Abstract
We report studies of the contribution of DNA structure, holding the sequence constant, to the affinity of calicheamicin γ1I and its aryltetrasaccharide moiety for DNA. We used polynucleotide chains as models of known protein-binding sequences [the catabolite activator protein (CAP) consensus sequence, AP-1 and cAMP response element (CRE) sites] in their free and protein-bound forms. The proteins were selected to provide examples in which the minor-groove binding site for the carbohydrate is (CAP) or is not (GCN4) covered by the protein. Additionally, peptides related to the GCN4 and CREB families, which have different bending effects on their DNA-binding sites, were used. We observe that proteins of the CREB class, which induce a tendency to bend toward the minor groove at the center of the site, inhibit drug-cleavage sites located at the center of the free AP-1 or CRE DNA sites. In the case of GCN4, which does not induce DNA bending, there is no effect on calicheamicin cleavage of the CRE site, but we observe a GCN4-induced rearrangement of the cutting pattern in the AP-1 site. This effect may arise from either a subtle local conformational rearrangement not accompanied by bending or a localized reduction in DNA flexibility. Whereas GCN4 binding is not inhibited by the calicheamicin aryltetrasaccharide, binding of CAP to its DNA target is significantly inhibited, and calicheamicin cutting of DNA at the center of the CAP–DNA complex site is strongly reduced by protein binding. This result probably reflects steric inhibition of drug binding by the protein.
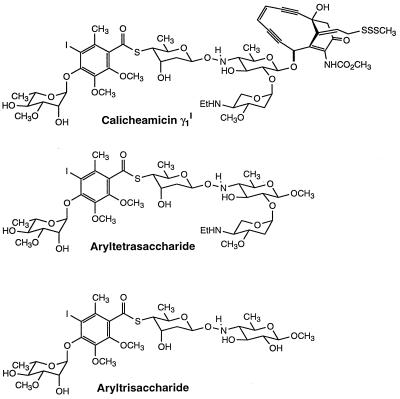
Among DNA-binding compounds, carbohydrates are a relatively newly investigated class (1). Many agents, including anticancer drugs, use sugar moieties to increase their affinity and selectivity for the nucleic acid. One of the most important representatives of this class is the enediyne calicheamicin γ1I (Fig. 1). It contains two “domains”: the enediyne portion, which, on reductive activation, is responsible for DNA cleavage, and the aryltetrasaccharide tail, which anchors the whole molecule to the DNA minor groove (2, 3). Although the reactive part of calicheamicin can play a role in the recognition process, previous studies reported that its carbohydrate tail is principally responsible for sequence specificity and tight DNA binding (4, 5). Moreover, it was recently reported that the aryltetrasaccharide moiety can efficiently inhibit the binding of transcription factors to a target DNA containing TCCT, generally considered to be the preferred canonical calicheamicin binding sequence. In this way, the drug can specifically interfere with DNA-related biological processes such as transcription (6, 7) and other biological events that depend on protein-DNA interactions.
Figure 1.
Molecular structures of calicheamicin γ1I and related carbohydrates.
It has been proposed that the specific calicheamicin γ1I–DNA-binding process depends not only on the base sequence of the double helix but also on its structure and flexibility (8, 9). To address this issue further, we have investigated the DNA structural requirements in the drug-recognition process by examining interference of the calicheamicin saccharide tail with DNA binding by two regulatory proteins, the catabolite activator protein (CAP) and GCN4. We also have examined the influence of protein binding on the cleavage preference of calicheamicin in the neighborhood of the protein-binding site.
CAP binds its target nucleic acid as an α-2 dimer (10). Each monomer consists of an N-terminal domain that binds cAMP (its allosteric effector) and a C-terminal region that binds DNA. The DNA-binding domain recognizes the major groove of two symmetrically related 5′ nucleotide sequences separated by 6 bp (5′-tgtgagttagctcact-3′). (Note that this consensus sequence lacks the tetrapyrimidine tract usually found at calicheamicin-cleavage sites.) Both monomers induce a 40° kink toward the protein, narrowing the major groove, at the dyad symmetric TG steps that are separated by 10 bp. This geometry results in an overall bending of ≈90° directed toward the minor groove in a coordinate frame at the center of the site (11, 12). When the protein is bound to DNA, even though it fits in the major groove, it covers the minor groove as well where it faces the protein (13, 14). Hence, the minor groove near the center of the site should no longer be accessible to the minor groove-binding aryltetrasaccharide.
