Abstract
Neuronal cell fate decisions are directed in Drosophila by NUMB, a signaling adapter protein with two protein–protein interaction domains: a phosphotyrosine-binding domain and a proline-rich region (PRR) that functions as an SH3-binding domain. Here we show that there are at least four human NUMB isoforms and that these serve two distinct developmental functions in the neuronal lineage: differentiation (but not proliferation) is promoted by human NUMB protein isoforms with a type I (short) PRR. In contrast, proliferation (but not differentiation) is directed by isoforms that have a type II (long) PRR. The two types of PRR may promote distinct intracellular signaling pathways downstream of the NOTCH receptor during mammalian neurogenesis.
During Drosophila peripheral nervous system development, a sensory organ precursor cell divides twice to produce the four cells that form a functional sensory organ (neuron, sheath, hair, and socket). NUMB functions in this lineage to direct specific binary cell fate choices: the IIb vs. IIa fate at the first division and at the second division, neuron vs. sheath as well as hair vs. socket (1). Absence of NUMB results in production of two IIa cells that then divide to give four sockets while ectopic expression of NUMB generates two IIb cells and, subsequently, four neurons and no hairs (2). The fate choices directed by Drosophila NUMB occur in a stereotypical lineage within which NUMB functions solely to control binary differentiation decisions and not the proliferation dynamics of the cells. NUMB has been hypothesized to function in directing cell fate choice by directly interacting with NOTCH and inhibiting NOTCH function (3).
The recent cloning of a mammalian NUMB homologue suggested an evolutionarily conserved function for mammalian NUMB (4, 5). We showed that ectopic expression of this mammalian NUMB protein (mNUMB) promotes neuronal commitment in both Drosophila and cultured mammalian cells (4). Recently, it has been observed that a human NUMB homologue (hNUMB) is transported to the nucleus and associates with the modulator of mitogenesis, MDM2 (6, 7). Our previous studies showed no mitogenic component to murine NUMB function (4). Here we report the existence of four hNUMB isoforms. Based on the structure of the proline-rich region (PRR), the NUMB isoforms regulate either cell fate or cellular proliferation, but not both, during mammalian neurogenesis.
MATERIALS AND METHODS
Library Screening.
Probe was synthesized by random priming of the phosphotyrosine-binding domain (PTB)-encoding domain of the published mNUMB cDNA (4) and was used to probe a human NT2 library (Stratagene) following the manufacturer’s instructions. The positive phage were plaque purified and converted into phagemid. Ten clones containing inserts greater than 3 kb were sequenced. All 10 clones contained a complete ORF of approximately 2.4–2.6 kb.
Northern Blot Analysis.
A 125-bp fragment corresponding to nucleotides 1046 to 1171 of the hNUMB PTBLPRRS mRNA isoform (Fig. 1) was random primed and used to probe a series of Northern blots containing 2 μg of poly-(A)+-selected mRNA from human cancer cell lines and multiple human tissues (CLONTECH) following the manufacturer’s procedures. The blots were exposed overnight and then stripped by incubating for 20 min in 0.1 × SSC/1% SDS at 100°C. After overnight exposure to ensure the blot was stripped, the blots were reprobed with a 191-bp radiolabeled PCR fragment containing the PRR insertion. Overnight exposure showed weak signals. The blots were reexposed for 1 wk. The 191-bp PRR insert-specific probe was also used to screen 2 μg of poly-(A)+ plus RNA from mice at various stages of embryonic development (CLONTECH) and multiple rat tissues (CLONTECH) according to the manufacturer’s instructions. Exposure times in both cases were overnight followed by a 1-wk reexposure.
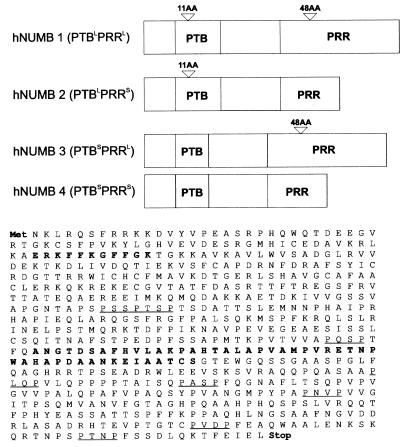
Figure 1.
