Coat color variations have long been invaluable for analyzing modes of genetic inheritance in mammals. In the mouse, for instance, Mendelian inheritance was first demonstrated with the albino mutation (Cuénot, 1902), and non-Mendelian inheritance was shown with the coat color allele Agouti-viable yellow (Avy; Morgan et al., 1999; Wolff, 1978). Now, Minoo Rassoulzadegan and collaborators up the ante with their report (Rassoulzadegan et al., 2006) that a targeted mutation of the melanocyte gene Kit can modify a wild type Kit allele in such a way that the wild type copy henceforth produces a Kit mutant-like coat color phenotype. If confirmed, these findings would add to the increasing evidence that genes control the fitness of offspring in more ways than simply by passing on DNA copies of themselves.
Trans-chromosomal modifications that lead to heritable changes in a phenotype but are not accompanied by DNA sequence changes are called ‘paramutations’ or ‘paramutation-like’ phenomena (Brink, 1956, reviewed in Chandler and Stam, 2004). To understand these phenomena, we have to distinguish between the ‘paramutagenic’ locus (the one capable of inducing a paramutation), the ‘paramutable’ locus (the one that can be paramutated), and the ‘paramutated’ locus (the paramutable locus after its modification). Two models that are not mutually exclusive have been proposed to explain paramutations at the molecular level. In one, the pairing model, a region of one chromosome with altered chromatin structure interacts with a homologous region of another and forces upon it a similar modification, perhaps by transfer of altered chromatin complexes from one sequence to the other. In a second model, the trans-RNA model, either long RNAs interfere with the function of a paramutable locus, or small interfering RNAs (siRNA) lead to degradation of RNAs transcribed from the paramutable locus, with or without associated chromatin changes (for a review of these models, see Chandler and Stam, 2004). Note that the interfering RNAs could originate from either the paramutagenic or the paramutable locus. Both the pairing and the trans-RNA model accommodate the fact that the paramutated locus becomes itself paramutagenic because in principle, altered chromatin structures can be transferred perpetually and the paramutated locus can provide interfering RNAs. Hence, the paramutated phenotype is transmitted in a non-Mendelian fashion.
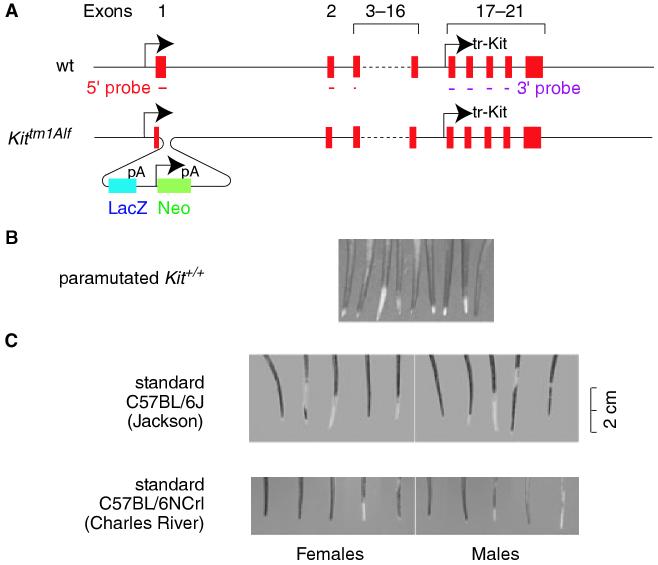
In Rassoulzadegan et al.’s case, the paramutagenic locus is the widely used targeted allele Kittm1Alf (formerly known as KitW-LacZ, Bernex et al., 1996), and the paramutable locus is a wild type Kit+ allele that encodes a receptor tyrosine kinase. In Kittm1Alf, a bacterial LacZ coding sequence is fused in frame with the first six codons of the KIT signal sequence, followed by a polyA signal and then, in the same transcriptional direction, a neomycin resistance cassette with its own promoter and polyA signal (see Figure 1A). This targeted allele produces nuclear βGAL in Kit-expressing cells but no functional KIT protein. Because KIT is needed for hematopoiesis, Kittm1Alf homozygotes die shortly after birth. Heterozygotes, however, survive, but because their reduced levels of KIT impair melanocyte development, they display a white belly spot of variable size, white feet, and a white tail tip usually several centimeters long.