GCN4, a transcriptional activator, is a modular protein that belongs to the basic leucine zipper family (15). The C-terminal domain is folded in a continuous α-helix, forming a leucine zipper where four heptad repeats are responsible for dimer formation in a parallel α-helical coiled coil. Rich in basic resides, the α-helical DNA-binding domain is found immediately adjacent and N-terminal to the dimerization domain (16, 17). The GCN4 natural consensus sequence is the pseudosymmetric AP-1 site (5′-ATGACTCAT-3′); the residues primarily responsible for the tight complex with the protein are the seven central base pairs, although more bases are involved in the process (18, 19). The protein interacts in the DNA major groove only and leaves the minor groove accessible, at least to hydroxyl radical cleavage (19). Thus, there is no minor-groove occupancy by the protein that could compete with the drug–DNA recognition process by steric exclusion.
In addition, it is known that GCN4 binds efficiently to the cAMP response element (CRE) site (20), which is derived from AP-1 by insertion of a G/C base pair in the middle of the sequence, converting it to the perfectly symmetric element 5′-ATGACGTCAT-3′. It has been observed that AP-1 and CRE exhibit different structural features before protein binding. In particular, solution measurements indicate that whereas AP-1 is in a nearly linear B form (21), CRE is bent toward the major groove (22), by an estimated 8° (23). Cocrystals of the DNA sequences with peptides containing the GCN4 DNA-binding domain reveal no appreciable DNA bending with the AP-1 site and a 20° bend toward the major groove at the center of the site for CRE (24, 25). Solution measurements indicate no significant change in DNA curvature at the AP-1 or CRE sites on binding either GCN4 or peptides that include the GCN4 dimerization and DNA-binding domains (22, 23). However, the contrary is observed for proteins of the CREB or activation transcription factor 2 (ATF-2) family, which have the dyad symmetric CRE site as their consensus sequence. They modify both CRE and AP1 structure to an appreciable extent, introducing a tendency to bend toward the minor groove in solution (22).
Because the structure of DNA depends on its sequence, it is difficult in principle to separate the contribution of sequence and structure to the drug–DNA discrimination process. The distortion of the double helix induced by CREB and ATF-2 proteins offers a powerful potential tool to induce different structural conformations in the same sequence. To exploit this concept, we used these DNA–protein complexes as models to gain insight into the role of DNA structure in the selective recognition of carbohydrate-containing drugs. In particular, we have investigated the calicheamicin γ1I-binding properties at GCN4 and CRE sites in their free and protein-bound forms. We compared the full-length protein GCN4 and its synthetic peptide corresponding to residues 228–281 (GGG) (22), and to the peptide CCC (22) containing residues 354–408 of CRE-BP1. Both GGG and CCC peptides comprise the basic and the zipper domains of the parent proteins linked through a 6-aa spacer region. In addition, we used the CGG hybrid peptide formed by the basic domain of CCC connected to the zipper region of GGG through the GGG-derived spacer, because its binding mode to AP1 and CRE sites resembles that of CRE-BP1.
In experiments complementary to those examining the influence of protein binding on drug cleavage, we have also studied competition effects between drug and protein binding. For this purpose, we used the synthetic calicheamicin aryltetrasaccharide to avoid unwanted cleavage effects. If, for example, protein binding blocks all drug-cleavage sites in a given region, binding of the aryltetrasaccharide should inhibit protein binding. However, if protein binding has no effect or prevents cleavage at a subset among overlapping sites, there should be little inhibition of protein binding by the aryltetrasaccharide.
MATERIALS AND METHODS
Calicheamicin γ1I was kindly provided by George Ellestad (Lederle, Pearl River, NY). The aryltetrasaccharide and aryltrisaccharide were prepared as described (26). Stock solutions (1 mg/ml) were prepared in ethyl acetate and stored at −20°C. Immediately before use, the correct amount was dried under vacuum and dissolved in 20% ethanol (calicheamicin) or 20% tetrahydrofuran (carbohydrate tail).
Peptides.
