Abstract
We have previously reported that the cabbage butterfly, Pieris rapae, contains a 98-kDa protein, named pierisin, that induces apoptosis in a variety of human cancer cell lines. In the present study, sequencing and cloning of a cDNA encoding pierisin was accomplished. PCR-direct sequencing showed that the gene encodes an 850-amino acid protein with a calculated molecular weight of 98,081. An intact clone at the amino acid level encompassing the entire coding region was obtained by recombination of two independent clones, and the molecular mass of its in vitro expressed protein was about 100 kDa on SDS/PAGE, the same as that of purified native pierisin. The expressed protein induced apoptosis in human gastric carcinoma TMK-1 and cervical carcinoma HeLa cells, like the native protein, indicating functional activity. The deduced amino acid sequence of pierisin showed 32% homology with a 100-kDa mosquitocidal toxin from Bacillus sphaericus SSII-1. In addition, pierisin showed regional sequence similarities with ADP-ribosylating toxins, such as the A subunit of cholera toxin. A glutamic acid residue at the putative NAD-binding site, conserved in all ADP-ribosylating toxins, was also found in pierisin. Substitution of another amino acid for glutamic acid 165 resulted in a great decrease in cytotoxicity and induction of apoptosis. Moreover, inhibitors of ADP-ribosylating enzymes reduced pierisin-induced apoptosis. These results suggest that the apoptosis-inducing protein pierisin might possess ADP-ribosylation activity that leads to apoptosis of the cells.
There are tremendous numbers of insect species throughout the world. The self-defense systems in insects are different from those of mammals (1–3). Thus, they could be good sources of new biologically active substances, including anticancer agents. However, reports describing anticancer agents from insect sources are very limited (4, 5).
Recently, our search for cytotoxic substances in various kinds of butterflies demonstrated the presence of agents active against human carcinoma cells in three butterflies, Pieris rapae, Pieris napi, and Pieris brassicae (6, 7), P. rapae and P. brassicae being cabbage butterflies commonly distributed over the globe. Among the three developmental stages, larvae, pupae, and adults of P. rapae, the pupae showed the strongest cytotoxic activity (6). The active protein principle in pupae, named pierisin, has been purified by various column chromatographies, and its molecular mass was estimated to be 98 kDa by mass spectrometry (8).
Pierisin exhibited potent cytotoxic effects against various kinds of human cancer cell lines with large differences in sensitivity, with IC50 values ranging from 0.043 to 150 ng/ml (8, 9). Among the cancer cells tested, the cervical carcinoma cell line, HeLa, was the most sensitive, followed by the gastric carcinoma cell line, TMK-1. Pierisin was found to clearly induce apoptotic cell death in most cancer cell lines, displaying characteristic morphological features, DNA fragmentation, and cleavage of poly(ADP-ribose) polymerase. The pathway was apparently distinct from anti-Fas, TNF-α, or p53-mediated apoptosis (9). Thus, pierisin derived from an insect source could offer informative data for development of novel anticancer agents.
To elucidate the biological function of pierisin, a cDNA encoding pierisin was cloned in the present study. Here, we report that the pierisin gene encodes an 850-amino acid protein whose deduced amino acid sequence shares regional sequence similarity with ADP-ribosylating toxins. Disruption of a putative NAD-binding site by site-directed mutagenesis and treatment with inhibitors of ADP-ribosyltransferases led to loss of apoptosis induction potential, suggesting that pierisin has ADP-ribosylation activity.
MATERIALS AND METHODS
Preparation of RNA for Cloning of Pierisin Gene.
Total RNA from different developmental stages of P. rapae was prepared by using a guanidinium thiocyanate method (10). RNA preparations were then subjected to in vitro translation using rabbit reticulocyte lysate (Ambion, Austin, TX) without radiolabeled amino acids. The cytotoxic activity of each translated sample was examined in human gastric carcinoma TMK-1 cells (11).
Internal Amino Acid Sequence Analysis of Pierisin.
Pierisin was purified from the pupae of P. rapae as described previously (8), reduced with DTT, modified in its cysteine residues with 4-vinylpyridine (12), and digested with lysylendopeptidase (13). The digested pierisin peptides were separated by reverse-phase-HPLC with a μ-Bondasphere column (Waters). Amino acid sequences of the isolated peptides were determined by using a PSQ-1 protein sequencer (Shimadzu).
PCR Amplification and Sequencing of cDNA Encoding Pierisin.