Figure 1.
Schematic representation of the wild type Kit gene and its targeted allele, Kittm1Alf, and tail phenotypes of paramutated and standard C57BL/6 mice. (A) Gene structure of wild type (top) and targeted (bottom) allele schematically representing the 21 Kit exons as well as the in-frame insertion of the LacZ-neo cassette in exon 1. The insertion removes codons 7-22 of exon 1 and 200 bp of intron 1. The remainder of the Kit gene remains intact. Also shown are the 5′ cDNA probe (covering exon 1, 2 and the 5′ end of exon 3) and the 3′ cDNA probe (covering exons 17, 18, 19, and part of 20). (B) White tail tips from genotypically wild type, paramutated offspring (published as part of supplementary Figure 1 in Rassoulzadegan et al., 2006, with permission from the authors). (C) White tail tips in standard 6 week-old C57BL/6J mice (Jackson, stock no. 000664) and C57BL/6NCrl (Charles River), purchased between August and September 2006.
Rassoulzadegan et al. observed that heterozygous × heterozygous or heterozygous × wild type crosses produced genotypically wild type (Kit+/+) offspring approximately at the expected frequencies, but that most of these showed ‘the white patches characteristic of the parental heterozygotes’. This surely is an exaggerated statement as there were no white feet or white belly spots. In fact, all that was seen were small white tail tips (see Figure 1B, reproduced with permission from the authors). According to an animal breeding facility (Jackson Laboratories, Bar Harbor, ME, USA), however, white tail tips are common among normal laboratory mice, including standard C57BL/6J mice (Jackson stock no. 000664). Indeed, in at least six of 10 such C57BL/6J mice recently shipped to our laboratory from Jackson, there were sizable white tail tips and patches (Figure 1C). Similar observations were made with mice of a different stock (C57BL/6NCrl) purchased from another breeding facility, Charles River (Lyon, France). One can safely assume, though, that none of the ancestors of these mice ever carried the Kittm1Alf allele.
Should we, therefore, dismiss Rassoulzadegan et al.’s study in its entirety? Perhaps. But the idea of a paramutation capable of influencing tail pigmentation remains intriguing and should be discussed, for several reasons. First, different stocks of wild type mice (including different C57BL/6 stocks) need not be identical phenotypically, and the authors assert that their stocks (129/Sv, C57BL/6 and C57BL/6 × DBA, precise origins not provided) are not prone to white tail tips. Second, and importantly, if inbred mice, contrary to expectation, do not all look alike (see Figure 1C), there must be a cause. This cause could be environmental, for instance nutritional during development, but one should not a priori exclude genomic explanations. Hence, studying the mechanisms of the observed tail phenotype among Kittm1Alf descendants could theoretically shed light on the generation of white tail tips in standard C57BL/6J mice. Third, while epigenetic DNA or chromatin changes leading to the observed tail phenotype cannot be excluded, Rassoulzadegan et al. show that Kittm1Alf/+ heterozygotes produce abnormal Kit RNAs, hinting at a mechanism involving RNA intermediates. As outlined below, this finding may be compared to other phenomena where abnormal RNAs play a role and may lead to deeper insights into the heritability of coat color variations.
