Abstract
Objective: The aim of this study was to investigate subgingival infection frequencies of Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans strains with genetic variation in Chinese chronic periodontitis (CP) patients and to evaluate its correlation with clinical parameters. Methods: Two multiplex polymerase chain reaction (PCR) assays were developed to detect the 16SrDNA, collagenase (prtC) and fimbria (fimA) genes of P. gingivalis and the 16SrDNA, leukotoxin (lktA) and fimbria-associated protein (fap) genes of A. actinomycetemcomitans in 60 sulcus samples from 30 periodontal healthy subjects and in 122 subgingival plaque samples from 61 patients with CP. The PCR products were further T-A cloned and sent for nucleotide sequence analysis. Results: The 16SrDNA, prtC and fimA genes of P. gingivalis were detected in 92.6%, 85.2% and 80.3% of the subgingival plaque samples respectively, while the 16SrDNA, lktA and fap genes of A. actinomycetemcomitans were in 84.4%, 75.4% and 50.0% respectively. Nucleotide sequence analysis showed 98.62%~100% homology of the PCR products in these genes with the reported sequences. P. gingivalis strains with prtC+/fimA+ and A. actinomycetemcomitans with lktA+ were predominant in deep pockets (>6 mm) or in sites with attachment loss ≥5 mm than in shallow pockets (3~4 mm) or in sites with attachment loss ≤2 mm (P<0.05). P. gingivalis strains with prtC+/fimA+ also showed higher frequency in gingival index (GI)=3 than in GI=1 group (P<0.05). Conclusion: Infection of P. gingivalis with prtC+/fimA+ and A. actinomycetemcomitans with lktA+ correlates with periodontal destruction of CP in Chinese. Nonetheless P. gingivalis fimA, prtC genes and A. actinomycetemcomitans lktA gene are closely associated with periodontal destruction, while A. actinomycetemcomitans fap gene is not.
Keywords: Porphyromonas gingivalis, Actinobacillus actinomycetemcomitans, Strain, Periodontitis, PCR
INTRODUCTION
Porphyromonas gingivalis, a black-pigmented gram-negative anaerobic coccobacillus, has been etiologically associated with various types of periodontal disease including chronic periodontitis (CP) (Chen et al., 2005; Cortelli et al., 2005a; Liu et al., 2003). This bacterium is frequently detected in deep periodontal pockets of CP patients and is occasionally found in healthy periodontal tissues without inflammation (Liu et al., 2003; van der Ploeg et al., 2004). There may be diversities in virulence among the organisms harbored by individuals who are periodontally healthy and those with periodontitis (Amano et al., 2000). Previous studies showed that the virulence of different P. gingivalis strains is closely associated with their ability to produce fimbria encoded by the fimbria (fimA) gene, collagenase encoded by the collagenase (prtC) gene and trypsin-like proteases (Amano et al., 2004; Imamura et al., 2003; Wittstock et al., 2000). However, a few P. gingivalis isolates lacking the prtC and/or fimA genes would result in virulence variation (Fujiwara et al., 1994; Slots et al., 1995; Wittstock et al., 2000). But little is known about the correlation between the P. gingivalis strains with virulence variation and the severity of CP in patients.
Actinobacillus actinomycetemcomitan is a gram-negative, facultative anaerobic coccobacillus. The presence of this bacterium and elevated serum antibody titers against the microbe is strongly correlated with the pathogenesis of aggressive periodontitis (Cortelli et al., 2005a; Leung et al., 2005; Nonnenmacher et al., 2001). Although it also exists in CP patients (Cortelli et al., 2005a; 2005b) and periodontally healthy individuals (Gafan et al., 2004; Yang et al., 2005), the proportions and serotypes of infected strains in different periodontal conditions are different (Lakio et al., 2002; Yang et al., 2005). Neutrophils are successful in eliminating most bacterial pathogens, but A. actinomycetemcomitans can produce a unique leukotoxin encoded by the leukotoxin (lktA) gene that contributes to the killing of human neutrophils and monocytes (Guthmiller et al., 2001; Poulsen et al., 2003). A. actinomycetemcomitans can also produce fimbria, which is closely associated with its ability to colonize various types of host cells. The fimbria-associated protein gene (fap) is strongly expressed in fimbriated A. actinomycetemcomitans strains but not in non-fimbriated ones (Ishihara et al., 1997). However, the relationship between A. actinomycetemcomitans heterogeneous strains and periodontal tissue destruction in CP seems to be rather complex and depends on the virulence of the bacteria (Johansson et al., 2005).
In order to evaluate the possible pathogenic role of different P. gingivalis and A. actinomycetemcomitans strains infecting Chinese CP patients, and to find the relationship between strains with genetic variation of the two microbes and periodontal tissue destruction of CP, two multiplex polymerase chain reaction (PCR) assays were established in this study to detect the fimA and prtC genes of P. gingivalis, and the fap and lktA genes of A. actinomycetemcomitans in subgingival plaque samples and in clinically isolated strains. The 16SrDNAs of P. gingivalis and A. actinomycetemcomitans were also included in the PCR assays as internal positive controls for both pathogens.
MATERIALS AND METHODS
Subjects
The subjects were 61 Chinese CP patients [23 males aged 31 to 66 years, mean age was (43.6±9.8) years; 38 females aged 29 to 65 years, mean age was (41.2±7.6) years] and 30 periodontally healthy individuals [13 males aged 27 to 49 years, mean age was (38.5±7.6) years; 17 females aged 32 to 51 years, mean age was (36.2±6.9) years] who were referred to the dental clinic in the Second Affiliated Hospital of School of Medicine of Zhejiang University for dental or periodontal treatment or health monitoring. All the subjects were non-smokers without any systemic disease, and with at least 14 teeth remaining. Those who had received professional cleaning or had history of antibiotic therapy during the preceding 3 months were excluded. All of the patients and the healthy individuals underwent full-mouth examination. The criteria of diagnosis for chronic periodontitis were based on the Classification of Periodontal Diseases issued by the American Academy of Periodontology in 1999 (Armitage, 1999). Briefly, the patients had >30% sites showing periodontal probing depth ≥3 mm, clinical attachment loss >1 mm and radiographic evidence of alveolar bone loss. These individuals were considered periodontally healthy with periodontal probing depth <3 mm, without clinical attachment loss, with no inflammation of gingiva and no alveolar bone absorption on X-ray examination. All the subjects received detailed information concerning the nature of the study and the procedures involved, and their informed consent was obtained.
