Abstract
Supramolecular self-assembly is an integral step in the formation of many biological structures. Here we report a DNA pentaplex that derives from a metal-assisted, hydrogen bond-mediated self-assembly process. In particular, cesium ions are found to induce pentameric assembly of DNA bearing the nonstandard nucleobase iso-guanine. The pentaplex was designed by using a simple algorithm to predict nucleobase structural requirements within a quintet motif. The design principles are general and should extend to complexes beyond pentaplex. Structures exhibiting molecularities of five or more were previously accessible to peptides, but not nucleic acids.
In abiologic systems, metal-mediated self-assembly has yielded arrays (1), nanometer-sized dendrimers (2), and other systems (3), notably, double- and triple-helical metal complexes that bear a formal resemblance to DNA (4). Recognition in the latter is driven by ligand constraints and metal coordination geometry. Recognition within DNA double and triple helices, in contrast, depends primarily on hydrogen bond complementarity. Quadruple helices of DNA based on the G-quartet motif are unusual, however, in that they rely on both hydrogen bonding and metal ion coordination (5, 6). Here we demonstrate the ability to expand DNA molecularity beyond quadruplexes by engineering nucleobases to fit dimensions required of higher-order motifs. Specifically, we design and characterize a DNA pentaplex.
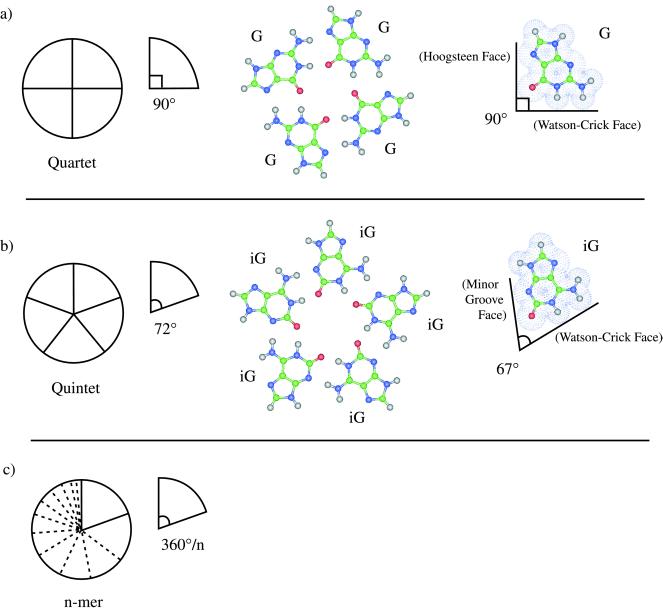
The iG-motif (iG refers to 2′-deoxy-iso-guanosine) may be predicted to yield the first nonquartet structure within the series shown in Fig. 1, which also illustrates working design principles to expand nucleobase motifs. Each of the generalized motifs in this figure is modular and comprised of components for which an ideal sector angle may be attributed, e.g., 360/4° and 360/5° for quartets and quintets, respectively. In the case of G-quartets, vectors along hydrogen bond donor/acceptor groups contributing to recognition result in the 90° ideal sector angle (Fig. 1a). In contrast to G, the 67° sector angle of iG approaches the quintet optimum (Fig. 1b). iG itself occurs naturally (7) and may be considered an elementary nucleobase in the sense that it derives from hydrogen cyanide, a prime prebiotic reagent (8). Thus, iG may have contributed to early biopolymer evolution (9) and has been used to encode genetic information during ribosomal translation in vitro (10). We (11) and others (12, 13) have established that higher-order self-pairing of iG depends on metal ions, similar to G-quartets (for iG self-pairing in another sugar system, see ref. 32). Our present findings show that monovalent cations can influence decisively the degree of strand association in a nonstandard DNA. Whereas tetraplexes are observed as the major outcome of incubating iG-rich DNA in the presence of potassium ions, pentaplex association dominates under the influence of cesium ions. As a consequence, the iG-motif fulfills geometric predictions.
Figure 1.