Under normal conditions, full-length Kit mRNAs are expressed in various tissues besides melanocytes, including the brain and the testis. In the mouse testis, Kit mRNAs can be found during gametogenesis from premeiotic, diploid germ cells up to, and including, the pairing stage of meiosis, and in fact, progression through meiosis depends on KIT protein, at least in in vitro assays (Vincent et al., 1998). Northern blots using a 5′ Kit probe that covers part of exon 1, all of exon 2, and a few bases of exon 3 (see Figure 1A) showed that brains of Kittm1Alf heterozygotes expressed about half as much full-length Kit mRNA as wild type, consistent with the fact that each cell carried only one intact copy of Kit. In whole testis, however, these levels were not decreased, and in fact, full-length Kit mRNAs accumulated to high levels in post-meiotic, haploid germ cells (spermatids) where they are not usually found. Surprisingly, the same probe also detected non-polyadenylated, short Kit RNAs in heterozygous brain and whole testis (though their presence in spermatids was not addressed). There was also a striking amplification of another short RNA (this time shown for spermatids) that was detectable with a 3′ probe. This RNA corresponds to a truncated transcript, called tr-Kit, that had previously been identified in normal spermatids, is initiated from a promoter in intron 16, and spans exons 17-21 (Albanesi et al., 1996, see Figure 1A). The authors argue that the 5′ short transcripts must come from the wild type Kit gene because the targeted allele is disrupted in the 5′ region and the transcripts did not hybridize with a LacZ probe (shown only for brain). A formal proof for the wild type origin of these transcripts, however, is missing as the 5′ probe fully covers exon 2, which is not deleted in the targeted gene (see Figure 1A). The same applies for the 3′ short transcripts because their genomic sequence also remains intact in the targeted allele.
The results showed, then, that spermatids from heterozygous animals expressed greater-than-normal amounts of both full-length and tr-Kit RNAs. Based on non-specific RNA staining techniques for light and electron microscopy and fluorescence-activated cell sorter analysis, it further appeared that the elevated RNAs were carried over into mature sperm, but it is not clear which specific RNAs gave rise to these elevated signals and why the product of a single gene should increase total RNA in sperm to a level detectable by these techniques. Be that as it may, a similar increase in fluorescence was observed in sperm cells derived from paramutated Kit+/+ progeny. This non-specific staining result, however, was the only evidence for elevated RNA levels in paramutated sperm, and, for that matter, in any paramutated tissue.
Nevertheless, if we accept that the white tail tips in paramutated mice are somehow linked to abnormalities in the expression of the wild type Kit gene, then we can principally think of two scenarios by which the tail phenotype may be generated. Both depend on partial silencing of the Kit gene in melanocytes or their precursors, indirectly inferred from the ca. 40% reduction in full-length Kit mRNA observed in paramutated Kit+/+ brain. In one scenario, regional chromatin changes in the paramutagenic allele might be transferred to the wild type allele, and this might lead to increased accumulation of short transcripts either from the targeted or the wild type allele, or both. In this scenario, chromatin changes are causal, and RNAs incidental. In the second scenario, abnormal RNAs, however produced, would accumulate and cause silencing of the wild type allele. In this scenario, the RNAs are causal, and, once they are generated, the paramutagenic DNA is no longer required for their action.
The authors provide the following experiment to distinguish between these possibilities. They reasoned that if the short RNAs were able to do the job by themselves, then the simple injection of such RNAs into zygotes should generate paramutated pups. Hence, they prepared total RNA from heterozygote-derived brain or testis, and (in Supplemental material in Rassoulzadegan et al., 2006) also from paramutated Kit+/+ brains (although, strangely, not testis). They then microinjected this RNA into fertilized eggs in amounts equivalent to what a sperm might carry into an egg [10-20 femtograms as estimated for human sperm, (Krawetz, 2005)], representing about 0.004% of total oocyte RNA), and let the eggs develop to term. Indeed, about half of the born pups had white tail tips. Injection of Kit micro-RNAs, thought to lead to comparable, short interfering Kit RNAs, gave similar results. Alas, white tail tips, although at much lower frequency, were also obtained when normal wild type brain or control LacZ RNAs were injected. Upon breeding to produce an F1 generation, however, these latter mice did not transmit the tail phenotype while the former ones did. Thus, the authors concluded that aberrant Kit RNAs alone can induce partial trans-silencing of a wild type Kit allele. One could imagine, then, that during gametogenesis in the paramutated mice, the synthesis of aberrant RNAs may get a boost, and the process would continue through the next and subsequent generations of mice until it would become weaker and eventually fade away.