Sample collection
For each patient, two subgingival plaque samples were taken from the bottom of periodontal pockets from the deepest sites of one front tooth and one molar with separate sterile Gracy curettes after supragingival plaque was gently removed. For each periodontally healthy individual, two samples from the bottom of gingival sulcular of one front tooth and one molar were collected with the same method. Each plaque sample was placed in 2 ml freshly prepared, pre-reduced fastidious anaerobe broth (Bioconnections, England, UK) for strain isolation and 200 µl lysis buffer (10 mmol/L Tris-HCl, 1.0 mmol/L EDTA, 1.0% Triton X-100, pH 8.0) for PCR assay. The broth containing the subgingival plaque sample was immediately transported to the laboratory while the lysis buffer was stored at −70 °C. The gingival index (GI), probing depth (PD) and attachment loss (AL) of each pocket were recorded.
Isolation and identification of P. gingivalis and A. actinomycetemcomitans strains
Broth (0.05 ml) containing subgingival plaque sample was inoculated into trypticase soy agar supplemented with haemin (5 µg/ml), vitamin K1 (1 µg/ml), 5% (v/v) sheep blood and menadione (1 µg/ml), or into tryptic soy medium containing 10% (v/v) horse serum, bacitracin (75 µg/ml) and vancomycin (5 µg/ml) respectively for selective recovery of P. gingivalis and A. actinomycetemcomitans. The bacteria from the subgingival plaque samples were grown in an anaerobic chamber with 10% H2, 10% CO2 and 80% N2 at 37 °C for 7 d. P. gingivalis and A. actinomycetemcomitans colonies were identified by Gram-staining and morphological examination, biochemical reaction and P. gingivalis 16SrDNA or A. actinomycetemcomitans 16SrDNA gene detection by PCR. P. gingivalis strain ATCC 33277 and A. actinomycetemcomitans strain Y4 were also cultured with the above media, respectively.
DNA extraction
Each of the subgingival plaque samples in the lysis buffer was boiled for 10 min, and 10 µl of the supernatant was directly used as template in PCR. P. gingivalis ATCC 33277 and A. actinomycetemcomitans Y4 removed from the plates were resuspended in 10 mmol/L PBS (pH 8.0). Then genomic DNA of the bacterial strain, used to optimize PCR conditions and as positive control for PCR, was obtained using the phenol-chloroform method. An OD (optical density) value at 260 nm of the P. gingivalis ATCC 33277 or A. actinomycetemcomitans Y4 DNA preparation was measured with an ultraviolet spectrophotometer, and was used to calculate the DNA concentration. DNA preparations at 10-fold dilutions (0.1~100 ng) were used as templates for determination of PCR sensitivity.
PCR primers and amplification
Two multiplex PCR assays were established to detect the P. gingivalis 16SrDNA, fimA and prtC genes and the A. actinomycetemcomitans 16SrDNA, lktA and fap genes in the subgingival plaque samples. To detect P. gingivalis, PCR amplification was performed in a volume of 100 µl containing 10 µl of the template, 10 µl PCR buffer (20 mmol/L Tris-HCl, 50 mmol/L KCl, pH 8.4) and 5 U Taq polymerase (Promega), 0.25 mmol/L of each dNTP, 2.5 mmol/L MgCl2, 25 pmol primers for the P. gingivalis 16SrDNA gene, 50 pmol primers for the fimA gene, and 25 pmol primers for the prtC gene. The sequences of primers specific for the P. gingivalis 16SrDNA gene are: 5′-AGG CAG CTT GCC ATA CTG CG-3′ (sense), and 5′-ACT GTT AGC AAC TAC CGA TGT-3′ (antisense) (Slots et al., 1995), for the fimA gene: 5′-ATA ATG GAG AAC AGC AGG AA-3′ (sense), and 5′-TCT TGC CAA CCA GTT CCA TTG C-3′ (antisense) (Watanabe and Frommel, 1993), and for the prtC gene: 5′-ACA ATC CAC GAG ACC ATC-3′ (sense), and 5′-TTC AGC CAC ACC GAG ACG-3′ (antisense) (Bodinka et al., 1994). Expected sizes of the PCR products amplified from the P. gingivalis 16SrDNA, fimA and prtC genes were 404 bp, 131 bp and 584 bp.
To detect A. actinomycetemcomitans, PCR amplification was conducted in a volume of 100 µl containing the same contents as mentioned above except for 25 pmol primers for the A. actinomycetemcomitans 16SrDNA gene, 50 pmol primers for the A. actinomycetemcomitans fap gene and 25 pmol primers for the A. actinomycetemcomitans lktA gene. The sequences of primers specific for the A. actinomycetemcomitans 16SrDNA gene are: 5′-GCT AAT ACC GCG TAG AGT CGG-3′ (sense), and 5′-ATT TCA CAC CTC ACT TAA AGG T-3′ (antisense) (Slots et al., 1995), for the A. actinomycetemcomitans lktA gene: 5′-TCG CGA ATC AGC TCG CCG-3′ (sense), and 5′-GCT TTG CAA GCT CCT CAC C-3′ (antisense) (Watanabe and Frommel, 1996), and for the A. actinomycetemcomitans fap gene: 5′-ATT AAA TAC TTT AAC TAC TAA AGC-3′ (sense), and 5′-GCA CTG TTA ACT GTA CTA GC-3′ (antisense) (Ishihara et al., 1997). Expected sizes of the PCR products amplified from the A. actinomycetemcomitans 16SrDNA, lktA and fap genes were 443 bp, 285 bp and 210 bp.
The PCR programs for detection of P. gingivalis and A. actinomycetemcomitans were identical, including an initial denaturation step at 94 °C for 5 min, followed by 35 cycles of denaturation at 94 °C for 1 min, primer annealing at 50 °C for 1 min and extension at 72 °C for 1.5 min, and then a final extension step at 72 °C for 7 min. In each of the PCR assays, 10 ng of P. gingivalis ATCC 33277 DNA preparation and 10 ng of A. actinomycetemcomitans Y4 DNA were co-amplified with the subgingival plaque samples as positive and negative control respectively for detection of P. gingivalis. Ten nanograms of A. actinomycetemcomitans Y4 DNA preparation and 10 ng of P. gingivalis ATCC 33277 DNA, used as positive and negative controls, were co-amplified with the subgingival plaque samples respectively for detection of A. actinomycetemcomitans. All PCR assays were repeated once more, if two results were not consistent then a third time assay was carried out.
Detection of PCR products
Ten microlitres of each reaction product were mixed with 10 µl of 2× loading buffer and fractionated in a 2% agarose gel containing ethidium bromide (1 µg/ml), using a 100 bp DNA ladder (Promega) as a size marker, and visualized under UV light.