Schematic of four hNUMB isoforms and amino acid sequence of the numb PTBL PRRL isoform (L, long; S, short). In bold are the amino acids inserted as a result of the alternative splicing of the human Numb transcript. Underlined are the potential SH3-binding sites denoted by the sequence PXXP.
PCR Amplification.
Primers corresponding to nucleotides 1046–1066 and 1293–1314 of the human PTBL PRRL coding region were used to PCR amplify 0.25 ng of adult brain and human fetal brain “marathon ready” cDNA (CLONTECH) under the following conditions: 94°C for 5 min (1 cycle); 94°C for 15 s, 55°C for 15 s, 72°C for 1 min (30 cycles); 72°C for 5 min (1 cycle). Twenty microliters of a 50-μl reaction was electrophoresed on a 2% agarose gel.
Two micrograms of total rat RNA was oligo-dT primed and reverse transcribed. One microliter of the resulting cDNA was used to amplify actin as a control. The cDNA concentrations were then adjusted such that equal starting amounts were present, and 1 μl was used for PCR amplification as above, by using primers corresponding to nucleotides 1020–1040 and 1180–1200 of the published rat coding sequence (4). These primers flank the potential PRR insertion. Samples were analyzed on a 2% agarose gel.
Cell Culture.
P19 cells were cultured according to the methods of Rudnicki and McBurney (8). To differentiate the P19 cells, cells were seeded at 1 × 105 cells per nontissue culture Petri plates and aggregated in the presence of 1 μM retinoic acid for 4 d. The cells were collected, trypsinized, and plated onto 60-mm Nunc plates in the absence of retinoic acid. Cultures were fixed 2 d later and stained for neurofilament (NF) (Sigma) (4).
Immortalized mouse neural crest cell line (MONC-1) cells were cultured according to the methods of Sommer (9). For analysis, hNUMB transformants were plated at clonal density and allowed to differentiate for 1 wk. Differentiation was induced by the addition of 10% FBS and 5 μM forskolin to the standard media (4, 9). Neural (NF staining) and smooth muscle differentiation (smooth muscle actin staining) occurred within 96 hr and glial differentiation (glial fibrillary acidic protein staining), within 4–7 d.
Primary cultures of rat neural crest stem cells were prepared and cultured as previously described (10). Briefly, trunk neural tubes were collected and excised by digestion with trypsin. The excised tubes were plated on fibronectin-coated dishes and the crest cells allowed to migrate overnight. The following morning, the tubes were removed and the cells infected with virus. After infection, the cells were trypsinized and plated at clonal density. Stem cells were identified by live immunostaining against the low-affinity nerve growth factor receptor. The positions of the positive cells were marked and the medium changed to differentiation medium for 1 wk (10). A similar immunohistochemical analysis was performed on primary crest as on MONC-1 cells.
Transfection.
P19 cells overexpressing each of the hNUMB isoforms were generated by the calcium phosphate coprecipitation method (4). The cells were washed, split, and 500 μg/ml G418 added for 1 wk. The resulting stable clones were pooled, expanded, and maintained in 250 μg/ml G418 until differentiated.
Generation of Virus and Infection.
BOSC23 cells (11) were maintained in GPT selection media. On the day before transfection, the cells were split and plated at 2 × 106 cells per 60-mm dish in DMEM plus 10% FBS. Calcium chloride transfection was used to introduce pBABE (12) constructs harboring each of the four Numb isoforms into the BOSC23 packaging cell line. Twenty micrograms of precipitate per plate was added to the cells for 10 hr in the presence of 25 μM chloroquin. The cells were then washed with media and 2 ml of medium was added. Virus was collected for the next 72 hr, with medium changes every 24 hr. At the end of 72 hr the virus-containing medium was divided in half and was concentrated according to the method of Cepko (13). The resulting viral pellet was resuspended in either MONC-1 medium or P19 medium.