So much for the reported results. Many issues would still need to be resolved to make the story convincing, however. Which RNA species in sperm, for instance, are responsible for the paramutated phenotype? Are they the elevated full-length or tr-Kit RNAs carried over from spermatids, or the short 5′ RNAs, which were documented, however, only for whole testis and not spermatids or sperm? At which stage in spermatogenesis do the abnormal RNAs start to accumulate, before, during, or after meiosis? In heterozygotes, is the only source of the abnormal RNAs the wild type allele? Why does the remaining intact Kit gene in heterozygous brain provide as much full-length Kit mRNA as the two copies of Kit in a paramutated Kit+/+ brain together, as Rassoulzadegan et al. show? Should the heterozygous brain not have at least as much interfering RNA per gene copy as the paramutated one? Are only Kit alleles that produce abnormal RNAs paramutagenic? What exactly are the Kit sequence requirements for paramutagenicity? Why are paramutations not more commonly observed with the thousands of transgene insertions that have been produced over the years, some of which certainly interrupting meiotically important genes? But, above all, why should the proposed mechanism exist in the first place?
In considering this last question, it may be helpful to contrast the Kit paramutation with a phenomenon called ‘meiotic silencing of unpaired DNA’ (MSUD) that was originally described in Neurospora (Shiu et al., 2001). Neurospora is a fungus that spends a large part of its life as a haploid but under certain environmental conditions can go through a diploid state from which haploid, melanized spores are produced through typical meiotic divisions. Mating between one haploid parent carrying a wild-type allele of a meiotically expressed gene (called Asm-1+, for ascus-maturation, ascus being the structure that gives rise to eight spores) and another carrying a deletion in the same gene (called Asm-1Δ) lead to diploids that eventually produce spores that are nearly all unpigmented and non-viable. One or even two copies of a wild type Asm-1+ gene placed in a different chromosomal location cannot correct the mutant phenotype, but the cross of two deletion mutants, each of them complemented with the rescue Asm-1+ gene at identical genomic positions, yields viable black spores. In fact, rescue still occurs when one of the two rescue copies is rendered non-functional by a frameshift mutation (Aramayo et al., 1996; Shiu et al., 2001). These observations led to the concept that the generation of viable spores depends on efficient meiotic pairing of all copies of the gene while mispairing of any one of them leads to silencing of the mispaired gene and, in trans, of all of its other copies. More recently, it was found that MSUD depends on an RNA-dependent RNA polymerase that likely leads to the generation of inhibitory dsRNA from which siRNA may be generated (Shiu et al., 2001).
It is thought that MSUD has evolved to silence transposons (rogue self-replicating DNA elements), which are usually meiotically unpaired and particularly prone to become active and spread during meiosis when they have an opportunity to invade a fresh genome. That a defense mechanism against transposons may also kill off all spores in situations where just half of them have a deleterious genomic deletion (as in the Asm-1 case) may be one ‘unintended consequence’ of an otherwise beneficial strategy. It is likely that similar defense mechanisms against transposons exist in other organisms as well, including in mammals. In fact, it has been speculated that MSUD serves as a general mechanism for sensing and silencing unpaired DNA and might even be involved in the elimination of ‘non-conforming’ gametes before they finish meiosis (Shiu et al., 2001).