Sequence analysis
After being ligated into the plasmid pUCm-T vector (Promega), each of the PCR amplicons were transformed into E. coli DH5α. Plasmid DNA of white colonies was extracted and then digested with restriction endonuclease Pst I (Promega) for examination of the size of the inserted fragment. The plasmid containing the inserted fragment with expected size was used for nucleotide sequencing. Nucleotide sequences of the inserted fragments were analyzed by the dideoxy-chain termination method using an ABI PRISM™ 377 sequencer. The homology of nucleotide and amino acid sequences from the PCR products was compared with the sequences registered in GenBank.
Statistical analysis
Chi-square test was employed to compare the positive rates of P. gingivalis and A. actinomycetemcomitans in samples from both healthy individuals and CP patients. Fisher’s exact test was used when the expected value was smaller than 1. Association between clinical parameters and genotypes was examined using multinomial logistic regression analysis with Stata 8.0 software. P-value equal to or below 0.05 was considered statistically significant.
RESULTS
Sensitivity and reproducibility of the PCR assay
Using as low as 0.1 ng DNA template of P. gingivalis ATCC 33277 or 10 ng DNA template of A. actinomycetemcomitans Y4, the amplification fragments with the expected sizes from the P. gingivalis 16SrDNA, prtC or fimA genes or the A. actinomycetemcomitans 16SrDNA, lktA or fap genes could be revealed in a 2% agarose gel, respectively (Figs.1 and 2). Repeated PCR assays showed 91.9% and 88.7% consistency between each detection of P. gingivalis 16SrDNA, prtC and fimA and A. actinomycetemcomitans 16SrDNA, lktA and fap genes, respectively.
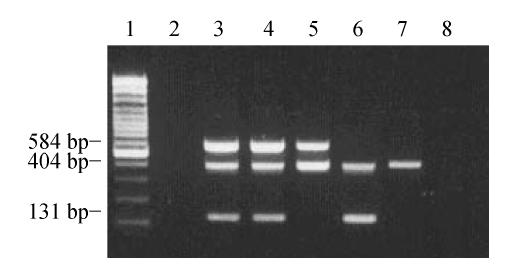
Fig. 1.
PCR detection of the P. gingivalis 16SrDNA, prtC and fimA genes in subgingival plaque samples
Lane 1: 100 bp marker; Lane 2: Negative control; Lane 3: Positive control; Lane 4: 3 amplification fragments from the 16SrDNA, prtC and fimA genes from a plaque sample; Lane 5: 2 amplification fragments from 16SrDNA and prtC from a plaque sample; Lane 6: 2 amplification fragments from 16SrDNA and fimA from a plaque sample; Lane 7: 1 amplification fragment from 16SrDNA; Lane 8: Blank control
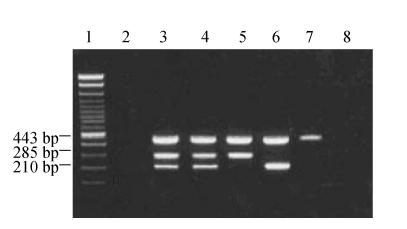
Fig. 2.
PCR detection of the A. actinomycetemcomitans 16SrDNA, lktA and fap genes in subgingival plaque samples
Lane 1: 100 bp DNA ladder; Lane 2: Negative control; Lane 3: Positive control; Lane 4: 3 amplification fragments from the 16SrDNA, lktA and fap genes from a plaque sample; Lane 5: 2 amplification fragments from 16SrDNA and lktA from a plaque sample; Lane 6: 2 amplification fragments from 16SrDNA and fap from a plaque sample; Lane 7: 1 amplification fragment from 16SrDNA; Lane 8: Blank control
Detection of P. gingivalis in subgingival plaque and sulcular samples and clinical isolates
In the 60 sulcular samples from the periodontally healthy individuals, 10.0% of the samples (6/60) were P. gingivalis 16SrDNA positive. Among the six 16SrDNA positive samples, one was prtC+/fimA+, three were prtC+/fimA− and two were prtC−/fimA− (Table 1). Two P. gingivalis strains (16SrDNA+/prtC+/fimA+ and 16SrDNA+/prtC+/fimA−) were isolated from the 60 sulcular samples.
Table 1.
Detection of different genotypes of P. gingivalis and A. actinomycetemcomitans in subgingival plaque samples and in clinical isolates
| Genotypes | Clinical isolated strains |
Subgingival plaque samples |
||
| Healthy | CP | Healthy | CP | |
| P. gingivalis | ||||
| 16SrDNA+/prtC+/fimA+ | 1 | 40 | 1 | 93 |
| 16SrDNA+/prtC+/fimA− | 1 | 5 | 3 | 11 |
| 16SrDNA+/prtC−/fimA+ | 0 | 3 | 0 | 5 |
| 16SrDNA+/prtC−/fimA− | 0 | 0 | 2 | 4 |
| A. actinomycetemcomitans | ||||
| 16SrDNA+/lktA+/fap+ | 1 | 17 | 1 | 58 |
| 16SrDNA+/lktA+/fap− | 1 | 4 | 3 | 34 |
| 16SrDNA+/lktA−/fap+ | 0 | 2 | 0 | 3 |
| 16SrDNA+/lktA−/fap− | 0 | 0 | 0 | 8 |
A hundred and thirteen out of the 122 subgingival plaque samples (92.6%) were P. gingivalis 16SrDNA positive while the rest of the 9 samples (7.4%) were not. As expected, none of the P. gingivalis 16SrDNA negative plaque samples were positive for prtC and/or fimA. Four genotypes (16SrDNA+/prtC+/fimA+, 16SrDNA+/prtC+/fimA−, 16SrDNA+/prtC−/fimA+ and 16SrDNA+/prtC−/fimA−) were found in the subgingival plaque samples (Table 1). In the 122 subgingival plaque samples, the positive rate of P. gingivalis 16SrDNA (92.6%) was significantly higher than that of fimA (98/122, 80.3%) (χ 2=7.890, P<0.01). However, there were no statistical differences between the positive rates of P. gingivalis 16SrDNA (92.6%) and prtC (104/122, 85.2%) (χ 2=3.370, P>0.05), nor between that of prtC and fimA (χ 2=1.04, P>0.05).
Fourty-eight strains of P. gingivalis were isolated from 38 out of the 61 patients (62.3%), all of which had detectable 16SrDNA. But three of the strains (6.3%) were negative for prtC, and five (10.4%) were negative for fimA. Anyhow the genotypes of all isolated strains of P. gingivalis were consistent with those detected directly in the corresponding subgingival plaque samples.
The positive rate of P. gingivalis 16SrDNA+/prtC+/fimA+ detected directly in subgingival plaque samples or in clinical isolates was significantly higher in CP than in periodontally healthy individuals (P=0.001; P=0.000), while the positive rates of the other three or two genotypes showed no statistical difference between CP and healthy controls (P=0.132~0.645; P=0.355, 0.299).