Working design for nucleobase motifs leading to expanded DNA molecularities. (a, from left to right) Schematic representation of a circularly modular quartet motif with ideal sector angle of 90°, ab initio optimized geometry of a G-quartet motif, and actual 90° sector angle for G defined by the intersection of vectors along the van der Waals donor-donor and acceptor-acceptor atom surfaces using the geometry from its optimized quartet. (b, from left to right) Schematic representation of a circularly modular quintet motif with ideal sector angle of 72°, ab initio optimized geometry of an iG-quintet motif, and actual 67° sector angle for iG defined by the intersection of vectors along the van der Waals donor-donor and acceptor-acceptor atom surfaces using the geometry from its optimized quintet. (c) Schematic representation of a circularly modular n-mer motif with ideal sector angle of 360°/n.
MATERIALS AND METHODS
Oligonucleotides.
Synthesis of iG phosphoramidite and oligonucleotides were performed as described (14).
PAGE Assays.
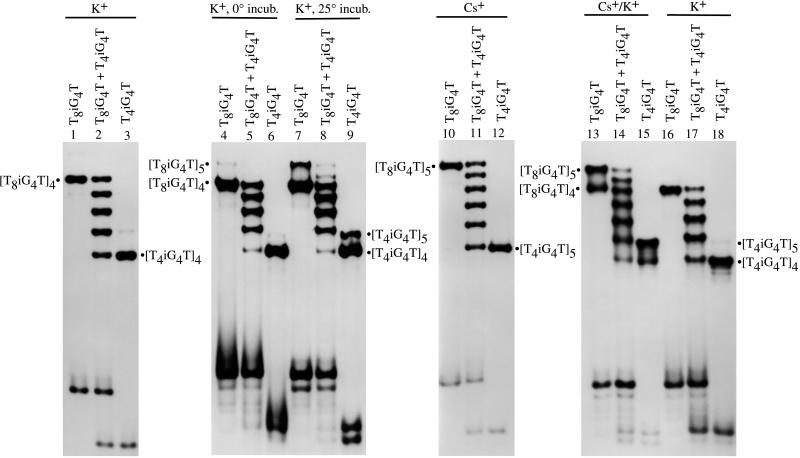
Oligonucleotide (400 pmol) was combined with 1 μl of 10× kinase buffer (700 mM Tris⋅HCl, pH 7.6/100 mM MgCl2/50 mM DTT), 1 μl of γ-32P-ATP (2 μCi), 6 μl of H2O, and 1 μl of T4 polynucleotide kinase (3 units, United States Biochemical). The sample was incubated for 30 min at 37°C, at which time 1 μl of 660 μM ATP was added, followed by incubation for an additional 30 min at 37°C. Kinased oligonucleotides were purified by denaturing 20% PAGE and desalted. Purified oligonucleotides (5 × 105 cpm) were combined with 10 μl of 500 mM CsCl or KCl, and the sample was heated at 90°C for 10 min, centrifuged, and then incubated at 4°C or 25°C for 48 hr, as indicated in Fig. 2. After incubation, samples were electrophoresed on a native 20% polyacrylamide gel kept at 4°C. Running buffer contained either 50 mM CsCl, or 50 mM KCl, along with 0.6× TBE (Tris/boric acid/EDTA, pH 8.6). After electrophoresis, gels were exposed to film for 0.5–2 hr. Ratios of 32P-T4iG4T to 32P-T8iG4T were determined for the set of five K+-complexes in lane 2 of Fig. 2 and the set of six Cs+-complexes shown in lane 11 of Fig. 2 by: (a) removing each individual complex from the above noted native gel, (b) placing individual complexes in individual lanes on a denaturing (7 M urea) 20% polyacrylamide gel, and (c) separation of the short and long iG-DNA strands by electrophoresis at 45°C in 1× TBE. PAGE-separated 32P-T4iG4T and 32P-T8iG4T were imaged with a PhosphorImager and quantitated by using imagequant software (Molecular Dynamics). The ratios are reported in Fig. 3.
Figure 2.
Autoradiograms of native 20% polyacrylamide gels demonstrating the number of electrophoretic bands that emanate from complexes formed by two different lengths of iG4-bearing DNA strands. Complexes were formed by incubating DNA strands separately or together in the presence of K+ and/or Cs+, as indicated at the top of each lane.