All PCR amplifications were performed with a DNA thermal cycler model 9600 (Perkin–Elmer Applied Biosystems, Foster City, CA). cDNA of mRNA from P. rapae was synthesized from the total RNA preparation (2 μg) from fifth-instar larvae by using a CapFinder PCR cDNA Library Construction Kit (CLONTECH). The synthesized cDNA was then subjected to PCR amplification (14) of pierisin-specific sequences by LA Taq DNA polymerase (Takara Shuzo, Otsu, Japan), using 1 μM sense and antisense degenerate primers corresponding to pierisin peptides, EMILRGYTNSGSNNQ and ESGNTWYLK, or a sense degenerate primer and 0.1 μM (dT)25 oligomer. For discrimination of PCR products, the cDNA fragments were cloned into pBluescript KS+ or pT7Blue and partially sequenced by using an Applied Biosystems PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit and an ABI377 PRISM DNA Sequencer (PE Applied Biosystems). Thus, internal and 3′-terminal cDNA fragments were obtained. To obtain a 5′-terminal cDNA fragment, PCR was performed with 0.3 μM each of the 5′ anchor primer (5′-TACGGCTGCGAGAAGACGACAGAA) and an antisense primer (5′-TGCCAGTATTGGTTATTGCT); the sequence was chosen from the internal cDNA fragment. The resulting 5′ cDNA fragment was cloned into pBluescript KS+ and partially sequenced. Full-length pierisin cDNA was obtained by PCR amplification using a 5′ primer (5′-GGACGGAATCAATCATTT), which is the sequence adjacent to the 5′ anchor primer, and a 3′ primer (5′-TTTAATAGTAAATAAAGTTTATTGACAT), which is the sequence adjacent to the poly(A) stretch. The amplified 4-kb fragment was subjected to PCR direct sequencing to determine the entire cDNA sequence.
Construction of a Clone of the Coding Region of the Pierisin Gene.
The coding region of the pierisin gene, with 293 bp of 3′-flanking region, was amplified and an EcoRI restriction site introduced at each end, using the primer pair 5′-CCGAATTCTCATGGCTGACCGTCAACCTTACATGACTAATGGTATTCAG and 5′-GGGAATTCGCCCTGTTTCACAATGTATG. The 4-kb PCR fragment described above was used as a template DNA. The amplified fragment was digested with EcoRI and inserted into λgt10. The recombinant clones were expressed in vitro, by the method described below, and sequenced to identify mutations at the nucleotide level. To construct an intact subclone at the amino acid level, two independent clones were recombined and subcloned into the EcoRI site of pMAL-p2 (New England Biolabs) in the opposite direction to the tac promoter. The subclone was verified by sequencing the insert.
In Vitro Expression of Pierisin cDNA.
A T7 promoter sequence attached a 5′ primer at the 5′ end of the coding region (5′-ACTTCGAAATTAATACGACTCACTATAGGGCGAATTGCCACCATGGCTGACCGTCAACCTTAC) and a 3′ primer corresponding to nucleotides 2,934–2,915 (shown in Fig. 1) were used for PCR to introduce a T7 promoter sequence upstream of the pierisin coding sequence. The amplified fragment of pierisin cDNA with the T7 promoter sequence was transcribed by using MEGAscript (Ambion) and then translated with rabbit reticulocyte lysate (Ambion) supplemented with 50 μM methionine and 5 μCi of [35S]methionine in 25 μl of translation reaction medium to produce nonradioactive and radioactive proteins, respectively. The labeled translation product was electrophoresed on an SDS/polyacrylamide gel and visualized by autoradiography.
Figure 1.
Nucleotide and deduced amino acid sequences of the cDNA for pierisin by PCR-direct sequencing. All determined internal amino acid sequences of purified native pierisin from pupae of P. rapae, including those used to make degenerate primers, existed in the deduced amino acid sequence of the gene, as underlined. Termination codons found at the 5′ end of the coding region are indicated by asterisks (∗). A possible polyadenylation signal, found near the 3′ end, is indicated by “#.”
Analysis of Cytotoxic and Apoptosis-Inducing Activities of in Vitro Expressed Protein.
Translated protein solution was added to human gastric carcinoma TMK-1 or human cervical carcinoma HeLa cells in culture (RIKEN cell bank, Tsukuba, Japan) in RPMI-1640 medium supplemented with 10% FBS (GIBCO/BRL). The culture was incubated at 37°C in 5% CO2 in air and subjected to morphological analysis and DNA fragmentation analysis, as described previously (8), to detect cytotoxicity and apoptosis-inducing activity of expressed protein from the isolated cDNA.