Retroviral Infections.
MONC-1 and primary neural crest stem cells overexpressing the full length hNUMB isoforms were generated by retroviral infection (4). Briefly, cells were incubated for a period of 4 hr with a viral supernatant containing 4 mg/ml polybrene obtained from the transient transfection of BOSC23 cells. The cells were washed, and an additional 2-hr infection ensued. The cells were allowed to recover for 12 hr before the addition of 350 μg/ml hygromycin and differentiation media for the duration of the experiment (4).
Production and Analysis of Transgenic Drosophila Strains.
The complete ORF of each of three hNUMB isoforms was directionally inserted into the EcoRI and XhoI sites of pUAST (14). P element-mediated germ line transformation was according to standard methods (15). The three hNUMB transgenic constructs (UAS:hNUMB2-4) were individually crossed to the GAL4 enhancer trap line C96 and analyzed as previously described (4). For quantification of hNUMB-induced cell type transformation, we counted the number of stout bristles in the wing margin medial triple row. Three to five independently derived transgenic lines for each construct were crossed and 10 flies from each cross were quantified. The number of stout bristles in wild-type wings (data not shown) was similar to those previously reported (16). All fly crosses were done at 25°C and 29°C to vary the level of GAL4-driven expression of transgenic constructs. Three new independent transgenic lines were isolated from a single original transformant of UAS:hNUMB3 by mobilizing the P element to a different chromosome with a transgenic source of Δ2–3 transposase. Images were analyzed and photographed with a Zeiss Axioplan microscope.
RESULTS AND DISCUSSION
Human Numb Transcripts Encode a Family of Four hNUMB Protein Isoforms.
To isolate human Numb homologs, a human neuronal precursor NT2 cDNA library was screened at reduced stringency. Analysis of 10 cDNA clones containing both predicted initiation, and termination codons revealed the existence of four classes of alternatively spliced hNUMB transcripts (Fig. 1). The alternative splicing generates variant ORFs that affect the two regions of NUMB, which direct interactions with signaling pathway components: the PTB and the SH3-binding domain defined by a PRR (4). Two of the four classes of transcripts contain a 33-nt (11-codon) insert in the PTB domain-encoding region relative to Drosophila NUMB, whereas two have a 144-nt (48-codon) insert in the PRR-encoding region (Fig. 1). Differential mRNA splicing leads to the production of NUMB isoforms representing all four possible combinations (Fig. 1): hNUMB1 PTBinsert(+) PRRinsert(+) = PTBL PRRL (two clones); hNUMB2 PTBinsert(+) PRRinsert(-) = PTBL PRRS (three clones); hNUMB3 PTBinsert(-) PRRinsert(+) = PTBS PRRL (two clones), and hNUMB4 PTBinsert(-) PRRinsert(-) = PTBS PRRS (three clones).
Transcripts Encoding hNUMB PRRS and PRRL Are Differentially Expressed in Adults.
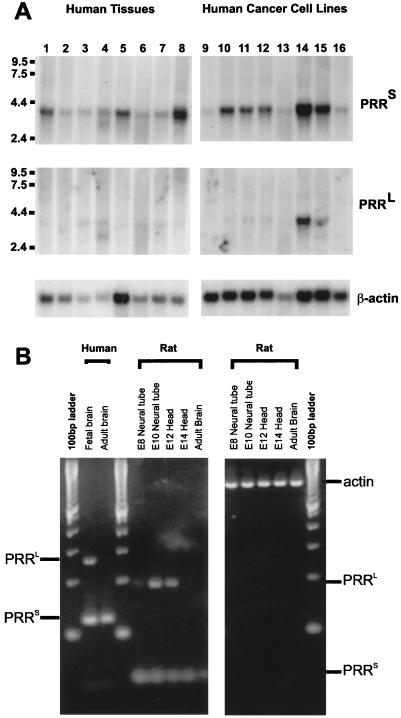
We next addressed whether these mRNA isoforms are differentially expressed in adult tissues. Northern blot analysis of multiple adult human tissues and cancer cell lines revealed that transcripts encoding hNUMB isoforms without insertions into the PRR domain (PRRS) are ubiquitously expressed (Fig. 2A), including in all human tumors examined. However, transcripts encoding isoforms with insertions into the PRR (PRRL) were detectable in significantly lower levels than PRRS and in only a subset of the tissues examined (prostate, testis, and intestine). The levels of both hNUMB isoforms were significantly elevated in the colon–rectal carcinoma sw480 cell line (Fig. 2A, lane 14).