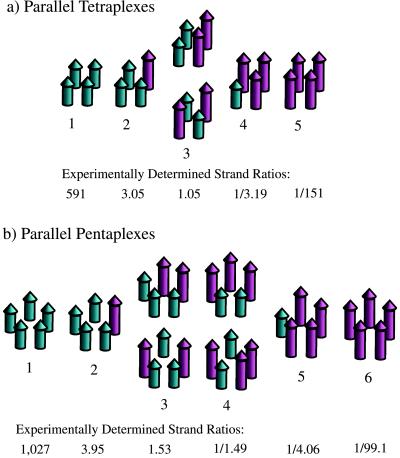
Figure 3.
Schematic representation of electrophoretically distinguishable tetraplexes and pentaplexes that may result when two different lengths of DNA strands are combined. (a) Five parallel-stranded tetraplexes that may be distinguished; (Lower) strand ratios that were experimentally determined. (b) Six parallel-stranded pentaplexes that may be distinguished; (Lower) experimentally determined strand ratios.
Photocrosslinking.
UV-crosslinking experiments were accomplished by irradiating with a low-pressure mercury lamp 10 μl of iG-DNA Cs+-complexes or K+-complexes in a solution containing 500 mM of the corresponding salt (complexes were preformed in CsCl or KCl solution by incubation at 4°C for 48 hr). The samples then were electrophoresed on a denaturing (7 M urea) 20% polyacrylamide gel at 45°C. After electrophoresis, gels were exposed to film for 12–24 hr. Additional details, including a discussion of “crossover” experiments, are noted in the legend to Fig. 4.
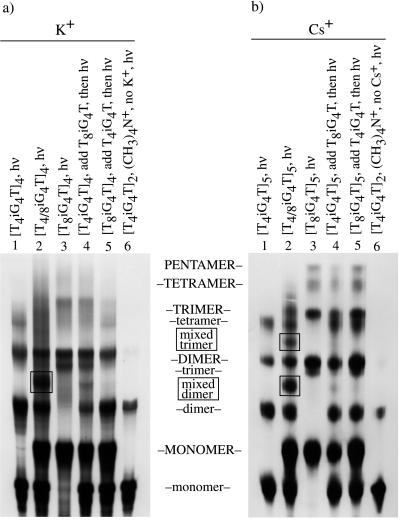
Figure 4.
Autoradiograms of denaturing polyacrylamide gels showing the result of UV photocrosslinking experiments with different lengths of iG4-bearing oligonucleotides in K+ or Cs+. (a) 32P-end-labeled oligonucleotides were incubated either alone, or combined together, in 0.5 M of KCl at 4°C for 2 days as indicated at the top of each lane. In “crossover” control lanes 4 and 5, a 32P-end-labeled single-strand “challenge oligomer” was added after the 2 days of incubation, just before irradiation, as indicated at the top of these lanes. Samples then were irradiated with UV light for 15 min at 4°C. Products of UV irradiation were electrophoresed on a 20% denaturing (7 M urea) polyacrylamide gel at 45°C with 1× TBE (Tris/boric acid/EDTA, pH 8.6) as running buffer. (b) The same procedure was followed as detailed for a except CsCl was used in place of KCl. Labels located in between the panels in all capital letters describe products of T8iG4T, labels in all lowercase letters refer to those of T4iG4T, and “mixed” refers to irradiation products having elements derived from both of these oligomers.
Computations.
Computations on the pentaplex of iG4 DNA strands were performed with Macromodel (Schrödinger, Portland, OR) by using the amber* force field. Partial atomic charges for iG reported by Leach and Kollman were used (15). Ab initio Hartree-Fock computations were performed on a Cray T90 at the San Diego Supercomputer Center using gaussian 94 (33) as described in earlier work (11, 14). Basis sets provided in gaussian 94 (3–21G) were supplemented with the 3–21G cesium basis set developed by Glendening and Feller (16).