Detection of A. actinomycetemcomitans in subgingival plaque and sulcular samples and clinical isolates
In the 60 sulcular samples from the periodontally healthy individuals, 6.7% of the samples (4/60) were A. actinomycetemcomitans 16SrDNA positive. One was positive for both lktA and fap and three were positive for lktA but negative for fap (Table 1). Two A. actinomycetemcomitans strains (16SrDNA+/lktA+/fap+ and 16SrDNA+/lktA+/fap−) were isolated from the 60 sulcular samples.
While 103 of the 122 subgingival plaque samples (84.4%) were A. actinomycetemcomitans 16SrDNA positive, only 19 samples (15.6%) showed no detectable A. actinomycetemcomitans 16SrDNA. As expected, none of the A. actinomycetemcomitans 16SrDNA negative samples were positive for either lktA or fap. There were four genotypes of A. actinomycetemcomitans: 16SrDNA+/lktA+/fap+, 16SrDNA+/lktA+/fap−, 16SrDNA+/lktA−/fap+, and 16SrDNA+/lktA−/fap− in the subgingival plaque samples (Table 1). In the 122 subgingival plaque samples, there was no statistical difference between the positive rates of A. actinomycetemcomitans 16SrDNA (84.4%) and lktA (92/122, 75.4%) (χ 2=3.090, P>0.05). However, the positive rate of fap was much lower than that of the other two genes (χ 2=16.820, 32.810; P<0.01).
Twenty-three strains of A. actinomycetemcomitans were isolated from 18 patients (29.5%). All of the strains were positive for A. actinomycetemcomitans 16SrDNA, except that 4 of the strains were negative for the fap gene and 2 was negative for lktA. Furthermore, the genotype detected directly in the subgingival plaque samples was consistent with those of all isolated strains of A. actinomycetemcomitans.
In subgingival plaque samples, the positive rates of A. actinomycetemcomitans 16SrDNA+/lktA+/fap+, 16SrDNA+/lktA+/fap− and 16SrDNA+/lktA−/fap− were significantly higher in CP than that in healthy controls (P=0.000, 0.000, 0.038), while that of 16SrDNA+/lktA−/fap+ was not statistically different between the two groups (P=0.299). However, in clinical isolates, only A. actinomycetemcomitans 16SrDNA+/lktA+/fap+ showed higher frequency in CP than in healthy controls (P=0.005), while the positive rates of the other two genotypes showed no significant difference between the two groups (P=0.466, 0.448).
Nucleotide sequence analysis
The PCR products of P. gingivalis 16SrDNA amplified from 10 representative subgingival plaque samples showed 98.62%~99.72% homology with the nucleotide sequence registered in GenBank (accession No. L16492), and the amplification fragments for the prtC and fimA genes of the same 10 samples showed between 99.09%~99.45% and between 98.89%~100% homology, respectively, with the nucleotide sequences registered in GenBank (accession Nos. AB006973 and AB004560).
In this study, the PCR products of A. actinomycetemcomitans 16SrDNA amplified from 10 subgingival plaque samples showed 98.75%~99.85% homology compared with the GenBank sequences (accession No. M75036) respectively, while that of the lktA and fap genes was between 99.46%~99.76% and between 99.45%~99.55%, respectively (accession Nos. X16829 and D83053).
Association between clinical parameters and P. gingivalis genotypes in subgingival plaque samples and clinically isolated strains
P. gingivalis with the 16SrDNA+/prtC+/fimA+ genotype was the predominant genotype detected in subgingival plaque samples and clinical isolates (93/113, 82.3%; 40/48, 83.8%). Multinomial logistic regression analysis suggested that both in the 122 subgingival plaque samples and in the 48 clinically isolated P. gingivalis strains, the 16SrDNA+/prtC+/fimA+ genotype might be closely associated with PD (P=0.023, OR=3.905; P=0.000, OR=17.953), AL (P=0.018, OR=5.408; P=0.000, OR=15.779), GI (P=0.004, OR=4.588; P=0.001, OR=3.202), and tooth site (P=0.026, OR=11.077; P=0.000, OR=5.996) while the other three genotypes did not show any association with these clinical parameters (P=0.395~0.925) (Table 2). The positive rate of P. gingivalis with the 16SrDNA+/prtC+/fimA+ genotype was higher in deep pockets (>6 mm) than that in shallow (3~4 mm) (P=0.000; P=0.001) or moderate depth ones (>4 mm and ≤6 mm) (P=0.002; P=0.003), yet without significant difference between shallow and moderate pockets (P=0.062; P=0.177). It was more frequently detected in AL≥5 mm sites than in AL≤2 mm sites (P=0.000; P=0.000) or in the 2 mm<AL<5 mm sites (P=0.012; P=0.000). But no statistical difference could be found between the AL≤2 mm and the 2 mm<AL<5 mm ones (P=0.299; P=0.035, α′=0.017). In the GI=3 groups, P. gingivalis with the 16SrDNA+/prtC+/fimA+ displayed much higher frequency than in GI=1 groups (P=0.000; P=0.013), but there was not discrepancy between GI=1 and GI=2 (P=0.024, α′=0.017; P=0.205) or between GI=2 and GI=3 groups (P=0.037, α′=0.017; P=0.123). In molars, its presence was also higher than that in front teeth (P=0.023; P=0.037).
Table 2.