Figure 2.
Tissue distribution of NUMB PRRS- and PRRL-encoding transcripts in adult tissues and human cancer cell lines. (A) Northern blots of rat tissues and human cancer cell lines were analyzed for PPRS and PRRL-specific mRNA expression. Lanes 1–8 are as follows: spleen, thymus, prostrate, testis, uterus, small intestine, colon, and peripheral blood leukocytes. Lanes 9–16 are Hl-60, HeLa, k-56, molt-4, Raji, sw480, a549, g361. Expression of a 4-kb transcript is observed in all lanes for PRRS isoforms, whereas PRRL transcripts are restricted to prostate, testis, uterus, and colon and heavily expressed in sw480 (colon–rectal adenocarcinoma) and A549 (lung carcinoma). (B) Transcripts encoding NUMB isoforms are differentially expressed in developing brain. PCR amplification of cDNA from human fetal brain, human adult brain, and neural tissue from developing embryonic rat was carried out using primers that flank the PRR insertion. Thirty cycles of amplification revealed the existence of PRRL transcripts in fetal but not adult brain. PCR products representing PRRL transcripts were observed in rat neural tube at day 8, decreasing in intensity through development. No PRRL transcripts were detected in embryonic day 6 rat head or human adult brain even after an additional five cycles of amplification.
That transcripts encoding hNUMB PRRL-isoforms are rarer than those encoding PRRS-isoforms was confirmed by reverse transcription–PCR (RT-PCR) analysis of human adult brain cDNA by using primers that flank the PRR-coding portion of the ORF (Fig. 2B). Further, a rat multiple tissue Northern blot was probed by using the 191-bp PRRL-specific insert from the human cDNA. Numb transcripts containing PRRL were detectable after a 1-wk exposure in adult brain, liver, testis, and kidney, but were absent in skeletal muscle, spleen, and heart even after prolonged exposure (data not shown).
Expression of Transcripts Encoding PRRL-Containing hNUMBs Is Developmentally Regulated.
To examine the expression of Numb transcripts during brain development, RT-PCR analysis of RNA from developing rat neural tissue was conducted by using primers that flank the potential PRRL insertion (Fig. 2B). PRRL-encoding transcripts are expressed at low levels throughout early neuronal development peaking at embryonic day 10 and decreasing thereafter. PRRL transcripts were undetectable after embryonic day 14. In contrast, PRRS transcript levels remained constant in developing and adult brain. The developmental profile of PRRL transcripts in mice was assayed by Northern blot analysis (data not shown), confirming the rat brain RT-PCR studies showing expression from very low levels at embryonic day 7 to modest levels at day 11. Numb transcripts encoding PRRL were not detectable after embryonic day 13 (data not shown). Together, these results demonstrate that transcripts encoding the PRRL vs. PRRS hNUMB isoforms are differentially expressed during neural development. In particular, only transcripts encoding the PRRL isoforms are dynamically expressed, peaking during the stages at which neuronal precursor cell proliferation is occurring and then decreasing to undetectable levels in adult brain.
hNUMB PRRS but Not PRRL Isoforms Mediate Neuronal Cell Fate Choice in Drosophila.