RESULTS AND DISCUSSION
Electrophoretic Assay for Strand Association. Tetramolecular association of G-rich RNA (17) and dimerization of G-rich DNA hairpins (18) were demonstrated earlier by comparing electrophoretically the number of complexes that emanate from two physically different oligomers, taken individually and together. Thus, incubating a mixture of T8iG4T and T4iG4T in K+ at 0°C for 48 hr yields five bands upon native gel electrophoresis (Fig. 2, lane 2). This outcome is consistent with tetramolecular association of parallel aligned strands, as reported earlier (11), and is modeled in Fig. 3a (11, 17). Numbered complexes in the figure are distinguishable electrophoretically.
The above finding opposes the prediction made in Fig. 1b. However, it may be assumed that if pentaplexes could possibly form, they would do so at a rate slower than other known helix-forming reactions of lesser molecularity, in analogy with the rate of tetraplex formation in comparison to triplexes or duplexes (19, 20). It follows that if pentaplex and tetraplex formation were to compete under kinetically controlled conditions, tetraplexes alone might result. To test this possibility, the iG-bearing oligomers were incubated with potassium ions at higher temperature than before to minimize kinetic biases. As can be observed in Fig. 2, lanes 7–9, increasing the incubation temperature gives rise to appreciable quantities of slower moving bands.
Strand Association as a Function of Monovalent Ion Size. To investigate whether the minor bands with diminished mobility may be pentaplexes, a means was sought to further suppress the main tetraplex reaction manifold. Computations (see below) suggested that Cs+ matches the dimensions found in the intersticial cavity created by succesive iG-quintets. In practice, use of the larger metal ion resulted in complete supression of the tetraplex association pathway (Fig. 2, lanes 10–12). At the same time, Cs+ produced only the putative pentaplex bands. Perhaps the most interesting outcome with Cs+ is the formation of six distinct electrophoretic bands instead of five from the mixture of long and short iG-DNA strands (Fig. 2, lane 11 versus lane 2).
As modeled in Fig. 3b, six electrophoretically distinguishable complexes are consistent with either: (i) the assortment of two different DNA strands within a parallel stranded pentaplex, or (ii) the assortment of two different DNA strands within an antiparallel stranded tetraplex. The second model is anticipated to be highly disfavored because it requires iG to adopt both syn and anti conformations. Apart from the anticipated increase in conformational energy as a result of positioning the nucleobase syn rather than anti, a syn conformation of iG also would be expected to preclude hydrogen bonding by the minor groove face (O2 and N3 acceptor atoms) within the iG-motif because of their proximity to ribose and attendant steric shielding. The situation is different for G in the G-motif as its acceptor atoms occur on the Hoogsteen face and therefore are far removed from the ribosyl group.
Distinguishing Parallel-Stranded Pentaplexes from Antiparallel-Stranded Tetraplexes.
Despite the above arguments favoring the pentaplex model of Fig. 3b, we sought to conduct an experiment whose outcome would unambiguously define the strand stoichiometry of iG-DNA Cs+-complexes. To achieve this end, individual electrophoretic bands for each complex of mixed length strands were isolated for both the Cs+-series (Fig. 2, lane 11) and the K+-series (Fig. 2, lane 2). The amounts of long and short strands within each individual complex then were determined by separating the two different-length strands via denaturing (7 M urea) PAGE, followed by quantification of 32P for each strand. The strand ratios determined in these experiments are reported in Fig. 3 a and b Lower. Strand ratios observed for both metal ion series are in very good agreement with the ratios predicted by the corresponding association model, parallel-stranded tetraplex for K+ and parallel-stranded pentaplex for Cs+. Significantly, the experimentally determined strand ratios for the Cs+-series (Fig. 3b Lower) are inconsistent with the strand ratios that would result from an antiparallel-stranded tetraplex.
Direct Electrophoretic Comparison of Cs+ and K+ Complexes.
Direct comparison of the different metal ion complexes was achieved by incubating T8iG4T and T4iG4T in Cs+ and K+ separately, and then electrophoresing the complexes in a native gel with running buffer that contained K+ but not Cs+. The result is shown in Fig. 2, lanes 13–18. Lanes 13–15 contain DNA strands incubated in Cs+ but electrophoresed in K+ and clearly show both the K+ and Cs+ species of complex in lanes 13 and 15. The Cs+ complexes in these two lanes may be discerned as moving slower than the K+ complexes through comparison to the pure K+ complexes in lanes 16 and 18 of the same gel. These observations are consistent with a more stable Cs+-pentaplex than K+-tetraplex when coupled with the mirror experiment (results not shown), in which the K+ complex upon electrophoresis in a native gel containing Cs+ running buffer yields only the Cs+ complex. Molecularity aside, one factor clearly in favor of Cs+ complex stability is the less unfavorable dehydration energy for Cs+ over K+ (21).