Distribution of P. gingivalis and A. actinomycetemcomitans genotypes in subgingival plaque samples and in clinical isolates from CP patients and its correlation with clinical parameters
| Groups | Cases (n) |
P. gingivalis 16SrDNA+ |
A. actinomycetemcomitans 16SrDNA+ |
||||||
| prtC+ fimA+ | prtC+ fimA− | prtC− fimA+ | prtC− fimA− | lktA+ fap+ | lktA+ fap− | lktA− fap+ | lktA− fap− | ||
| PD | |||||||||
| ≤4 mm | 49 | 29 (1) | 7 (1) | 4 (2) | 3 (0) | 18 (5) | 12 (0) | 2 (1) | 5 (0) |
| >4 mm and ≤6 mm | 39 | 30 (10) | 4 (4) | 1 (1) | 1 (0) | 18 (5) | 10 (2) | 1 (1) | 3 (0) |
| >6 mm | 34 | 34* (29*) | 0 (0) | 0 (0) | 0 (0) | 22* (7) | 12* (2) | 0 (0) | 0 (0) |
| AL | |||||||||
| ≤2 mm | 51 | 31 (1) | 6 (2) | 4 (2) | 3 (0) | 19 (5) | 13 (0) | 2 (1) | 6 (0) |
| >2 mm and <5 mm | 38 | 29 (11) | 5 (3) | 1 (1) | 1 (0) | 19 (6) | 9 (1) | 1 (1) | 2 (0) |
| ≥5 mm | 33 | 33* (28*) | 0 (0) | 0 (0) | 0 (0) | 20* (6) | 12* (3) | 0 (0) | 0 (0) |
| GI | |||||||||
| 1 | 18 | 9 (2) | 3 (2) | 1 (1) | 2 (0) | 6 (2) | 4 (1) | 1 (0) | 3 (0) |
| 2 | 46 | 33 (9) | 3 (2) | 2 (1) | 2 (0) | 22 (5) | 14 (2) | 1 (1) | 2 (0) |
| 3 | 58 | 51* (29*) | 5 (1) | 2 (1) | 0 (0) | 30 (10) | 16 (1) | 1 (1) | 3 (0) |
| Tooth site | |||||||||
| Front tooth | 61 | 39 (9) | 7 (3) | 4 (2) | 3 (0) | 33 (11) | 18 (3) | 1 (1) | 3 (0) |
| Molar | 61 | 54* (31*) | 4 (2) | 1 (1) | 1 (0) | 25 (6) | 16 (1) | 2 (1) | 5 (0) |
Data in the parenthesis are from P. ginglvalis or A. actinomycetemcomtitans clinical isolates
P<0.05
Further analysis indicated that in subgingival samples and clinical isolates, P. gingivalis with fimA+ genotype (16SrDNA+/prtC+/fimA+ and 16SrDNA+/prtC−/fimA+) might correlate with PD (P=0.007, OR=3.580; P=0.017, OR=5.215), AL (P=0.017, OR=2.813; P=0.008, OR=7.014), and GI (P=0.027, OR=2.339; P=0.013, OR=5.526) compared to fimA− genotype (16SrDNA+/prtC+/fimA− and 16SrDNA+/prtC−/fimA−). No statistical difference could be found in the distribution of P. gingivalis with fimA+ genotype in different tooth sites (P=0.108, OR=2.558; P=0.132, OR=4.364). In subgingival samples, P. gingivalis with fimA+ was more frequently detected in deep pockets than in shallow ones (P=0.002), yet without significant difference between moderate and deep ones (P=0.031, α′=0.017) or between shallow and moderate ones (P=0.222). It also showed a higher frequency in AL≥5 mm sites than that in AL≤2 mm (P=0.004) or the 2 mm<AL<5 mm sites (P=0.016). But its positive rates between the AL≤2 mm and the 2 mm<AL<5 mm ones were not statistically different (P=0.666). In GI=3 group, the frequency of P. gingivalis with fimA+ was higher than that in GI=1 group (P=0.013), with no statistical difference between GI=3 and GI=2 (P=0.533) or between GI=1 and GI=2 groups (P=0.674). However, in clinical isolates, post hoc tests did not show any significant difference in the positive rates of P. gingivalis with fimA+ in different PD (P=0.121, 0.946), AL (P=0.019~0.366, α′=0.017) or GI groups (P=0.045~0.330, α′=0.017), except that between deep pockets and moderate depth pockets (P=0.010).
Both in subgingival samples and clinical isolates, P. gingivalis with prtC+ genotype (16SrDNA+/prtC+/fimA+ and 16SrDNA+/prtC+/fimA−) might associate with PD (P=0.023, OR=5.038; P=0.014, OR=16.420) and AL (P=0.026, OR=4.842; P=0.021, OR=13.072), compared to prtC− genotype (16SrDNA+/prtC−/fimA+ and 16SrDNA+/prtC−/fimA−). P. gingivalis with prtC+ genotype was more frequently detected in deep pockets than in shallow pockets (P=0.013; P=0.011), but was no discrepancy between deep and moderate ones (P=0.261; P=0.341) or between shallow and moderate ones (P=0.127; P=0.097). In subgingival plaque samples, the positive rate of the prtC+ genotype in AL≥5 mm sites was higher than that in AL≤2 mm sites (P=0.016), with no difference between AL≥5 mm and the 2 mm<AL<5 mm ones (P=0.269) or between AL≤2 mm and the 2 mm<AL<5 mm ones (P=0.135). However, in clinical isolates post hoc tests indicated that the frequency of the prtC+ genotype in the different AL groups showed only slight difference (P=0.019~0.349, α′=0.017). Although multinomial logistic regression analysis of subgingival plaque samples suggested that P. gingivalis with prtC+ genotype might also correlate with GI (P=0.035, OR=2.756), but further analysis showed that its positive rate was not statistically different among the three GI groups (P=0.056~0.284). Compared to the prtC− genotype, no significant difference was found in the distribution of P. gingivalis with prtC+ genotype in different tooth sites in subgingival plaque samples and in clinical isolates (P=0.072, OR=4.413; P=0.180, OR=5.500) or that in the three GI groups in clinical isolates (P=0.147, OR=3.052).
Association between clinical parameters and A. actinomycetemcomitans genotypes in subgingival plaque samples and clinically isolated strains
Multinomial logistic regression analysis suggested that in the 122 subgingival plaque samples A. actinomycetemcomitans with 16SrDNA+/lktA+/fap+ or 16SrDNA+/lktA+/fap− genotypes might associate with PD (P=0.002, OR=3.575; P=0.008, OR=3.215) and AL (P=0.015, OR=2.510; P=0.035, OR=2.340) rather than the other two genotypes (P=0.415~0.980). No correlation was found between any one of the four genotypes or the lktA+ genotypes and GI or tooth site (P=0.061~0.952). A. actinomycetemcomitans with lktA+ genotype (16SrDNA+/lktA+/fap+ and 16SrDNA+/lktA+/fap−) was more frequently detected in deep pockets than that in shallow pockets (P=0.008), with no significant difference between deep and moderate ones (P=0.050) or between moderate and shallow ones (P=0.348). In sites with AL≥5 mm, the positive rate of A. actinomycetemcomitans with lktA+ was higher than that in AL≤2 mm sites (P=0.006), while no statistical difference was shown between AL≥5 mm and the 2 mm<AL<5 mm sites (P=0.113) or between the 2 mm<AL<5 mm and AL≤2 mm ones (P=0.196). However, in the 23 clinically isolated A. actinomycetemcomitans strains, no association could be established between any one of the four genotypes with PD, AL, GI or tooth site (P=0.065~0.921). Moreover, in subgingival plaque samples and clinical isolates, statistical difference did not appear in the positive rate of A. actinomycetemcomitans with fap+ genotype (16SrDNA+/lktA+/fap+ and 16SrDNA+/lktA−/fap+) in different PD, AL, GI or tooth site groups (P=0.139~0.654) compared with that of the fap− genotype (16SrDNA+/lktA+/fap− and 16SrDNA+/lktA−/fap−).