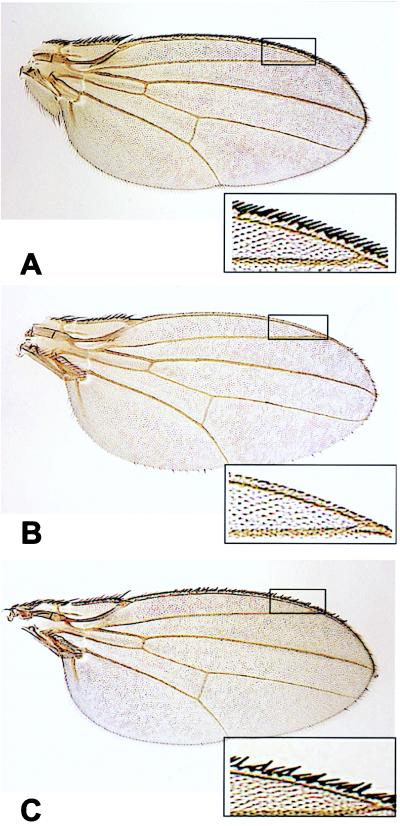
We next asked whether the variations within the two protein–protein interaction domains of the molecule affect their biological function. We initially assayed functional differences by ectopic expression of hNUMB isoforms in transgenic Drosophila. Our previous misexpression of rat NUMB (PTBL PRRS) at the developing wing margin by using the GAL4C96 driver resulted in transformation of the sensory organ precursor lineage throughout the entire margin toward neuronal fate (4). For the present assays, because hNUMB4 (PTBS PRRS) is structurally homologous to Drosophila NUMB, we compared hNUMB4 to hNUMB2 (PTBL PRRS; to uncover differences in PTB domain function) and hNUMB4 to hNUMB3 (PTBS PRRL; to uncover differences in PRR domain function). Insertions within the hNUMB PRR dramatically abrogated the ability of hNUMB to direct neuronal cell fate choice, whereas insertions into the PTB domain had no detectable effect (Fig. 3, Table 1). Ectopic expression of hNUMB4 (PTBS PRRS; Fig. 3B) and hNUMB2 (PTBL PRRS) caused the strongest transformation toward neuronal fate (>87% loss of hairs from each wing margin, Table 1), whereas hNUMB3 (PTBS PRRL; Fig. 3C) caused a significantly weaker transformation (<20% loss of hairs from each wing margin, Table 1). We conclude that, in the Drosophila assay, both types of PTB-domains are equally functional, but the insert into the PRR (PRRL) reduces the ability to direct both the IIa to IIb transformation and neuronal fate specification.
Figure 3.
Misexpression of hNUMB isoforms during sensory organ precursor development in Drosophila. (A) Wild-type wing margin. (B) Expression of hNUMB PRRS at the dorsoventral margin of P[UAS-hNumbPRRS] transgenic larval wing imaginal discs results in a dramatic loss of marginal hairs (>87%). (C) Expression of hNUMB PRRL at the dorsoventral margin results in a significantly lower loss of marginal hairs (<20%). (Insets) Enlargements of the wing margin.
Table 1.
Overexpression of PRRS but not PRRL hNUMB isoforms leads to wing margin balding in Drosophila
| numb | Percent balding*
|
|
|---|---|---|
| 25 degrees | 29 degrees | |
| PTB | ||
| PTBSPRRS | 87 ± 3.6 | 96 ± 0.9 |
| PTBLPRRS | 84 ± 10 | 93 ± 6.6 |
| PRR | ||
| PTBSPRRS | 87 ± 3.6 | 96 ± 0.9 |
| PTBSPRRL | 7.5 ± 5.8 | 20 ± 14 |
*Mean percent balding of the wing margin ± SEM (see Materials and Methods).
hNUMB PRRS -Containing Isoforms Promote Differentiation, Whereas PRRL-Containing Isoforms Promote Proliferation During Mammalian Neurogenesis.