Results with G-DNA.
In a set of experiments analogous to those described in Fig. 2, the behavior of G-bearing DNA strands T8G4T and T4G4T also has been investigated (data not shown). In contrast to iG, the G-oligomers give nearly identical results for both Cs+ and K+. This observation is in keeping with the earlier findings of Venczel and Sen (22).
Photocrosslinking Assay for Molecularity.
Photocrosslinking of iG-DNA in Cs+ also supports pentaplex formation. Experiments were conducted for both K+ and Cs+- driven assembly as illustrated in Fig. 4. Lanes 1–3 of Fig. 4 involve irradiation of the complexes shown at the top of each lane, followed by denaturing electrophoresis at 45°C. Discernible in both autoradiograms are monomer, dimer (one crosslink), trimer (two crosslinks), and tetramer (three crosslinks) for all complexes. In addition, pentamer (four crosslinks) is seen for T8iG4T in the presence of Cs+ (Fig. 4b, lane 3). Photocrosslinked pentamer is consistent with Cs+ driven assembly of T8iG4T into a pentaplex. The fact that pentamer is seen only with T8iG4T is furthermore consistent with crosslinking through thymidine residues, in that this sequence bears four additional such residues over T4iG4T, and therefore would possess an increased likelihood of reaction. Lanes 4–6 of Fig. 4 are control experiments. Lanes 4 and 5 are “crossover” experimental variations of lanes 1 and 3, respectively. The purpose of this particular experiment is to detect whether any appreciable strand exchange among complexes occurs under the conditions of the photocrosslinking experiment. In practice, preformed tetraplex or pentaplex of one strand type (long or short) is challenged to exchange during the photocrosslinking by addition of the other strand type (short or long) just before irradiation. If such exchange were to occur, crosslinking as an assay for molecularity would be invalidated. Lanes 2 of Fig. 4 define crossover bands, which appear boxed. In neither lanes 4 nor 5 does there appear appreciable crossover product. Finally, lanes 6 show that a monovalent cation alone is not sufficient to support formation of tetraplexes or pentaplexes, as replacement of the alkali metal cations with tetramethylammonium ion yields only dimer crosslinked products.
Pentaplex and Quintet Structure. Several lines of evidence support the iG-quintet motif structure as represented in Fig. 1b. Tetraplexes of iG-DNA do not depend on Hoogsteen pairing, as replacement of the Hoogsteen-face N7 by a C-H group does not impede their formation (13). Examination of iG-quartets by 1HNMR and UV spectroscopies confirm the occurrence of the N1-H tautomeric form (11, 12) and also the direct involvement of the N1-H in cation-dependent hydrogen bonding (12). Computations are consistent with the iG-quintet model, the results of which are displayed in Figs. 5 and 6.
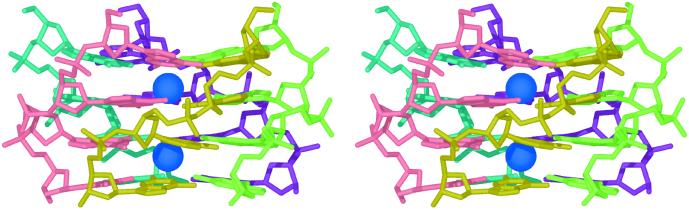
Figure 5.
Pentaplex of iG4 DNA strands incorporating two cesium ions. The structure shown results from unrestrained minimization of the complex by using the amber* forcefield implemented in Macromodel. Two cesium ions are incorporated into the model because a three-ion model with an ion between each layer proved unstable. The average intrastrand phosphate-phosphate distance is observed to be 6.8 Å.
Figure 6.