Site-Directed Mutagenesis.
A DNA fragment containing a mutation at Glu-165 was amplified from the subclone by using modified 5′ primers, 5′-CAGATGCATCGCCTTGGCCTAATCAAATGGATGTTGC for the mutation to aspartic acid and 5′-CAGATGCATCGCCTTGGCCTAATCAAATGCAGGTTGC for the mutation to glutamine, and a 3′ primer corresponding to the nucleotides 902–881 in Fig. 1. To confirm the appropriate recombination, the 5′ primers also contained another, silent, sequence alteration that disrupted a NcoI site. The amplified fragment was digested with NsiI and BsrGI, and recombined into NsiI–BsrGI digest of the subclone. After verification of the nucleotide sequence of the replaced NsiI–BsrGI region, the mutated subclones were used for protein expression in vitro.
Effects of Inhibitors of ADP-Ribosyltransferases on the Cytotoxicity of Pierisin.
3-Aminobenzamide and benzamide were obtained from Tokyo Kasei Kogyo (Tokyo). 3-Aminobenzoic acid and 5-amino-2,3-dihydro-1,4-phthalazinedione were from Wako Junyaku Kogyo (Osaka) and Sigma, respectively. TMK-1 or HeLa cells were incubated with pierisin and various doses of these chemicals for 72 h at 37°C, and then subjected to 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay. The dose of pierisin was 1 ng/ml for TMK-1 cells and 0.1 ng/ml for HeLa cells.
RESULTS
Sequence of the Pierisin Gene. Total RNA was prepared from various developmental stages of P. rapae, larvae (third and fifth instar), pupae (pre, early, and late stages), and adults, and the levels of pierisin mRNA were estimated by in vitro translation. The samples from fifth-instar larvae contained the highest level, followed by those from prestage pupae. The observed mRNA expression preceded the protein accumulation, which was greatest in prestage pupae, then in early-stage pupae. Pierisin mRNA was not detectable in the adults, and the protein content was approximately 100-fold less than that in the prestage pupae.
Using RNA from fifth-instar larvae and internal amino acid sequences of digested peptides, we obtained a cDNA encoding the entire pierisin. The nucleotide sequence of the isolated cDNA is shown in Fig. 1. The full-length cDNA consisted of 3,973 bp accompanied by a 5′-anchor sequence and a polyadenylate stretch. The longest ORF began with the first ATG at nucleotide 92 and ended with a TAA termination codon at nucleotide position 2,642. The predicted initiation codon for the ORF was preceded by termination codons in all three reading frames. There was a polyadenylation signal (AATAAA) at nucleotide 3,954, close to the poly(A) sequence. The putative protein encoded by the ORF comprised 850 amino acids with a calculated molecular weight of 98,081. Moreover, the 13 peptide sequences of digests of purified pierisin from pupae of P. rapae were found to be identical to parts of the deduced amino acid sequence from the ORF (Fig. 1). From these results, the ORF in the isolated cDNA was concluded to be the coding region of pierisin.
Cloning and Expression of the Pierisin Gene.
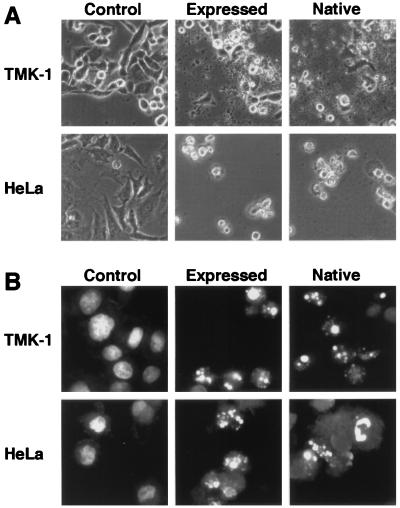
To obtain a clone, the PCR-amplified pierisin gene was inserted into λgt10, and 24 clones were isolated. Five active clones, judged by in vitro translation, were selected and sequenced. Finally, recombination of two clones was carried out to construct a subclone without any nonsilent sequence alterations. After confirmation of the subclone by sequencing, the gene was expressed and the resultant protein was analyzed. Its molecular mass was about 100 kDa on SDS/PAGE (Fig. 2), the same as that of purified native pierisin. The expressed protein induced morphological change in TMK-1 and HeLa cells, as observed by phase-contrast microscopy (Fig. 3A). Fluorescent micrography revealed chromatin condensation and nuclear fragmentation in the cells (Fig. 3B). Oligonucleosomal DNA fragmentation also occurred (Fig. 4). Thus, the expressed protein apparently induced apoptosis, as with the native pierisin. The results indicate that functionally active protein was expressed in vitro from the isolated cDNA.