The murine P19 embryonic carcinoma cell line serves as an excellent tissue culture model for mammalian neurogenesis (17, 18). To confirm functional differences between PRRL and PRRS isoforms, we overexpressed the hNUMB isoforms in P19 cells. We have previously shown that overexpression of a rat NUMB (mNUMB = PTBL PRRS) dramatically biases undifferentiated P19 cells toward neuronal fate, whereas overexpression of a dominant-negative form of NUMB (only the PTB domain) biases cells away from neuronal fate (4). Pooled stable lines of P19 cells overexpressing type I (PTBL PRRS or PTBS PRRS) human NUMB isoforms showed a dramatic increase in the number of neurofilament-positive cells after 4 d of aggregation and retinoic acid treatment (2.6- and 2.2-fold, respectively; Table 2). In contrast, pooled stable lines of P19 cells expressing type II isoforms (PTBS PRRL or PTBL PRRL) showed an increase in the total number of cells, although the fraction of cells bearing neuronal processes was similar to controls (Table 2). This observation confirmed that hNUMB PRRS-containing isoforms bias cells toward neuronal fate. Unexpectedly, however, the data also suggested that hNUMB isoforms containing insertions into the PRR (PRRL) either increased the survival of undifferentiated cells or increased the proliferation rate of undifferentiated cells.
Table 2.
PRRS but not PRRL hNUMB isoforms promote neuronal differentiation of P19 cells
| Isoform | NF+ cells | Total cells | % Neurons | Composite |
|---|---|---|---|---|
| Control | 828 | 6284 | 13.2 | 13.6 ± 0.8 |
| hNUMB4 PTBS PRRS | 2232 | 6248 | 35.5 | 35.9 ± 3.3 |
| hNUMB2 PTBL PRRS | 1844 | 6756 | 27.3 | 29.0 ± 3.6 |
| hNUMB3 PTBS PRRL | 1156 | 8544 | 13.5 | 16.4 ± 3.6 |
| hNUMB1 PTBL PRRL | 1780 | 9140 | 19.5 | 18.9 ± 4.0 |
The first three columns present the results of the initial experiment. In column four is the mean ± SEM of four such experiments resulting from two distinct transfections.
To test whether PRRL-containing hNUMB isoforms promote proliferation, BrdU incorporation assays were conducted on unaggregated P19 cell lines harboring each of the four hNUMB isoforms (19, 20). No differences in BrdUrd incorporation were observed from control values for cells not undergoing overt cellular differentiation (Table 3). However, on differentiation (aggregation in the presence of retinoic acid) P19 lines expressing type II isoforms (PTBL PRRL and PTBS PRRL) showed a 1.5-fold increase in BrdUrd-positive cells after an 8-hr pulse (Table 3). BrdU incorporation into lines expressing the type I isoforms (PTBS PRRS and PTBL PRRS) showed no increase in proliferation, and perhaps even a slight decrease, relative to controls (Table 3). These results suggest distinct developmental functions for PRRL- vs. PRRS-containing hNUMB isoforms: PRRS-containing isoforms promote neuronal differentiation. In contrast, PRRL-containing isoforms do not direct differentiation, but rather promote cell proliferation.
Table 3.
PRRL hNUMB isoforms promote cell proliferation
| Isoform | BrdU incorporation, %
|
|
|---|---|---|
| Unaggregated | Aggregated + RA | |
| Control | 22.8 ± 2.6 | 29.2 ± 1.9 |
| hNUMB4 PTBS PRRS | 21.8 ± 1.4 | 29.9 ± 1.1 |
| hNUMB2 PTBL PRRS | 23.9 ± 3.3 | 27.1 ± 1.6 |
| hNUMB3 PTBS PRRL | 23.2 ± 2.4 | 43.8 ± 4.3 |
| hNUMB1 PTBL PRRL | 24.7 ± 1.5 | 37.6 ± 3.8 |
Presented are the mean plus SEM for four experiments resulting from two independent transfections.