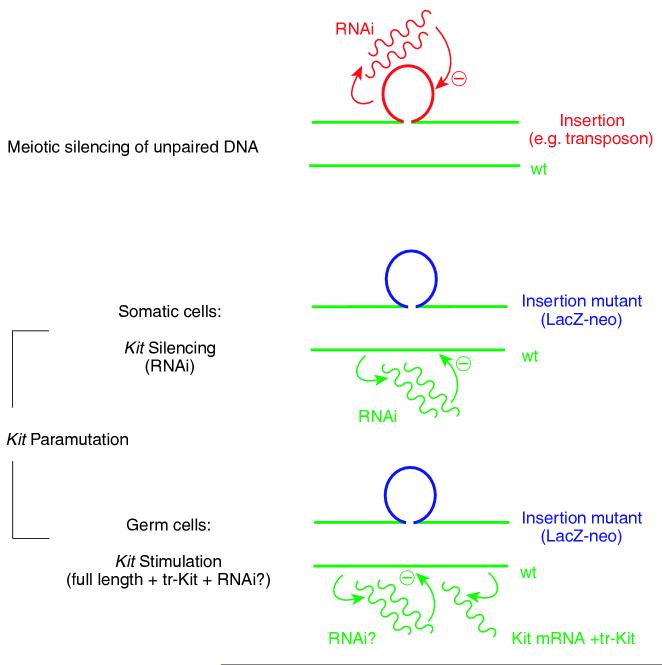
The Kit paramutation and MSUD resemble each other in several aspects. Both phenomena are epigenetic. Both involve genes that are expressed during, and important for, gametogenesis, both are seemingly induced during meiosis and initiated when there is extra DNA unable to pair, and both involve RNA intermediates. In other aspects, however, the two phenomena differ. MSUD, although critically depending on transchromosomal communication, does not involve the transfer of a silenced state from one chromosome to the next. A meiotically silenced transposon might lose its silenced state through mitoses, and only if it were to meet up again meiotically with an unpaired stretch of DNA would silencing resume. In contrast, paramutations, however induced, do not depend on mispairing for further transmission. MSUD and the Kit paramutation also differ in another mechanistic detail. In MSUD, it is the extra DNA, which leads to its own silencing, while in Kit, it is the deleted DNA (in this case the wild type DNA which relative to the insertion contains a ‘deletion’), which is proposed to be silenced (Figure 2). But does this make evolutionary sense? Can we explain why there should be a mechanism that silences a wild type Kit gene sitting opposite an insertion? Are we not accustomed to think that insertions are the bad boys, and not the pristine unmodified genes?
Figure 2.
Comparison of the mechanisms underlying meiotic silencing of unpaired DNA and the Kit paramutation. In meiotic silencing, the unpaired (extra) DNA (for instance, a transposon insertion, or the looped-out DNA opposite a genomic deletion) leads to excess RNA that serves as interfering RNA (RNAi) to silence the extra DNA. The establishment of the Kit paramutation, as proposed by Rassoulzadegan et al., is different. Here, the targeted Kit allele that contains the LacZ-neo insertion is thought to remain untouched and normally active, but it would induce the DNA that does not loop out (the wild type DNA) to produce abnormal short RNAs that then serve as a source for interfering RNAs (RNAi). In somatic cells (including melanocytes), the presence of these RNAs would lead to (partial) silencing of the wild type Kit gene. In germ cells, the situation is more complex as there is also an increase of the wild type, full-length transcript, along with increased levels of a truncated Kit RNA (tr-Kit, seen in spermatids). To explain the heritability of the paramutated tail phenotype, it is thought that gametes (sperm or egg) would carry abnormal RNAs over into the next generation, thereby leading to a continued reduction of full-length transcripts from wild type Kit alleles in somatic tissues. Whether a Kit paramutation that works in the proposed way really exists, however, still awaits confirmation (see text).
A solution to this paradox may perhaps come from a shift of the emphasis away from the presumed reduced activity of the Kit gene during melanogenesis to its documented increased activity during gametogenesis. Because Kit is important for meiosis (Vincent et al., 1998), we can view the increased accumulation of full-length Kit mRNAs around a meiotic stage of gametogenesis as the organism’s (over) compensatory response to the deleterious gene dosage effect of an insertion into one of its two copies. What happens after fertilization and during the build up of the somatic parts of the body may then be a different story altogether, potentially caused by different mechanisms and subject to different selection criteria. If so, we have to ask the following questions: during gametogenesis in a paramutated animal, is there again a transcriptional stimulation of full-length mRNA from the paramutated gene, followed by silencing in somatic tissues of the offspring? In other words, are stimulation and silencing coupled, or are they now, in the paramutated generation, uncoupled, and somatic silencing is all that is left? In either case, is the perpetuation of the paramutated state evolutionarily selected, or is it also an ‘unintended consequence’ of a strategy selected for a different purpose?