DISCUSSION
There is a growing body of evidence linking P. gingivalis and A. actinomycetemcomitans with pathogenesis of periodontitis (Cortelli et al., 2005a; Liu et al., 2003; Yuan et al., 2001). In previous studies, P. gingivalis isolates lacking prtC and/or fimA and A. actinomycetemcomitans isolates lacking fap were found (Fujiwara et al., 1994; Slots et al., 1995; Ishihara et al., 1997). However, the clinical significance of this virulence variation for both microbes was not understood. Routine diagnosis assays such as culture methods and biochemical reactions, enzyme-linked immunosorbent assays (ELISA) and immunofluorescence had been used to identify P. gingivalis and A. actinomycetemcomitans in clinical samples; but none of these diagnostic methods had focused on potential virulence factors. In the present study, multiplex PCR assays were adopted to detect the toxic genes of P. gingivalis and A. actinomycetemcomitans, using 16SrDNA as internal positive control. Multiplex PCR assays revealed clear PCR products with expected sizes (Figs.1 and 2). Repeated PCR amplification also demonstrated high reproducibility of the assays (91.9% and 88.7%). The genotypes of all cultured strains of P. gingivalis and A. actinomycetemcomitans were consistent with those detected directly in the corresponding subgingival plaque samples. Moreover, nucleotide sequence analysis showed high homology (98.62%~100%) of the PCR products for these toxic genes from partial subgingival plaque samples with the reported sequences. All the results from this study indicated that multiplex PCR could be used as a reliable method for diagnosis of the toxic genes of these two microbes in clinical samples.
The correlation between the infection of P. gingivalis and A. actinomycetemcomitans with different genotypes and periodontal destruction of CP is an interesting and important subject for investigation. Previous reports indicated that P. gingivalis was frequently detected in serious periodontal destruction sites with deep pockets (Noiri et al., 2001; Darout et al., 2003; Takeuchi et al., 2001), whereas the presence of A. actinomycetemcomitans in deep pockets was only occasionally (Noiri et al., 2001; Hamlet et al., 2001). But in this study, it was shown that P. gingivalis with prtC+/fimA+ and A. actinomycetemcomitans with lktA+ were predominant in teeth with deep pockets or serious attachment loss, suggesting that these genotypes might exhibit greater periodontal destruction potential than strains lacking either one or two of the toxic genes. P. gingivalis strains with prtC+/fimA+ were also found to be correlated with GI and tooth site. It was more frequently detected in sites with GI=3 and in molars, which implied its possible role in inflammation of periodontal tissue and potential inclination for colonization in molars rather than front teeth. A. actinomycetemcomitans with lktA+ was not associated with GI or tooth site, which correlated with previous reports (Tan et al., 2001). These studies indicated P. gingivalis’s presence in periodontal pockets was related with higher gingival index or deeper pocket depth (Yuan et al., 2001; Chen et al., 2005), while A. actinomycetemcomitans’s association with GI was not found (Tan et al., 2001; Nogueira et al., 2004). However, unlike the same results obtained from clinically isolated P. gingivalis strains and from subgingival plaque samples, in A. actinomycetemcomitans clinical isolates, no correlation could be established between lktA+ strains and PD or AL. This might be explained by the relatively low culture frequency of this microbe in this study.
Fimbriae were crucial pathogenic factors for P. gingivalis during adherence and colonization of periodontal epithelial cells (Amano et al., 1999; Amano et al., 2004). P. gingivalis expressed two distinct fimbria molecules, major and minor fimbriae, on its cell surfaces (Amano et al., 2004; Amano, 2003). The fimA gene, which encoded the major fimbriae—fimbrillin, has been classified into six variants (type I through V and Ib) on the basis of their nucleotide sequences. It was demonstrated that the fimbria variations might have an influence on the development of periodontal disease (Amano et al., 2004; Beikler et al., 2003a; Tamura et al., 2005), and that type II fimA was predominant in deep pockets (Amano et al., 1999; 2004). In the present study, PCR primers for fimA were designed based on DNA alignment of type I fimA gene by Watanabe and Frommel (1993), which could not detect types IV and V fimA. It was found that P. gingivalis with fimA+ was more frequently detected in deep pockets or in serious attachment loss sites than P. gingivalis strains with fimA−, indicating that the fimA gene of P. gingivalis was associated with periodontal destruction. It showed higher frequency in GI=3 sites which also suggested its correlation with inflammation of gingiva. As collagen is an important component of the periodontium, collagenase activity might play a role in tissue destruction and progression of periodontitis (Wittstock et al., 2000; Potempa et al., 2000; Beikler et al., 2003b). The prtC gene described by Kato et al.(1992) was 1002 bp in length and encoded a protein with 333 amino acids and a calculated mass of 37.8 kDa. Specific cleavage of type I collagen had been attributed to the function of the prtC gene product, which was referred to as collagenase (Kato et al., 1992). But the relationship between prtC and severity of the disease remained controversial (Bodinka et al., 1994; Wittstock et al., 2000; Potempa et al., 2000). This study showed that P. gingivalis with prtC+ was more frequently found in deep pockets or in serious attachment loss sites than P. gingivalis strains with prtC−, suggesting that the prtC gene of P. gingivalis was also associated with periodontal destruction.
Leukotoxin, a unique toxic factor produced by A. actinomycetemcomitans, could kill polymorphonuclear leukocytes in periodontal tissue, thus enabling the bacterium to escape an important part of the innate host immune response and was thus considered a significant virulence factor in periodontitis (Guthmiller et al., 2001; Poulsen et al., 2003). Greater mean attachment loss was found in subjects with highly leucotoxic A. actinomycetemcomitans than in subjects with minimally leucotoxic or subjects not infected (Cortelli et al., 2005a). In this study, it was found that A. actinomycetemcomitans with lktA+ genotype associated with periodontal destruction and that the frequency of lktA+ genotypes was significantly higher in deep pockets or in serious periodontal destruction sites, which demonstrated the clinical importance of the lktA of A. actinomycetemcomitans in the pathogenesis of CP. The fimbriae were generally critical for the bacteria to adhere to cells, which was an important step in pathogenesis. However, no correlation could be established between A. actinomycetemcomitans strains with fap+ (encoding fimbriae) and periodontal destruction in the present study. Hence it suggested that rather than directly adhere to cells and exert destructive effect on periodontal tissue, A. actinomycetemcomitans might inhibit local immune response by producing leukotoxin and thus enable the host prone to infection by other periodontal pathogens to develop chronic peridontitis.