Figure 2.
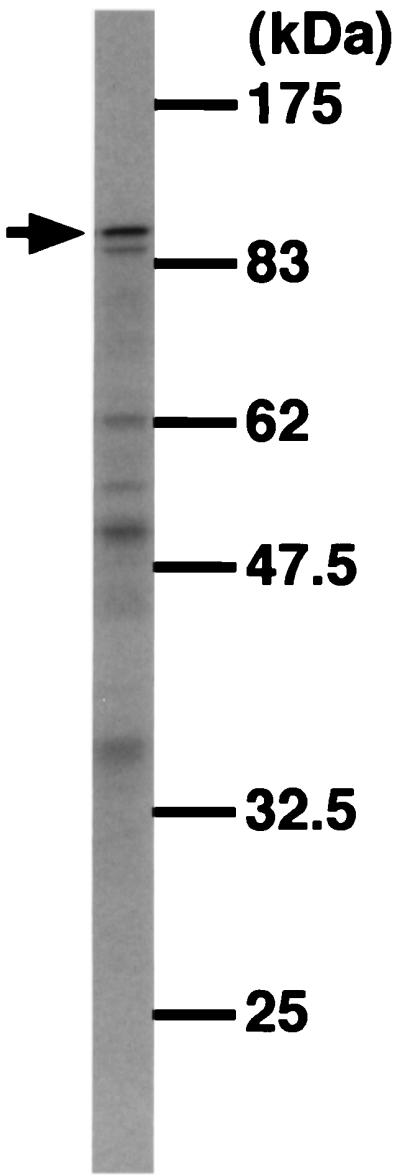
SDS/PAGE of [35S]methionine-labeled proteins, expressed from the subclone in the pMAL-p2 plasmid. The arrow indicates the molecular size of the native pierisin.
Figure 3.
Morphological analysis of TMK-1 and HeLa cells treated with the in vitro-expressed protein from the subclone. The cells were treated for 18 h with 3% (TMK-1) or 0.6% (HeLa) of rabbit reticulocyte lysate alone (control), or containing synthesized protein (expressed), or 2.5 ng/ml (TMK-1) or 0.5 ng/ml (HeLa) purified native pierisin (native). (A) Phase-contrast micrographs. (B) Fluorescent micrographs.
Figure 4.
DNA fragmentation in TMK-1 and HeLa cells treated with the in vitro-expressed protein. The cells were treated with synthesized protein (expressed) or purified native pierisin (native) under the same conditions described in Fig. 3, and their DNA was extracted and run on agarose gels.
Possible Involvement of ADP-Ribosylation in the Activity of Pierisin. Gene and protein sequences homologous with pierisin were searched for by using the National Center for Biotechnology Information blast program. As a result, a 100-kDa mosquitocidal toxin (Mtx) of Bacillus sphaericus SSII-1 exhibited sequence similarity throughout the coding region (Fig. 5A) (15). The overall sequence homology was 32% at the amino acid level. In addition, pierisin exhibited regional sequence similarities with some ADP-ribosylating toxins, including cholera toxin and Escherichia coli heat-labile toxin (Fig. 5B) (16–20). A glutamic acid residue, known to be an NAD-binding site and conserved among all ADP-ribosylating toxins (21–23), corresponded to Glu-165 in pierisin (Fig. 5B).
Figure 5.
Sequence similarity of pierisin with Mtx and ADP-ribosylating toxins. The conserved glutamic acid residue is boxed. (A) Alignment of the deduced amino acid sequence of pierisin with that of Mtx (15). P, pierisin; M, Mtx. Identical amino acids are marked by asterisks (∗). (B) Homologous region of pierisin with ADP-ribosylating toxins. CT-A, the A subunit of cholera toxin (16); LTH-A and LTP-A, the A subunit of E. coli heat-labile toxin pathogenic for humans and pigs, respectively (17–19); PT-S1, the S1 subunit of pertussis toxin (20). Completely conserved amino acids are marked by asterisks (∗).
