [PubMed] [Google Scholar]
- 28.Weaver LM, Swiderski MR, Li Y, Jones JDG. Plant J. 2006;47:829–840. doi: 10.1111/j.1365-313X.2006.02834.x. [DOI] [PubMed] [Google Scholar]
- 29.Tao Y, Yuan F, Leister RT, Ausubel FM, Katagiri F. Plant Cell. 2000;12:2541–2554. doi: 10.1105/tpc.12.12.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moffett P, Farnham G, Peart J, Baulcombe DC. EMBO J. 2002;21:4511–4519. doi: 10.1093/emboj/cdf453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mackey D, Holt BF, III, Wiig A, Dangl JL. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 32.Axtell MJ, Staskawicz BJ. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- 33.Mucyn TS, Clemente A, Andriotis VM, Balmuth AL, Oldroyd GE, Staskawicz BJ, Rathjen JP. Plant Cell. 2006;18:2792–2806. doi: 10.1105/tpc.106.044016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rairdan GJ, Moffett P. Plant Cell. 2006;18:2082–2093. doi: 10.1105/tpc.106.042747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyers BC, Shen KA, Rohani P, Gaut BS, Michelmore RW. Plant Cell. 1998;10:1833–1846. doi: 10.1105/tpc.10.11.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farnham G, Baulcombe DC. Proc Natl Acad Sci USA. 2006;103:18828–18833. doi: 10.1073/pnas.0605777103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frost D, Way H, Howles P, Luck J, Manners J, Hardham A, Finnegan J, Ellis J. Mol Plant–Microbe Interact. 2004;17:224–232. doi: 10.1094/MPMI.2004.17.2.224. [DOI] [PubMed] [Google Scholar]
- 38.Day B, Dahlbeck D, Huang J, Chisholm ST, Li D, Staskawicz BJ. Plant Cell. 2005;17:1292–1305. doi: 10.1105/tpc.104.030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aoyama T, Chua N-H. Plant J. 1997;11:605–612. doi: 10.1046/j.1365-313x.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
- 40.Wroblewski T, Tomczak A, Michelmore RW. Plant Biotechnol J. 2005;3:259–273. doi: 10.1111/j.1467-7652.2005.00123.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.