A. actinomycetemcomitans was thought to be a major pathogen of aggressive periodontitis (Leung et al., 2005; Nonnenmacher et al., 2001). Although the primer set for 16SrDNA of A. actinomycetemcomitans designed by Slots et al.(1995) is known to cross-react with 16SrDNA of Haemophilus species, with the lktA specific primers, a high infection rate of A. actinomycetemcomitans was shown in CP patients by this study, which suggested that this microbe might also be an important factor in the pathogenesis of Chinese CP patients. The positive culture rate of A. actinomycetemcomitans from subgingival plaque samples was much lower when compared with that of PCR. This discordance between the positive rates of the PCR assay and the culture method might be due to the fact that a positive culture required a larger number of bacteria in clinical samples than in PCR. Further study would be needed to define its role in periodontal tissue destruction in CP. Although a long time study is necessary to establish whether P. gingivalis with prtC+/fimA+ or A. actinomycetemcomitans with lktA+ is a definitive factor for establishing periodontitis, this genotype is most likely to be an infectious agent contributing to the etiology. Future study should focus on the genotypic diversity of the fimA and prtC genes of P. gingivalis and the lktA gene of A. actinomycetemcomitans to elucidate its association with the pathogenesis of periodontitis.
Footnotes
Project (No. 30471888) supported by the National Natural Science Foundation of China
References
- 1.Amano A. Molecular interaction of Porphyromonas gingivalis with host cells: implication for the microbial pathogenesis of periodontal disease. J Periodontol. 2003;74(1):90–96. doi: 10.1902/jop.2003.74.1.90. [DOI] [PubMed] [Google Scholar]
- 2.Amano A, Nakagawa I, Kataoka K, Morisaki I, Hamada S. Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients. J Clin Microbiol. 1999;37(5):1426–1430. doi: 10.1128/jcm.37.5.1426-1430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amano A, Kuboniwa M, Nakagawa I, Akiyama S, Morisaki I, Hamada S. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J Dent Res. 2000;79(9):1664–1668. doi: 10.1177/00220345000790090501. [DOI] [PubMed] [Google Scholar]
- 4.Amano A, Nakagawa I, Okahashi N, Hamada N. Variations of Porphyromonas gingivalis fimbriae in relation to microbial pathogenesis. J Periodontal Res. 2004;39(2):136–142. doi: 10.1111/j.1600-0765.2004.00719.x. [DOI] [PubMed] [Google Scholar]
- 5.Armitage GC. Development of a classification system for periodontal disease and conditions. Ann Periodontol. 1999;4(1):1–6. doi: 10.1902/annals.1999.4.1.1. [DOI] [PubMed] [Google Scholar]
- 6.Beikler T, Peters U, Prajaneh S, Prior K, Ehmke B, Flemmig TF. Prevalence of Porphyromonas gingivalis fimA genotypes in Caucasians. Eur J Oral Sci. 2003;111(5):390–394. doi: 10.1034/j.1600-0722.2003.00065.x. [DOI] [PubMed] [Google Scholar]
- 7.Beikler T, Ehmke B, Wittstock M, Schmidt H, Karch H, Flemmig TF. Serum antibody reactivity against recombinant PrtC of Porphyromonas gingivalis following periodontal therapy. J Periodontal Res. 2003;38(3):276–281. doi: 10.1034/j.1600-0765.2003.01405.x. [DOI] [PubMed] [Google Scholar]
- 8.Bodinka A, Schmidt H, Henkel B, Flemmig TF, Klaiber B, Karch H. Polymerase chain reaction for the identification of Porphyromonas gingivalis collagenase genes. Oral Microbiol Immunol. 1994;9(3):161–165. doi: 10.1111/j.1399-302x.1994.tb00053.x. [DOI] [PubMed] [Google Scholar]
- 9.Chen LL, Wu YM, Yan J, Sun WL, Sun YZ, Ojcius D. Association between coinfection of Porphyromonas gingivalis, Actinobacillus actinomycetemcomitans and Treponema denticola and periodontal tissue destruction in chronic periodontitis. Chin Med J (Engl) 2005;118(11):915–921. [PubMed] [Google Scholar]
- 10.Cortelli JR, Cortelli SC, Jordan S, Haraszthy VI, Zambon JJ. Prevalence of periodontal pathogens in Brazilians with aggressive or chronic periodontitis. J Clin Periodontol. 2005;32(8):860–866. doi: 10.1111/j.1600-051X.2005.00777.x. [DOI] [PubMed] [Google Scholar]
- 11.Cortelli SC, Feres M, Rodrigues AA, Aquino DR, Shibli JA, Cortelli JR. Detection of Actinobacillus actinomycetemcomitans in unstimulated saliva of patients with chronic periodontitis. J Periodontol. 2005;76(2):204–209. doi: 10.1902/jop.2005.76.2.204. [DOI] [PubMed] [Google Scholar]
- 12.Darout IA, Skaug N, Albandar JM. Subgingival microbiota levels and their associations with periodontal status at the sampled sites in an adult Sudanese population using miswak or toothbrush regularly. Acta Odontol Scand. 2003;61(2):115–122. doi: 10.1080/00016350310002784. [DOI] [PubMed] [Google Scholar]
- 13.Fujiwara T, Nakagawa I, Morishima S, Takahashi I, Hamada S. Inconsistency between the fimbrillin gene and the antigenicity of lipopolysaccharides in selected strains of Porphyromonas gingivalis . FEMS Microbiol Lett. 1994;124(3):333–341. doi: 10.1111/j.1574-6968.1994.tb07305.x. [DOI] [PubMed] [Google Scholar]
- 14.Gafan GP, Lucas VS, Roberts GJ, Petrie A, Wilson M, Spratt DA. Prevalence of periodontal pathogens in dental plaque of children. J Clin Microbiol. 2004;42(9):4141–4146. doi: 10.1128/JCM.42.9.4141-4146.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guthmiller JM, Lally ET, Korostoff J. Beyond the specific plaque hypothesis: are highly leukotoxic strains of Actinobacillus actinomycetemcomitans a paradigm for periodontal pathogenesis? Crit Rev Oral Biol Med. 2001;12(2):116–124. doi: 10.1177/10454411010120020201. [DOI] [PubMed] [Google Scholar]
- 16.Hamlet SM, Cullinan MP, Westerman B, Lindeman M, Bird PS, Palmer J, Seymour GJ. Distribution of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis and Prevotella intermedia in an Australian population. J Clin Periodontol. 2001;28(12):1163–1171. doi: 10.1034/j.1600-051X.2001.281212.x. [DOI] [PubMed] [Google Scholar]