To examine whether ADP-ribosylation is involved in the cytotoxicity and apoptosis induction activity of pierisin to TMK-1 and HeLa cells, the glutamic acid residue at position 165 was replaced with aspartic acid or glutamine by site-directed mutagenesis, and the mutated clones were transcribed and translated in vitro. The efficiencies of protein expression from the nonmutated control subclone and the subclones with an aspartic acid (E165D) or glutamine (E165Q) substitution were about the same, as judged by SDS/PAGE after translation with [35S]methionine. However, the cytotoxic activity of the expressed proteins against HeLa cells was greatly decreased or even entirely lost (Fig. 6). The IC50 value of in vitro-expressed protein from mutated clone E165D was more than 10-fold higher than that from original clone, and the value of E165Q clone was the same as that of control lysate without expressed protein. As in the case of HeLa cells, similar cytotoxic activity of the expressed proteins was observed in TMK-1 cells (data not shown). Apoptosis-inducing activities of the mutated proteins, determined by observation of nuclear condensations and fragmentations of HeLa and TMK-1 cells, were also decreased concordantly (data not shown). Thus, the amino acid substitutions in pierisin strongly affected the cytotoxicity and apoptosis induction activity against human cells, suggesting possible functional involvement of ADP-ribosylation activity.
Figure 6.
Dose-dependent cytotoxic effects of nonmutated and mutated proteins on HeLa cells. Cells were incubated with various concentrations of rabbit reticulocyte lysate: control lysate (○); lysate containing nonmutated protein (●), E165D protein (■), or E165Q protein (▴). After incubation for 72 h at 37°C, MTT assay was carried out.
Consistent with the above results, the cytotoxic activity of pierisin against TMK-1 and HeLa cells was reduced to around half by treatment with 3-aminobenzamide (5 mM), benzamide (5 mM), or 5-amino-2,3-dihydro-1,4-phthalazinedione (0.1 mM), inhibitors of ADP-ribosyltransferases (24–26), but not with 3-aminobenzoic acid, a noninhibitory analog (25, 27).
DISCUSSION
In the present study, we determined the nucleotide sequence of a cDNA for the pierisin gene by PCR amplification and direct sequencing. Insertion of the PCR-amplified pierisin gene into λgt10 allowed a clone to be obtained whose in vitro-expressed protein was functionally active, like the purified native pierisin. Site-directed mutagenesis at a putative NAD-binding site, glutamic acid 165, further demonstrated that ADP-ribosylation activity could be essential for the cytotoxicity and apoptosis-inducing activity of pierisin against cancer cells.
Because pierisin might be toxic to E. coli, clones encoding intact pierisin gene might be lethal. Therefore, preferential selection of defective clones (28) could be a critical issue in this study. Thus, we first determined the sequence of the gene without cloning, and then carefully selected several clones by using a nonexpression vector, λgt10, and an in vitro expression system. Based on the sequence from PCR direct sequencing, an intact subclone was constructed by recombination of two clones. The four sequence alterations in the subclone—A124T, A1513G, A2155G, and T2778C—did not change the amino acid sequence of pierisin. As we expected, the E. coli strains transformed by the cloned pierisin gene in plasmid pET-32a(+) or pBAD/His were easily lysed when the gene was expressed (data not shown). This suggests that pierisin induces death of E. coli, even at low concentrations. From these observations, our experimental approach used in the present study can be considered appropriate, although further study is required to efficiently obtain large amounts of the recombinant protein.
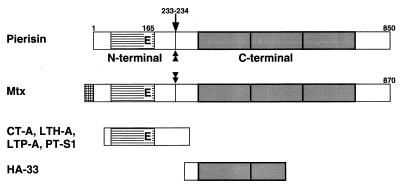
The present study showed pierisin to share sequence similarity with a 100-kDa Mtx produced in Bacillus sphaericus SSII-1. The mtx gene is reported to encode an 870-amino acid protein containing a possible signal sequence (15), but sequence alignment suggested that pierisin does not contain this signal sequence (Fig. 5A). On the other hand, treatment with a variety of proteases revealed that pierisin possesses a protease-sensitive region, shown in Fig. 7, like Mtx (29). This fact provides additional evidence of similarity in the structure of both proteins. However, Mtx is reported to be nontoxic to HeLa cells (30), while pierisin is markedly toxic (9).
Figure 7.