GGG, CCC, and CGG (22, 27) were kindly provided by Alanna Schepartz (Yale University), CAP by Tom Steitz (Yale University), and GCN4 by Rosa Beltran (Yale University).
DNA Fragments.
The CAP-binding site, AP-1, and CRE sites were inserted in pBluescript II KS +/− (Stratagene) and cloned in competent Escherichia coli cells. The DNA-labeling procedure was performed essentially as described (28). Briefly, the DNA was first linearized with the appropriate restriction enzyme under the conditions recommended by the supplier (New England Biolabs). For 3′ labeling, DNA was incubated with [α-32P]ATP and Klenow DNA polymerase fragment; for 5′ labeling, the DNA was dephosphorylated with calf alkaline phosphatase (Boehringer Mannheim) and then labeled with [γ-32P]ATP. After phenol/chloroform extraction and ethanol precipitation, DNA was digested with the second restriction enzyme. The uniquely 3′-end-labeled or 5′-end-labeled DNA fragments were then separated by 8% native PAGE, identified by autoradiography, cut, and eluted. After precipitation, they were washed and dissolved in 10 mM Tris/1 mM EDTA.
Cleavage and Sequencing Reactions.
Uniquely end-labeled DNA (5,000 cpm) was dissolved in 12 μl of PBS (2.7 mM KCl/137 mM NaCl/3 mM Na2HPO4/1.4 mM KH2PO4, pH 7.3) in the presence of varying amounts of peptides or GCN4. For CAP, the reaction buffer was 50 mM Tris (pH 8.0)/100 mM cAMP. Calicheamicin was then added to a final concentration of 5 μM, and after 5 min of incubation, the reaction was started by adding DTT to a final concentration of 2 mM. The mixture was incubated for 15 min, and the reaction was quenched by adding 7 μl of stop mixture [1.2 M NaOAc/0.4 μg/μl yeast tRNA/15 mM Mg(OAc)2]. After ethanol precipitation, the fragments were washed, dried, resuspended in sequencing loading buffer (80% formamide/10 mM NaOH/1 mM EDTA/0.1% xylene cyanol/0.1% bromphenol blue), heated for 2 min at 90°C, chilled in ice, and loaded onto 8–12% denaturing polyacrylamide gels (19:1) in 1× TBE (89 mM Tris base/89 mM boric acid/2 mM Na2EDTA). Gels were transferred to Whatman 3MM paper, dried under vacuum at 80°C, and autoradiographed (Amersham Hyperfilm MP) at −70°C with an intensifying screen.
Gel Shift Assay.
Labeled DNA was equilibrated at 4°C in the presence or absence of arylsaccharides and mixed with different amounts of protein or peptide in PBS containing 5% glycerol/1 mM EDTA/1 mM DTT (peptides and GCN4) or 10 mM Tris (pH 8.8)/1 mM EDTA/100 mM cAMP/40 mM NaCl (CAP). The mixture was incubated for 15 min at 20°C (CAP and GCN4) or 4°C (peptides) and then stopped with 1.5 μl of 50% glycerol containing bromophenol. Products were immediately loaded onto an 8% native polyacrylamide gel (75:1) and run at low voltage (35 V/cm) at 20°C (CAP and GCN4) or 4°C (peptides) in 0.5× TBE. Bands were visualized by using autoradiography and quantified with a Betascope 603 Blot Analyzer (Betagen, Waltham, MA).
RESULTS
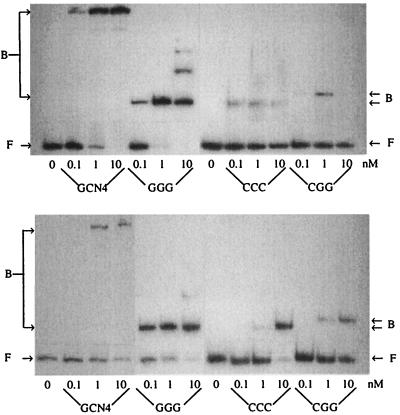
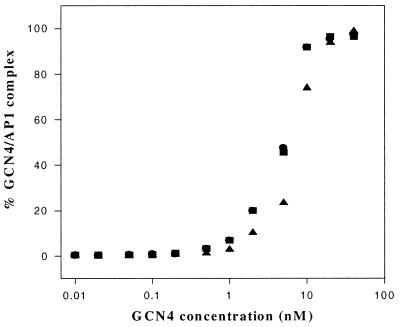
DNA Binding. Our experiments compare the selected DNA sites when free and when bound to the tested peptides and proteins, by using assays for tetrasaccharide inhibition of protein binding and of calicheamicin cleavage of free and complexed DNA. In agreement with literature data, very efficient DNA binding was observed with GCN4 (or its derived peptide GGG) on both AP1 and CRE sites, whereas complex formation was remarkably less efficient by using the peptides CCC or CGG, particularly on the AP1 site (27, 29–30). To evaluate the best experimental conditions for full formation of the DNA–peptide complex, DNA gel-shift binding titrations were performed with the peptides GGG, CCC, and CGG and with the protein GCN4 (Fig. 2) or CAP (data not shown). In agreement with previously reported results (31), C50, the protein concentration for half-dissociation of CAP from its consensus sequence, was in the nanomolar range (C50 ≈ 6 nM).