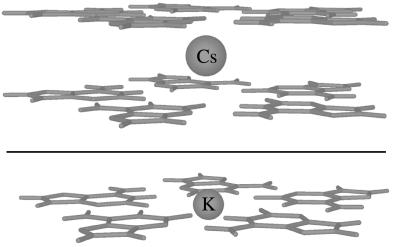
Hartree-Fock optimized nucleobase quintet-metal cation geometries. (a) iG-quintet/Cs+/iG-quintet 3–21 optimized geometry; the Cs+ is positioned between quintet layers. (b) iG-quintet/K+ 3–21G* optimized geometry; the K+ lies in the quintet plane.
Unrestrained force field calculations indicate that DNA readily accommodates a pentaplex structure (Fig. 5). Ab initio computation yields a stable (iG-quintet/Cs+/iG-quintet) sandwich complex (Fig. 6, Upper). Computations were performed by using structures with C5H symmetry imposed to maximize efficiency. Noteworthy features of the optimized geometry for the Cs+ sandwich are: (i) a viable quintet layer distance of 3.8 Å, a dimension that exceeds the layer distance for B-DNA base pairs by only 0.4 Å, and (ii) a Cs-O bond distance of 3.5 Å, a dimension that closely approximates the average 3.4 Å Cs-O distance predicted by crystallographic data (23) for deca-coordinate Cs+. An intersticial metal cation such as the model incorporates also fits with structural data for the positioning of potassium and sodium ions in G-tetraplexes (5, 24). An in-plane rather than interstitial model is proposed for the K+-pentaplex structure (Fig. 6, Lower). The in-plane model allows an appropriate metal-oxygen coordination distance to be maintained. The 2.8-Å K-O distance calculated for the in-plane model differs by 0.1 Å from the 2.9-Å average K-O distance for penta-coordinate K+ (23).
Conclusions.
DNA pentaplexes are expected to be morphologically similar to pentameric, coiled-coil alpha-helical peptides implicated as ion channels (25). Engineering of activities beyond the already diverse set known for DNA (26–28) and RNA (29–31) may be possible with these structures. It also may be possible to create supramolecular helices with orders beyond 5.
Acknowledgments
This work was supported by the Astrobiology Institute of the National Aeronautics and Space Administration.
ABBREVIATION
- iG
2′-deoxy-iso-guanosine
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Drain C M, Nifiatis F, Vasenko A, Batteas J D. Angew Chem Int Ed Engl. 1998;37:2344–2347. doi: 10.1002/(SICI)1521-3773(19980918)37:17<2344::AID-ANIE2344>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 2.Huck W T S, van Veggel F C J M, Reinhoudt D N. Angew Chem Int Ed Engl. 1996;35:1213–1215. [Google Scholar]
- 3.Linton B, Hamilton A D. Chem Rev. 1997;97:1669–1680. doi: 10.1021/cr960375w. [DOI] [PubMed] [Google Scholar]
- 4.Hasenknopf B, Lehn J M, Boumediene N, Leize E, Dorsselaer A V. Angew Chem Int Ed Engl. 1998;37:3265–3268. doi: 10.1002/(SICI)1521-3773(19981217)37:23<3265::AID-ANIE3265>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 5.Kang C H, Zhang X, Ratliff R, Moyzis R, Rich A. Nature (London) 1992;356:126–131. doi: 10.1038/356126a0. [DOI] [PubMed] [Google Scholar]
- 6.Smith F W, Feigon J. Nature (London) 1992;356:164–168. doi: 10.1038/356164a0. [DOI] [PubMed] [Google Scholar]
- 7.Cherbuliez E, Bernhard K. Helv Chim Acta. 1932;15:978. [Google Scholar]
- 8.Eschenmoser A, Loewenthal B. Chem Soc Rev. 1992;21:1–16. [Google Scholar]