Structural similarity of pierisin with other proteins. Cross-hatched bar, possible signal sequence observed in Mtx; striped bars, homologous region among pierisin, Mtx, and ADP-ribosylating toxins; shaded bars, repeat unit of 150 amino acids. The conserved glutamic acid residue is indicated as “E”. The trypsin cleavage site of pierisin, indicated by an arrow, was determined by using a Shimadzu PSQ-1 protein sequencer after separation and blotting of the digested C-terminal 70-kDa fragment on an SDS/polyacrylamide gel. Pronase E and proteinase K also produced the 70-kDa fragment, suggesting that there is a protease-sensitive region close to the trypsin cleavage site. Mtx possesses a protease-sensitive region (29). The protease-sensitive regions in pierisin and Mtx are indicated by double-headed arrows. HA-33, hemagglutinin component of botulinus toxin; other protein abbreviations as in Fig. 5.
Part of the N-terminal region of both pierisin and Mtx shares regional sequence similarity with several ADP-ribosylating toxins (Fig. 5B). ADP-ribosyltransferase is an enzyme that catalyzes transfer of ADP-ribosyl moieties of NAD to cellular proteins, and various bacterial exotoxins and eukaryotic endogenous enzymes possess this activity (31, 32). Although the amino acid sequence homology among various ADP-ribosyltransferases is poor, there is a highly conserved glutamic acid residue, like the Glu-112 of the cholera toxin A subunit and the Glu-148 of diphtheria toxin (16, 23, 33). A number of mutagenesis assays have indicated that such glutamic acid residues are essential for exertion of toxicity and ADP-ribosylating activity (34–38). X-ray crystallographic and photocrosslinking analyses suggest that the glutamic acid residue serves as an NAD-binding site (21, 39, 40). The presence in pierisin of Glu-165 suggests that this protein also has an ADP-ribosylation activity, as supported by site-directed mutagenesis. In addition, inhibitors of mono- and poly-ADP-ribosylating enzymes suppressed pierisin-induced apoptosis. These results thus strongly suggest that pierisin possesses enzymatic activity of an ADP-ribosyltransferase that is responsible for the apoptosis of human cancer cells. However, it should be noted that several toxins other than ADP-ribosylating enzymes also contain an essential glutamic acid residue. Site-directed mutagenesis has shown that ricin-A and Shiga toxin require glutamic acid residues to exert toxic effects and enzymatic activity (41–43). Thus, it is important to clarify the target protein of ADP-ribosylation by pierisin and its function toward the cells.
The C-terminal regions of both pierisin and Mtx share sequence similarity with HA-33 (or HA-34, -35), a subcomponent of hemagglutinin of botulinus toxin (44–47), which enhances toxicity by protecting components against proteolysis under acidic conditions, such as in gastric juice (48, 49). Interestingly, repeat units of about 150 amino acids were observed for pierisin and the HA-33 moiety of botulinus toxin with significant sequence similarity. The C-terminal region of both pierisin and Mtx consists of three repeat units, accompanied by short sequences on both sides (Fig. 7). HA-33 also consists of a single unit with another incomplete unit. Thus, there may be an 150-amino acid ancestral protein or domain possibly possessing functions such as binding ability to the cells.
The gut of larvae of P. rapae contains a variety of microorganisms, such as bacteria. In addition, P. rapae is known to be a host of a parasitic hymenopteran, Apanteles glomeratus. However, the pierisin gene cloned here must be derived from the butterfly’s own genome, because the nucleotide sequence of the cloned gene showed a typical eukaryotic structure with a polyadenylation signal, and the materials used for preparation of RNA and protein were free from the parasitic wasp. The mRNA expression level of pierisin was highest in fifth-instar larvae, and the protein was accumulated just prior to pupation. Pierisin protein constitutes about 0.4% of the total protein in the pupae (8). Taken together, these results suggest that pierisin may play some important role in insect development, such as pupation. At the same time, its strong cytotoxicity might be effective as a protective agent, in line with the compatible dual function of Sarcophaga lectin (50). Northern blot analysis demonstrated that expression of pierisin-like gene at the mRNA level is restricted to Pieris butterflies. Larvae of both P. rapae and P. brassicae expressed the gene, observed as 4 kb and smaller bands, whereas it was lacking in Hebomoia glaucippe (belonging to the same family, Pieridae) and other families of butterfly and moth, Papilio xuthus and Bombyx mori. This is consistent with the results of screening of a variety of butterflies and moths for cytotoxicity (6). Understanding the roles of pierisin should be instructive with respect to our knowledge of insect biology and also helpful in elucidation of mechanisms of apoptosis of human cancer cells mediated by pierisin.
Acknowledgments
This study was supported by a Grant-in Aid for Cancer Research from the Ministry of Health and Welfare, Japan, and a grant from the Nishi Cancer Research Fund.
ABBREVIATION
- Mtx
Mosquitocidal toxin
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB030305).