Figure 2.
Gel shift assay for peptides binding on AP1 (Upper) and CRE (Lower) sites in PBS containing 5% glycerol, 1 mM EDTA, and 1 mM DTT at 4°C. F, free DNA; B, DNA bound to the indicated protein.
A quantitative analysis of the resolved bands showed that on the CRE site, GCN4 and GGG have similar affinities (C50 ≈ 5 nM), whereas CCC and CGG bind less efficiently (50 nM). Interaction with AP-1 was strong with GCN4 (5 nM) and GGG (1 nM). On the other hand, by using CCC or CGG, it was not possible to obtain > 50% of complex formation because of low affinity constant and peptide-aggregation problems at high concentration. Of these two, we focused on the CGG peptide in this study because of its slightly higher binding constant on the AP-1 site compared with CCC and its similar ability to induce structural changes in DNA (22, 27).
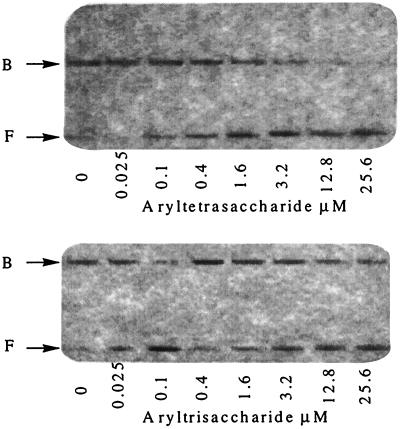
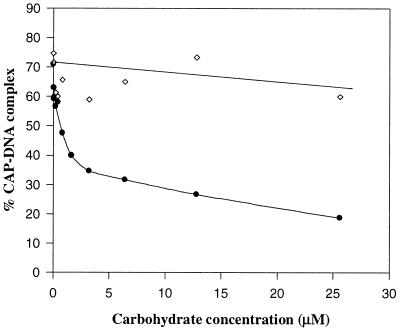
Carbohydrate Competition. Because both CAP and GCN4 reduce DNA electrophoretic mobility on binding, interactions of the aryltetrasaccharide with the protein–DNA complex were tested by a competitive gel-shift assay, with the nonbinding (5) truncated aryltrisaccharide (Fig. 1) as a control. Carbohydrate titrations were performed in the presence of a constant amount of CAP (Fig. 3). The aryltetrasaccharide decreases the fraction of DNA bound to CAP at equilibrium, even when the protein concentration was sufficiently high to bind all of the DNA in the absence of carbohydrate. Fig. 4 shows the results obtained with [CAP] = 10 nM and increasing concentrations of the two carbohydrates. The aryltetrasaccharide showed a 50% inhibition at 2.6 μM, whereas the control aryltrisaccharide, even at much higher concentrations, did not significantly affect the CAP–DNA complex.
Figure 3.
Effect of calicheamicin-related carbohydrates on CAP DNA binding in 10 mM Tris (pH = 8.8)/1 mM EDTA/100 mM cAMP/40 mM NaCl at 20°C, [CAP] = 10 nM. F, free DNA; B, CAP–DNA complex.
Figure 4.
Plot of CAP–DNA complex formation as a function of carbohydrate concentration in 10 mM Tris (pH = 8.8)/1 mM EDTA/100 mM cAMP/40 mM NaCl at 20°C, [CAP] = 10 nM. ●, In the presence of increasing amounts of aryltetrasaccharide; ⋄, in the presence of increasing amounts of aryltrisaccharide.