To verify the distinct functions of PRRS- and PRRL-containing hNUMB isoforms, we expressed these in an immortalized neural crest stem cell line (MONC-1) (4, 9) and in primary neural crest stem cells (10). Neural crest stem cells offer a wider possibility of phenotypic cell fate choices in culture (neurons, glia, and smooth muscle) than do P19 cells (neurons and glia) (9, 10). Moreover, the development of individual cells can be followed during the assay to monitor the effects on proliferation, differentiation, and apoptosis. As in our previous analyses of mNUMB (rat PTBL PRRS), overexpression in MONC-1 cells of PRRS-containing hNUMB isoforms forced the majority of the resulting clones into a “neuron only” phenotype (Table 4). However, when PRRL-containing hNUMB isoforms were expressed, there was no difference relative to control clones (i.e., there was no bias toward or away from neuronal differentiation; Table 4). MONC-1 and primary neural crest stem cells overexpressing the same isoforms also showed a strong bias toward the neuronal lineage when carrying PRRS-containing hNUMB isoforms, whereas no neuronal bias was seen when PRRL-containing hNUMB isoforms were expressed. Strikingly, the resulting terminal clone size in both MONC experiments and primary crest experiments for PRRL hNUMB expressing clones was two to three times greater (MONC-1 PRRL clones: 20.9 ± 5.4; primary PRRL clones: 17.9 ± 4.0) than that of PRRS hNUMB-expressing cells (MONC-1 PRRS clones: 7.8 ± 2.8; primary PRRS clones: 8.8 ± 3.0) or control clones (MONC-1 hygromycin clones: 9.1 ± 3.3; primary Hygromycin clones: 10.0 ± 3.2). These results confirm that PRRL-containing hNUMB isoforms promote mitosis of undifferentiated progenitors.
Table 4.
PRRS but not PRRL hNUMB isoforms direct neuronal fate choice in primary and immortalized neural crest stem cells
| hNUMB isoform class | Neuron-only clones | Neurons + other phenotypes |
|---|---|---|
| MONC-1 cells | ||
| PRRS | 47 (46%) | 54 (54%) |
| PRRL | 1 (1.0%) | 86 (88%) |
| Control | 1 (1.3%) | 70 (89%) |
| Primary neural crest stem cells | ||
| PRRS | 38 (60%) | 13 (21%) |
| PRRL | 1 (1.4%) | 63 (90%) |
| Control | 1 (1.5%) | 59 (87%) |
Presented is the composite of two independent experiments originating from independent infections. The total number of clones containing only neurons is shown in the first column. In the second column, the percentage of mixed clones (Neuron + other phenotype) is shown. The percentage of each type of clone is presented in parentheses.
CONCLUSIONS
We have shown that the PRRS- and the PRRL-containing hNUMB isoforms are likely to implement distinct functions during mammalian neurogenesis by promoting either neuronal differentiation (PRRS) or proliferation (PRRL). The recent observation that hNUMB is translocated into the nucleus and interacts with a critical member of the mitotic index machinery MDM2 (6, 7) is consistent with this conclusion. What is the mechanism underlying these distinct hNUMB functions? The Drosophila NUMB protein is known to interact with Drosophila NOTCH through its PTB domain, thus inhibiting NOTCH function (3). If a similar hNUMB–NOTCH interaction occurs in mammalian cells, then PRRS and PRRL could serve to link NOTCH to distinct SH3 domain-containing proteins. The PRRS class would promote neuronal differentiation, whereas the PRRL class would direct proliferation. At present, we have no data that address whether additional modulation of NOTCH signaling might be accomplished through differential NOTCH binding of the PTBS and the PTBL domains. Because there are at least four vertebrate NOTCH isoforms (21), it is possible that distinct hNUMB isoforms interact with distinct NOTCH isoforms to signal either differentiation or proliferation during neurogenesis as well as in other lineages (22, 23).
Acknowledgments
We thank Pushpa Sharma, Aly Cassam, Cuiping Zhao (Robarts Research Institute), Bill Fisher, and Angelo Karaiskakis (Hospital for Sick Children) for technical assistance. This research was funded in part by grants from the Medical Research Council of Canada (grants MT-14144 to J.M.V. and MT-13208 to H.D.L.).