References
- 1.Boman H G, Hultmark D. Annu Rev Microbiol. 1987;41:103–126. doi: 10.1146/annurev.mi.41.100187.000535. [DOI] [PubMed] [Google Scholar]
- 2.Hultmark D. Trends Genet. 1993;9:178–183. doi: 10.1016/0168-9525(93)90165-e. [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann J A. Curr Opin Immunol. 1995;7:4–10. doi: 10.1016/0952-7915(95)80022-0. [DOI] [PubMed] [Google Scholar]
- 4.Itoh A, Iizuka K, Natori S. Jpn J Cancer Res (GANN) 1985;76:1027–1033. [PubMed] [Google Scholar]
- 5.Moore A J, Devine D A, Bibby M C. Pept Res. 1994;7:265–269. [PubMed] [Google Scholar]
- 6.Koyama K, Wakabayashi K, Masutani M, Koiwai K, Watanabe M, Yamazaki S, Kono T, Miki K, Sugimura T. Jpn J Cancer Res (GANN) 1996;87:1259–1262. doi: 10.1111/j.1349-7006.1996.tb03141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kono T, Watanabe M, Koyama K, Sugimura T, Wakabayashi K. Proc Jpn Acad. 1997;73B:192–194. [Google Scholar]
- 8.Watanabe M, Kono T, Koyama K, Sugimura T, Wakabayashi K. Jpn J Cancer Res (GANN) 1998;89:556–561. doi: 10.1111/j.1349-7006.1998.tb03297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kono T, Watanabe M, Koyama K, Kishimoto T, Fukushima S, Sugimura T, Wakabayashi K. Cancer Lett. 1999;137:75–81. doi: 10.1016/s0304-3835(98)00346-2. [DOI] [PubMed] [Google Scholar]
- 10.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 11.Ochiai A, Yasui W, Tahara E. Jpn J Cancer Res (GANN) 1985;76:1064–1071. [PubMed] [Google Scholar]
- 12.Friedman M, Krull L H, Cavins J F. J Biol Chem. 1970;245:3868–3871. [PubMed] [Google Scholar]
- 13.Masaki T, Fujihashi T, Nakamura K, Soejima M. Biochim Biophys Acta. 1981;660:51–55. doi: 10.1016/0005-2744(81)90107-8. [DOI] [PubMed] [Google Scholar]
- 14.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 15.Thanabalu T, Hindley J, Jackson-Yap J, Berry C. J Bacteriol. 1991;173:2776–2785. doi: 10.1128/jb.173.9.2776-2785.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mekalanos J J, Swartz D J, Pearson G D, Harford N, Groyne F, de Wilde M. Nature (London) 1983;306:551–557. doi: 10.1038/306551a0. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto T, Tamura T, Yokota T. J Biol Chem. 1984;259:5037–5044. [PubMed] [Google Scholar]
- 18.Yamamoto T, Gojobori T, Yokota T. J Bacteriol. 1987;169:1352–1357. doi: 10.1128/jb.169.3.1352-1357.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spicer E K, Noble J A. J Biol Chem. 1982;257:5716–5721. [PubMed] [Google Scholar]
- 20.Nicosia A, Perugini M, Franzini C, Casagli M C, Borri M G, Antoni G, Almoni M, Neri P, Ratti G, Rappuoli R. Proc Natl Acad Sci USA. 1986;83:4631–4635. doi: 10.1073/pnas.83.13.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroll S F, Collier R J. Proc Natl Acad Sci USA. 1984;81:3307–3311. doi: 10.1073/pnas.81.11.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cockle S A. FEBS Lett. 1989;249:329–332. doi: 10.1016/0014-5793(89)80652-0. [DOI] [PubMed] [Google Scholar]
- 23.Bazan J F, Koch-Nolte F. Adv Exp Med Biol. 1997;419:99–107. doi: 10.1007/978-1-4419-8632-0_12. [DOI] [PubMed] [Google Scholar]
- 24.Purnell M R, Whish W J. Biochem J. 1980;185:775–777. doi: 10.1042/bj1850775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rankin P W, Jacobson E L, Benjamin R C, Moss J, Jacobson M K. J Biol Chem. 1989;264:4312–4317. [PubMed] [Google Scholar]
- 26.Banasik M, Komura H, Shimoyama M, Ueda K. J Biol Chem. 1992;267:1569–1575. [PubMed] [Google Scholar]
- 27.Griffin R J, Curtin N J, Newell D R, Golding B T, Durkacz B W, Calvert A H. Biochimie. 1995;77:408–422. doi: 10.1016/0300-9084(96)88154-5. [DOI] [PubMed] [Google Scholar]