For the GCN4–DNA complex, even at aryltetrasaccharide concentrations up to 500 μM, it was not possible to observe the inhibitory effect that we found for the CAP–DNA complex. The DNA gel shift on adding increasing amounts of GCN4 in absence or presence of a known concentration of calicheamicin carbohydrate (1 μM) confirmed that little or no competition is occurring (Fig. 5). The same experiments performed on the CREB site also showed that GCN4 binding is insensitive to the presence of both the tri- and tetrasaccharide (data not shown).
Figure 5.
GCN4 titration experiments in the absence or presence of calicheamicin-related carbohydrates in PBS containing 5% glycerol, 1 mM EDTA, and 1 mM DTT at 20°C, [GCN4] = 0–40 nM. ●, control without arylsaccharides; ▴, in the presence of aryltetrasaccharide (1 μM); ■, in the presence of aryltrisaccharide (1 μM).
Drug Distribution Along the DNA Chain. Characterization of the calicheamicin–DNA complex (3, 9, 32–33) shows that the localization of the drug sugar moiety in the minor groove positions the enediyne function to generate DNA double-strand breaks by hydrogen abstraction at sequences that contain tetranucleotide tracts with a preponderance of pyrimidines on one strand and purines on the other. The tetrapyrimidine tract is cleaved at the second pyrimidine from its 5′ end, but if the strand is 3′-labeled, it runs 2 nt longer than the corresponding Maxam–Gilbert marker because of the 5′-aldehyde function that results from 5′-hydrogen abstraction (34). The strand carrying the tetrapurine tract is cleaved by either 1′-H or 4′-H abstraction 2 nt from the 3′ end of the tract and runs together with the Maxam–Gilbert marker if 3′-labeled (34, 35).
Taking advantage of the correspondence between aryltetrasaccharide binding and calicheamicin-cutting sites (36), we used the reactive whole drug as a probe to investigate the presence of sugar-binding sites in the consensus sequences of the tested proteins. Under the experimental conditions used for the gel-shift assay, all tested sequences were cut by the drug. Sequencing both strands of the fragments corresponding to the protein-binding sites allowed us to locate the calicheamicin-binding/cleavage sites as reported in Figs. 6–8 (2). Moreover, in agreement with previous data (5), the presence of the aryltetrasaccharide in the reaction mixture was shown to induce an almost unselective suppression of each drug-associated cutting site (data not shown). Because the protein-binding sites in question do not generally contain tetrapyrimidine sequences, the cleavage sites observed are not the strongest in the whole fragment. However, there was nevertheless sufficient reactivity at these secondary cleavage sites to characterize them.
Figure 6.
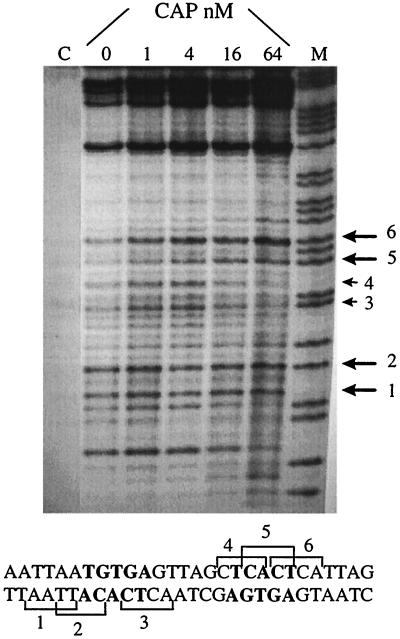
Sequencing gel of calicheamicin induced cuts on the selected DNA fragment (5′-CATGGCGCAACGCAATTAATGTGAGTTAGCTCACTCATTAGGCACCCTAGGTCTAG-3′) containing the CAP-binding sequence in the presence of increasing amounts of CAP. DNA is labeled 3′ on the bottom strand. [calicheamicin] = 5 μM, [CAP] = 0–64 nM; A + G, purine marker. The large arrows indicate drug-binding sites stimulated by the DNA–protein complex formation; the small arrows indicate inhibited sites. The DNA bases in bold are in direct contact with CAP.
Figure 8.