- 17.Imamura T, Travis J, Potempa J. The biphasic virulence activities of gingipains: activation and inactivation of host proteins. Curr Protein Pept Sci. 2003;4(6):443–450. doi: 10.2174/1389203033487027. [DOI] [PubMed] [Google Scholar]
- 18.Ishihara K, Honma K, Miura T. Cloning and sequence analysis of the fimbriae associated protein (fap) gene from Actinobacillus actinomycetemcomitans . Microb Pathog. 1997;23(2):63–69. doi: 10.1006/mpat.1997.0137. [DOI] [PubMed] [Google Scholar]
- 19.Johansson A, Buhlin K, Koski R, Gustafsson A. The immunoreactivity of systemic antibodies to Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in adult periodontitis. Eur J Oral Sci. 2005;113(3):197–202. doi: 10.1111/j.1600-0722.2005.00218.x. [DOI] [PubMed] [Google Scholar]
- 20.Kato T, Takahashi N, Kuramitsu HK. Sequence analysis and characterization of the Porphyromonas gingivalis prtC gene, which expresses a novel collagenase activity. J Bacteriol. 1992;174(12):3889–3895. doi: 10.1128/jb.174.12.3889-3895.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lakio L, Kuula H, Dogan B, Asikainen S. Actinobacillus actinomycetemcomitans proportion of subgingival bacterial flora in relation to its clonal type. Eur J Oral Sci. 2002;110(3):212–217. doi: 10.1034/j.1600-0447.2002.201238.x. [DOI] [PubMed] [Google Scholar]
- 22.Leung WK, Ngai VK, Yau JY, Cheung BP, Tsang PW, Corbet EF. Characterization of Actinobacillus actinomycetemcomitans isolated from young Chinese aggressive periodontitis patients. J Periodontal Res. 2005;40(3):258–268. doi: 10.1111/j.1600-0765.2005.00805.x. [DOI] [PubMed] [Google Scholar]
- 23.Liu L, Wen X, He H, Shi J, Ji C. Species-specific DNA probe for the detection of Porphyromonas gingivalis from adult Chinese periodontal patients and healthy subjects. J Periodontol. 2003;74(7):1000–1006. doi: 10.1902/jop.2003.74.7.1000. [DOI] [PubMed] [Google Scholar]
- 24.Nogueira Moreira A, Chiappe V, Fernandez Caniggia L, Alonso C, Piovano S. Clinical and microbiological associations in chronic periodontitis. Acta Odontol Latinoam. 2004;17(1-2):15–21. [PubMed] [Google Scholar]
- 25.Noiri Y, Li L, Ebisu S. The localization of periodontal-disease-associated bacteria in human periodontal pockets. J Dent Res. 2001;80(10):1930–1934. doi: 10.1177/00220345010800101301. [DOI] [PubMed] [Google Scholar]
- 26.Nonnenmacher C, Mutters R, de Jacoby LF. Microbiological characteristics of subgingival microbiota in adult periodontitis, localized juvenile periodontitis and rapidly progressive periodontitis subjects. Clin Microbiol Infect. 2001;7(4):213–217. doi: 10.1046/j.1469-0691.2001.00210.x. [DOI] [PubMed] [Google Scholar]
- 27.Potempa J, Banbula A, Travis J. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontology 2000. 2000;24(1):153–192. doi: 10.1034/j.1600-0757.2000.2240108.x. [DOI] [PubMed] [Google Scholar]
- 28.Poulsen K, Ennibi OK, Haubek D. Improved PCR for detection of the highly leukotoxic JP2 clone of Actinobacillus actinomycetemcomitans in subgingival plaque samples. J Clin Microbiol. 2003;41(10):4829–4832. doi: 10.1128/JCM.41.10.4829-4832.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slots J, Ashimoto A, Flynn MJ, Li GL, Chen C. Detection of putative pathogens in subgingival specimens by 16S ribosomal DNA amplification with the polymerase chain reaction. Clin Infect Dis. 1995;20(Suppl. 2):304–307. doi: 10.1093/clinids/20.supplement_2.s304. [DOI] [PubMed] [Google Scholar]
- 30.Takeuchi Y, Umeda M, Sakamoto M, Benno Y, Huang Y, Ishikawa I. Treponema socranskii, Treponema denticola, and Porphyromonas gingivalis are associated with severity of periodontal tissue destruction. J Periodontol. 2001;72(10):1354–1363. doi: 10.1902/jop.2001.72.10.1354. [DOI] [PubMed] [Google Scholar]
- 31.Tamura K, Nakano K, Nomura R, Miyake S, Nakagawa I, Amano A, Ooshima T. Distribution of Porphyromonas gingivalis fimA genotypes in Japanese children and adolescents. J Periodontol. 2005;76(5):674–679. doi: 10.1902/jop.2005.76.5.674. [DOI] [PubMed] [Google Scholar]
- 32.Tan KS, Woo CH, Ong G, Song KP. Prevalence of Actinobacillus actinomycetemcomitans in an ethnic adult Chinese population. J Clin Periodontol. 2001;28(9):886–890. doi: 10.1034/j.1600-051x.2001.028009886.x. [DOI] [PubMed] [Google Scholar]
- 33.van der Ploeg JR, Giertsen E, Ludin B, Morgeli C, Zinkernagel AS, Gmur R. Quantitative detection of Porphyromonas gingivalis fimA genotypes in dental plaque. FEMS Microbiol Lett. 2004;232(1):31–37. doi: 10.1016/S0378-1097(04)00064-3. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe K, Frommel TO. Detection of Porphyromonas gingivalis in oral plaque samples by use of the polymerase chain reaction. J Dent Res. 1993;72(6):1040–1044. doi: 10.1177/00220345930720060801. [DOI] [PubMed] [Google Scholar]
- 35.Watanabe K, Frommel TO. Porphyromonas gingivalis, Actinobacillus actinomycetemcomitans and Treponema denticola detection in oral plasque samples using the polymerase chain reaction. J Clin Periodontol. 1996;23(3):212–219. doi: 10.1111/j.1600-051X.1996.tb02078.x. [DOI] [PubMed] [Google Scholar]
- 36.Wittstock M, Schmidt H, Flemmig TF, Karch H. Heterogeneity of the prtC gene of Porphyromonas gingivalis . Oral Microbiol Immunol. 2000;15(1):33–39. doi: 10.1034/j.1399-302x.2000.150106.x. [DOI] [PubMed] [Google Scholar]
- 37.Yang HW, Huang YF, Chan Y, Chou MY. Relationship of Actinobacillus actinomycetemcomitans serotypes to periodontal condition: prevalence and proportions in subgingival plaque. Eur J Oral Sci. 2005;113(1):28–33. doi: 10.1111/j.1600-0722.2004.00192.x. [DOI] [PubMed] [Google Scholar]
- 38.Yuan K, Chang CJ, Hsu PC, Sun HS, Tseng CC, Wang JR. Detection of putative periodontal pathogens in non-insulin-dependent diabetes mellitus and non-diabetes mellitus by polymerase chain reaction. J Periodontal Res. 2001;36(1):18–24. doi: 10.1034/j.1600-0765.2001.90613.x. [DOI] [PubMed] [Google Scholar]