- 28.Forns X, Bukh J, Purcell R H, Emerson S U. Proc Natl Acad Sci USA. 1997;94:13909–13914. doi: 10.1073/pnas.94.25.13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thanabalu T, Hindley J, Berry C. J Bacteriol. 1992;174:5051–5056. doi: 10.1128/jb.174.15.5051-5056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thanabalu T, Berry C, Hindley J. J Bacteriol. 1993;175:2314–2320. doi: 10.1128/jb.175.8.2314-2320.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ueda K, Hayaishi O. Annu Rev Biochem. 1985;54:73–100. doi: 10.1146/annurev.bi.54.070185.000445. [DOI] [PubMed] [Google Scholar]
- 32.Krueger K M, Barbieri J T. Clin Microbiol Rev. 1995;8:34–47. doi: 10.1128/cmr.8.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greenfield L, Bjorn M J, Horn G, Fong D, Buck G A, Collier R J, Kaplan D A. Proc Natl Acad Sci USA. 1983;80:6853–6857. doi: 10.1073/pnas.80.22.6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Douglas C M, Collier R J. J Bacteriol. 1987;169:4967–4971. doi: 10.1128/jb.169.11.4967-4971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsuji T, Inoue T, Miyama A, Okamoto K, Honda T, Miwatani T. J Biol Chem. 1990;265:22520–22525. [PubMed] [Google Scholar]
- 36.Wilson B A, Reich K A, Weinstein B R, Collier R J. Biochemistry. 1990;29:8643–8651. doi: 10.1021/bi00489a021. [DOI] [PubMed] [Google Scholar]
- 37.Cieplak W, Jr, Mead D J, Messer R J, Grant C C. J Biol Chem. 1995;270:30545–30550. doi: 10.1074/jbc.270.51.30545. [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto S, Takeda Y, Yamamoto M, Kurazono H, Imaoka K, Yamamoto M, Fujihashi K, Noda M, Kiyono H, McGhee J R. J Exp Med. 1997;185:1203–1210. doi: 10.1084/jem.185.7.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sixma T K, Kalk K H, van Zanten B A, Dauter Z, Kingma J, Witholt B, Hol W G. J Mol Biol. 1993;230:890–918. doi: 10.1006/jmbi.1993.1209. [DOI] [PubMed] [Google Scholar]
- 40.Bell C E, Eisenberg D. Biochemistry. 1996;35:1137–1149. doi: 10.1021/bi9520848. [DOI] [PubMed] [Google Scholar]
- 41.Frankel A, Welsh P, Richardson J, Robertus J D. Mol Cell Biol. 1990;10:6257–6263. doi: 10.1128/mcb.10.12.6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ready M P, Kim Y, Robertus J D. Proteins. 1991;10:270–278. doi: 10.1002/prot.340100311. [DOI] [PubMed] [Google Scholar]
- 43.Hovde C J, Calderwood S B, Mekalanos J J, Collier R J. Proc Natl Acad Sci USA. 1988;85:2568–2572. doi: 10.1073/pnas.85.8.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsuzuki K, Kimura K, Fujii N, Yokosawa N, Indoh T, Murakami T, Oguma K. Infect Immun. 1990;58:3173–3177. doi: 10.1128/iai.58.10.3173-3177.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henderson I, Whelan S M, Davis T O, Minton N P. FEMS Microbiol Lett. 1996;140:151–158. doi: 10.1016/0378-1097(96)00172-3. [DOI] [PubMed] [Google Scholar]
- 46.Inoue K, Fujinaga Y, Watanabe T, Ohyama T, Takeshi K, Moriishi K, Nakajima H, Inoue K, Oguma K. Infect Immun. 1996;64:1589–1594. doi: 10.1128/iai.64.5.1589-1594.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.East A K, Stacey J M, Collins M D. Syst Appl Microbiol. 1994;17:306–312. [Google Scholar]
- 48.Fu F N, Sharma S K, Singh B R. J Protein Chem. 1998;17:53–60. doi: 10.1023/a:1022590514771. [DOI] [PubMed] [Google Scholar]
- 49.Sharma S K, Singh B R. J Nat Toxins. 1998;7:239–253. [PubMed] [Google Scholar]
- 50.Natori S. Res Immunol. 1990;141:938–939. doi: 10.1016/0923-2494(90)90198-8. [DOI] [PubMed] [Google Scholar]