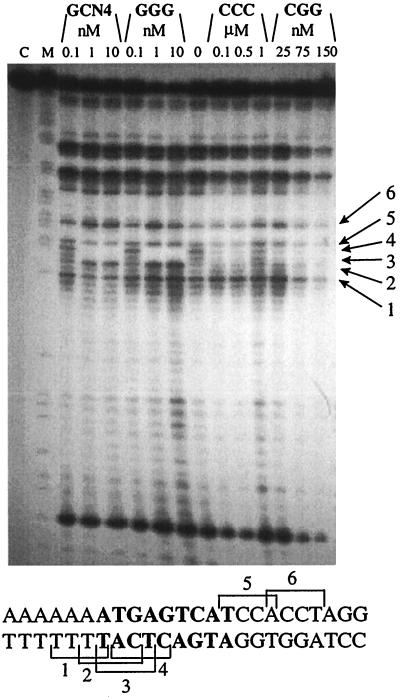
Sequencing gel of calicheamicin induced cuts on the selected DNA fragment (5′-AAGCTTGGCCATGGCAGCACGTTGTACTCGAGCAAAAAAAAATGACGTCATCCACCTAGGTCTAGAATTCAGCTGTACGGATC-3′) containing CRE site in the presence of increasing amounts of GCN4 or the tested synthetic peptide. DNA is labeled 3′ on the bottom strand. M, purine marker; C, control labeled DNA; 0, no protein in the reaction mixture. The sequence reported on the bottom corresponds to the portion of sequenced DNA fragment containing the protein-binding site. The DNA residues in bold correspond to the CRE site. Arrows indicate the breaks corresponding to the binding sites shown in the sequence.
Cleavage at the Occupied CAP Site.
Mapping of calicheamicin-cutting sites on the CAP consensus sequence reveals several sites within the region where protein–DNA contacts occur. In the presence of increasing amounts of protein, the most interesting feature is the inhibition of drug-induced breaks in the central portion of the CAP-binding site (Fig. 6). Because in this region the minor grove is covered by the protein (13, 14), this result means either that the drug is not able to overcome the steric hindrance because of CAP or possibly that the DNA structure is altered by the protein to prevent binding. Sequencing experiments performed by using increasing amounts of CAP revealed that high protein concentration is required to observe a modulation in the cleavage pattern, as expected from the observed competition effect. It is worthy of note that where the major groove faces CAP, a modest enhancement of the cleavage can be observed. A likely explanation is that the minor groove becomes wider and thus more accessible to the drug because of the protein-induced bend toward the major groove.
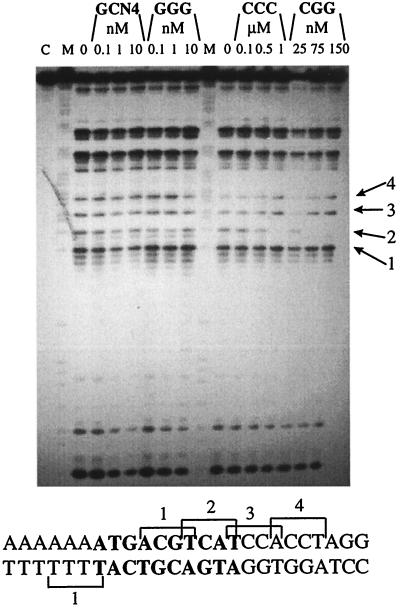
Cleavage at the Occupied AP1 Site. GCN4 or the tested synthetic peptides have significant effects on the enediyne cleavage pattern at the AP-1 site. As expected, the modifications induced by GCN4 and GGG are similar, whereas the effects of CCC and CGG resemble each other but are different from those observed with the former peptides (Fig. 7). Incubation of AP-1 with GCN4 or its related peptide GGG induces a selective inhibition of calicheamicin cutting, with a correlated enhancement of the breaks in the flanking areas. In particular, a strong inhibition of cleavage at the two central drug-binding sites TCAT (band 3, site 3, Fig. 7) and CTCA (site 4) (reporting the pyrimidine strand in the 5′ → 3′ direction) occurs, whereas a new cutting site CATT (site 2) appears near the middle of the GCN4-binding site, even at low protein concentration. The flanking sites 1, 5, and 6 are largely unaffected. In the case of CCC and CGG, the primary effect observed is inhibition of sites 3 and 4; any stimulation of site 2 that may be produced is certainly less pronounced than for GCN4 and GGG. Once again the flanking sites 1, 5, and 6 are largely unaffected.