- 9.Rich A. In: Horizons in Biochemistry. Kasha M, Pullman B, editors. New York: Academic; 1962. p. 103. [Google Scholar]
- 10.Bain J D, Switzer C, Chamberlin A R, Benner S A. Nature (London) 1992;356:537–539. doi: 10.1038/356537a0. [DOI] [PubMed] [Google Scholar]
- 11.Roberts C, Chaput J C, Switzer C. Chem Biol. 1997;4:899–908. doi: 10.1016/s1074-5521(97)90298-2. [DOI] [PubMed] [Google Scholar]
- 12.Tirumala S, Davis J T. J Am Chem Soc. 1997;119:2769–2776. [Google Scholar]
- 13.Seela F, Wei C F. Chem. Commun. 1997. 1869–1870. [Google Scholar]
- 14.Roberts C, Bandaru R, Switzer C. J Am Chem Soc. 1997;119:4640–4649. [Google Scholar]
- 15.Leach A R, Kollman P A. J Am Chem Soc. 1992;114:3675–3683. [Google Scholar]
- 16.Glendening E D, Feller D. J Phys Chem. 1995;99:3060–3067. [Google Scholar]
- 17.Kim J, Cheong C, Moore P B. Nature (London) 1991;351:331–332. doi: 10.1038/351331a0. [DOI] [PubMed] [Google Scholar]
- 18.Sundquist W I, Klug A. Nature (London) 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 19.Wyatt J R, Davis P W, Freier S M. Biochemistry. 1996;35:8002–8008. doi: 10.1021/bi960124h. [DOI] [PubMed] [Google Scholar]
- 20.Sarma M H, Luo J, Umemoto K, Yuan R-D, Sarma R H. J Biomol Struct Dyn. 1992;9:1131–1153. doi: 10.1080/07391102.1992.10507984. [DOI] [PubMed] [Google Scholar]
- 21.Hud N V, Smith F W, Anet F A L, Feigon J. Biochemistry. 1996;35:15383–15390. doi: 10.1021/bi9620565. [DOI] [PubMed] [Google Scholar]
- 22.Venczel E A, Sen D. Biochemistry. 1993;32:6220–6228. doi: 10.1021/bi00075a015. [DOI] [PubMed] [Google Scholar]
- 23.Shannon R D. Acta Crystallogr A. 1976;32:751. [Google Scholar]
- 24.Laughlan G, Murchie A I H, Norman D G, Moore M H, Moody P C E, Lilley D M J, Luisi B. Science. 1994;265:520–524. doi: 10.1126/science.8036494. [DOI] [PubMed] [Google Scholar]
- 25.Malashkevich V N, Kammerer R A, Efimov V P, Schulthess T, Engel J. Science. 1996;274:761–765. doi: 10.1126/science.274.5288.761. [DOI] [PubMed] [Google Scholar]
- 26.Cuenoud B, Szostak J W. Nature (London) 1995;375:611–614. doi: 10.1038/375611a0. [DOI] [PubMed] [Google Scholar]
- 27.Li Y F, Sen D. Nat Struct Biol. 1996;3:743–747. doi: 10.1038/nsb0996-743. [DOI] [PubMed] [Google Scholar]
- 28.Braun E, Eichen Y, Sivan U, Ben-Yoseph G. Nature (London) 1998;391:775–778. doi: 10.1038/35826. [DOI] [PubMed] [Google Scholar]
- 29.Lehman N, Joyce G F. Nature (London) 1993;361:182–185. doi: 10.1038/361182a0. [DOI] [PubMed] [Google Scholar]
- 30.Ekland E H, Bartel D P. Nature (London) 1996;382:373–376. doi: 10.1038/382373a0. [DOI] [PubMed] [Google Scholar]
- 31.Convery M A, Rowsell S, Stonehouse N J, Ellington A D, Hirao I, Murray J B, Peabody D S, Philips S E, Stockley P G. Nat Struct Biol. 1998;5:133–139. doi: 10.1038/nsb0298-133. [DOI] [PubMed] [Google Scholar]
- 32.Krishnamurthy R, Pitsch S, Minton M, Miculka C, Windhab N, Eschenmoser A. Angew Chem Int Ed. 1996;35:1537–1541. [Google Scholar]
- 33.Frisch M J, Trucks G W, Schlegel H B, Gill P M W, Johnson B G, Robb M A, Cheeseman J R, Keith T, Petersson G A, Montgomery J A, et al. gaussian 94, Revision B.1. Pittsburgh: Gaussian; 1995. [Google Scholar]