ABBREVIATIONS
- PTB
phosphotyrosine-binding domain
- PRR
proline-rich region
- MONC-1
immortalized mouse neural crest cell line
Footnotes
References
- 1.Uemura T, Sheperd S, Ackerman L, Jan L Y, Jan Y N. Cell. 1989;58:349–360. doi: 10.1016/0092-8674(89)90849-0. [DOI] [PubMed] [Google Scholar]
- 2.Rhyu M S, Jan L Y, Jan Y N. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 3.Lily M G, Jan L Y, Jan Y N. Neuron. 1996;17:27–41. doi: 10.1016/s0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 4.Verdi J M, Schmandt R, Bashirullah A, Jacob S, Salvino R, Craig C G, Lipshitz H, McGlade C J Amgen est program. Curr Biol. 1996;6:1134–11345. doi: 10.1016/s0960-9822(02)70680-5. [DOI] [PubMed] [Google Scholar]
- 5.Zhong W, Feder J N, Jiang M-M, Jan L Y, Jan Y N. Neuron. 1995;17:43–53. doi: 10.1016/s0896-6273(00)80279-2. [DOI] [PubMed] [Google Scholar]
- 6.Juven-Gershon T, Shifman O, Unger T, Elkeles A, Haupt Y, Oren M. Mol Cell Biol. 1998;7:3974–3982. doi: 10.1128/mcb.18.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freedman D A, Wu L, Levine A J. Cell Mol Life Sci. 1999;1:96–107. doi: 10.1007/s000180050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones-Villeneuve E M, Rudnicki M A, Harris J F, McBurney M W. Mol Cell Biol. 1983;12:2271–2279. doi: 10.1128/mcb.3.12.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sommer L, Shaw N, Rao M, Anderson D J. Neuron. 1995;15:1245–1258. doi: 10.1016/0896-6273(95)90005-5. [DOI] [PubMed] [Google Scholar]
- 10.Stemple D L, Anderson D J. Cell. 1992;71:973–985. doi: 10.1016/0092-8674(92)90393-q. [DOI] [PubMed] [Google Scholar]
- 11.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgenstern J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cepko C L, Ryder E, Austin C, Golden J, Fields-Berry S, Lin J. Methods. 1998;14:393–406. doi: 10.1006/meth.1998.0594. [DOI] [PubMed] [Google Scholar]
- 14.Brand A H, Manoukian A S, Perrimon N. Methods in Cell Biology (Academic, San Diego) 1994;44:635–654. doi: 10.1016/s0091-679x(08)60936-x. [DOI] [PubMed] [Google Scholar]
- 15.Lawrence P A, Johnston P, Morata G. Methods of Marking Cells, In: Roberts D B, editor. Drosophila: A Practical Approach. Oxford: IRL; 1986. pp. 229–242. [Google Scholar]
- 16.Hartenstein V, Posakony J W. Development (Cambridge, UK) 1989;107:389–405. doi: 10.1242/dev.107.2.389. [DOI] [PubMed] [Google Scholar]
- 17.Nye J S, Kopan R, Axel R. Development (Cambridge, UK) 1994;120:2421–2430. doi: 10.1242/dev.120.9.2421. [DOI] [PubMed] [Google Scholar]
- 18.McBurney M W. Int J Dev Biol. 1993;37:135–140. [PubMed] [Google Scholar]
- 19.Verdi J M, Anderson D J. Neuron. 1994;13:1359–1372. doi: 10.1016/0896-6273(94)90421-9. [DOI] [PubMed] [Google Scholar]
- 20.Raff M C. Nature (London) 1992;356:397–400. doi: 10.1038/356397a0. [DOI] [PubMed] [Google Scholar]
- 21.Weinmaster G. Mol Cell Neurosci. 1997;9:91–102. doi: 10.1006/mcne.1997.0612. [DOI] [PubMed] [Google Scholar]
- 22.Carlesso N, Aster J C, Sklar J, Scadden D T. Blood. 1999;93:838–848. [PubMed] [Google Scholar]
- 23.Lewis J. Semin Cell Dev Biol. 1998;9:583–589. doi: 10.1006/scdb.1998.0266. [DOI] [PubMed] [Google Scholar]