Figure 7.
Sequencing gel of calicheamicin-induced cuts on the selected DNA fragment (5′-AAGCTTGGCCATGGCAGCACGTTGTAGCTCGAGCAAAAAAAAATGAGTCATCCACCTAGGTCTAGAATTCAGCTGTACGGATC-3′) containing the AP-1 site in the presence of increasing amounts of GCN4 or the tested synthetic peptides. DNA is labeled 3′ on the bottom strand. M, purine marker; C, control labeled DNA; 0, no protein in the reaction mixture. The sequence reported on the bottom corresponds to the portion of sequenced DNA fragment containing the protein-binding site. The DNA residues in bold correspond to the AP1 site. Arrows indicate the breaks corresponding to the binding sites shown in the sequence.
We note that we were able to identify strong cutting sites not containing G bases in the purine strand such as TTTT and TTGT. This confirms the previous suggestion that polynucleotide sequence is not a simple discriminating parameter for the specific drug–DNA interaction (8, 37, 38).
Cleavage at the Occupied CRE Site.
The tested peptides induce different modification of the enediyne distribution in the CRE site (Fig. 8). Even at high protein concentrations, neither GCN4 nor GGG affect the calicheamicin cutting pattern. In contrast, when the CRE site is bound to CCC or CGG, there is complete inhibition of the cutting occurring at the central G/C base pair, probably because of the binding sequence TCAT (site 2, Fig. 8).
DISCUSSION
Calicheamicin γ1I is a moderately sequence-selective drug. Early studies reported the tetranucleotide TCCT as its “consensus” sequence (2), but other sequences were subsequently discovered that were firmly bound and cut by the drug. Because all of them contained G/C base pairs, the first explanation for this behavior invoked the presence of at least one guanine group on the purine strand (39). However, the discovery that calicheamicin binds also to a TTTT sequence (8, 37) undermines this model. It is difficult to explain a specificity for both A/T- or G/C-rich sequences because they are substantially different in shape and hydrogen-bonding array. Because the carbohydrate tail of calicheamicin seems to be a rigid structure (40), binding the drug should induce steric pressure on DNA, suggesting a definition of “sequence specificity” as an ability to make the necessary adjustment for the best fit at the lowest energy cost (38, 41–44).
We have addressed the question of the role of DNA structure in recognition by the aryltetrasaccharide by investigating the effect of protein binding on calicheamicin cleavage patterns in the region of the DNA bound by the protein. It should be noted that the extent of cleavage can be modulated both by altered binding and by changes in the cleavage efficiency of the bound drug. Our experiments are, in general, not able to distinguish between these two effects, although in the case of CAP, the inhibition of protein binding by the aryltetrasaccharide indicates clearly that reduced cleavage in the CAP complex must be at least partly a consequence of reduced binding of calicheamicin. However, because the probable source of cleavage inhibition in this case is steric inhibition by the protein, we can draw no conclusions concerning modulation of binding by alteration of DNA structure.
For the basic leucine zipper proteins, however, our results reveal modulation of cutting that is not accompanied by binding competition between protein and the aryltetrasaccharide. The carbohydrate is not able to inhibit GCN4–DNA complex formation significantly, even though the protein causes a redistribution of calicheamicin-cleavage sites on the AP-1 sequence (Fig. 7). This result implies that GCN4 and calicheamicin can bind simultaneously to DNA, just as 1-methylimidazole-2-carboxamide netropsin can bind to the GCN4–DNA complex (45). Because previous studies reported that the calicheamicin carbohydrate moiety binds DNA with almost the same sequence selectivity as the whole drug, even if with a slightly lower affinity (5, 46, 47), we infer that the tested aryltetrasaccharide also binds the DNA–protein complex.