Finally, if we can come up with an evolutionary reason for MSUD, and another one for, let us call it, ‘meiotic stimulation opposite an insertion’ (MSOI), could the two mechanisms co-exist, each affecting the corresponding stretch of mismatched DNA at the same time? There is no obvious reason that would speak against this possibility, but the situation has to be considered separately for insertions and deletions. As mentioned, the wild type Kit gene sitting opposite the insertion in Kittm1Alf is stimulated, MSOI-style, but the inserted Kit-LacZ gene is not totally silenced, MSUD-style, although partial silencing cannot be excluded in the absence of a proper comparison. In similar ways, it is conceivable that in Neurospora, meiotically expressed, intact genes opposite MSUD-silenced insertions are actually stimulated. With respect to deletions, we can generally assume that MSUD is active on the opposite, intact chromosome, but the presence of MSOI on the chromosome that contains the deletion might depend on the extent of this deletion, as the absence of functional regulatory regions may preclude a transcriptional stimulation of the gene portions that remain intact.
One hundred and one years after the first demonstration of Mendelian inheritance in the mouse (Cuénot, 1902), the editor of Pigment Cell Research, in a comment on Bennett and Lamoreux’ review of coat color loci of mice (Bennett and Lamoreux, 2003), wrote that the ‘usefulness (of the collection of coat color genes) seems destined to expand even further in the future’. Indeed, the tail’s tale of Rassoulzadegan et al., and, by extension, that of standard laboratory mice, shows that coat color variations still hold secrets which, once uncovered, might impact how we think about the generation and heredity of a phenotype, whether it is one of health or one of disease.
Acknowledgements
I would like to thank Drs Matzinger, Metzenberg, Panthier, Larue, Pavan, Steingrímsson, Dubois-Dalcq, Bharti and Nodari for many helpful comments. This work was supported by the Intramural Research Program of the NIH, NINDS.
References
- Albanesi C, Geremia R, Giorgio M, Dolci S, Sette C, Rossi P. A cell- and developmental stage-specific promoter drives the expression of a truncated c-kit protein during mouse spermatid elongation. Development. 1996;122:1291–1302. doi: 10.1242/dev.122.4.1291. [DOI] [PubMed] [Google Scholar]
- Aramayo R, Peleg Y, Addison R, Metzenberg R. Asm-1+, a Neurospora crassa gene related to transcriptional regulators of fungal development. Genetics. 1996;144:991–1003. doi: 10.1093/genetics/144.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DC, Lamoreux ML. The color loci of mice - a genetic century. Pigment Cell Res. 2003;16:333–344. doi: 10.1034/j.1600-0749.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- Bernex F, De Sepulveda P, Kress C, Elbaz C, Delouis C, Panthier JJ. Spatial and temporal patterns of c-kit-expressing cells in WlacZ/+ and WlacZ/WlacZ mouse embryos. Development. 1996;122:3023–3033. doi: 10.1242/dev.122.10.3023. [DOI] [PubMed] [Google Scholar]
- Brink RAA. A genetic change associated with the R locus in maize which is directed and potentially reversible. Genetics. 1956;41:872–889. doi: 10.1093/genetics/41.6.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler VL, Stam M. Chromatin conversations: mechanisms and implications of paramutation. Nat. Rev. 2004;5:532–544. doi: 10.1038/nrg1378. [DOI] [PubMed] [Google Scholar]
- Cuénot L. La loi de Mendel et l′hérédité de la pigmentation chez les souris. C. R. Acad. Sci. Gen. 1902;134:779–781. [Google Scholar]
- Krawetz SA. Paternal contribution: new insights and future challenges. Nat. Rev. 2005;6:633–642. doi: 10.1038/nrg1654. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- Shiu PK, Raju NB, Zickler D, Metzenberg RL. Meiotic silencing by unpaired DNA. Cell. 2001;107:905–916. doi: 10.1016/s0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- Vincent S, Segretain D, Nishikawa S, Nishikawa SI, Sage J, Cuzin F, Rassoulzadegan M. Stage-specific expression of the Kit receptor and its ligand (KL) during male gameto-genesis in the mouse: a Kit-KL interaction critical for meiosis. Development. 1998;125:4585–4593. doi: 10.1242/dev.125.22.4585. [DOI] [PubMed] [Google Scholar]
- Wolff GL. Influence of maternal phenotype on metabolic differentiation of agouti locus mutants in the mouse. Genetics. 1978;88:529–539. doi: 10.1093/genetics/88.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]