When GCN4 or GGG bind to the AP-1 site, a clear modulation of drug-associated cutting is observed: the central cutting sites (TCAT and CTCA) are disfavored relative to a flanking site (CATT), despite its lower reactivity in the absence of protein. In this case, no bending or twisting of the DNA structure has been demonstrated on protein binding. Hence, the observed modulation of cutting efficiency must be a consequence of a subtle, localized conformational change or alteration in flexibility. Recent cyclization kinetic experiments indicate that GCN4 can stiffen AP-1 DNA on binding (23). This effect could make DNA in the central cutting sites less adaptable to the structural change required for calicheamicin binding.
In the case of the CRE site, binding of GCN4 or its related peptide GGG does not affect its intrinsic curvature (22), which correlates with an unmodified calicheamicin cutting pattern in the peptide–DNA complex. On the other hand, when this site is complexed with CREB-related peptides (CCC, CGG), it becomes bent toward the minor groove (away from the leucine zipper). The result is complete inhibition of drug-induced cleavage occurring in the middle of the tested sequence. These findings support the view that minor-groove width plays a key role in calicheamicin–DNA interaction.
In conclusion, the results presented here confirm experimentally the importance of DNA shape in the calicheamicin-binding process. Any time the minor groove is reduced in its accessibility or width, we observe suppression of drug-induced DNA cleavage. Additionally, we emphasize the potential importance of DNA flexibility in accommodating drug binding. Suppression of drug cutting consequent on CRE–CREB and AP-1–CREB complex formation could arise from the combined narrowing of the minor groove and increased rigidity of the DNA chain. Our data support a model in which calicheamicin is not simply sensitive to DNA sequence but to double-helix structural features as well. We infer that calicheamicin and its related aryltetrasaccharide can interfere with biological processes not simply by cleaving free DNA but also by displacing a DNA-binding protein through competition or modulation of DNA structure. Correspondingly, protein binding can modulate calicheamicin cleavage patterns, leading to novel sites of action in vivo.
Acknowledgments
We thank J. Kahn for clone preparation and M. Palumbo for helpful reading of the manuscript. This work was supported by National Institutes of Health Grant GM 21966 and the Associazione Italiana per la Ricerca sul Cancro.
ABBREVIATIONS
- CAP
catabolite activator protein
- CRE
cAMP response element
References
- 1.Kahne D. Chem Biol. 1995;2:7–12. doi: 10.1016/1074-5521(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 2.Zein N, Sinha A M, McGahren W J, Ellestad G A. Science. 1988;240:1198–1201. doi: 10.1126/science.3240341. [DOI] [PubMed] [Google Scholar]
- 3.Zein N, Poncin M, Nilacantan R, Ellestad G A. Science. 1989;244:697–699. doi: 10.1126/science.2717946. [DOI] [PubMed] [Google Scholar]
- 4.Nicolaou K C, Tsay S-C, Suzuki T, Joyce G F. J Am Chem Soc. 1992;114:7555–7557. [Google Scholar]
- 5.Aiyar J, Danishefsky S J, Crothers D M. J Am Chem Soc. 1992;114:7552–7554. [Google Scholar]
- 6.Ho S N, Boyer S H, Schreiber S L, Danishefsky S J, Crabtree G R. Proc Natl Acad Sci USA. 1994;91:9197–9199. doi: 10.1073/pnas.91.20.9203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu C, Smith B M, Ajito K, Komatsu H, Gomez-Paloma L, Li T, Theodorakis E A, Nicolaou K C, Vogt P L. Proc Natl Acad Sci USA. 1996;93:940–944. doi: 10.1073/pnas.93.2.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mah S C, Price M A, Townsend C A, Tullius T D. Tetrahedron. 1994;50:1361–1378. [Google Scholar]
- 9.Walker S, Murnick J, Kahne D J. J Am Chem Soc. 1993;115:7954–7961. [Google Scholar]
- 10.Kolb A, Busby S, Buc H, Garges S, Adhya S. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- 11.Heyduk T, Lee J C. Biochemistry. 1992;31:5165–5171. doi: 10.1021/bi00137a011. [DOI] [PubMed] [Google Scholar]
- 12.Zinkel S S, Crothers D M. Biopolymers. 1990;29:29–38. doi: 10.1002/bip.360290106. [DOI] [PubMed] [Google Scholar]
- 13.Schultz S C, Shields G C, Steitz T A. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 14.Parkinson G, Wilson C, Gunasekera A, Ebright Y W, Ebright E, Berman H M. J Mol Biol. 1996;260:395–408. doi: 10.1006/jmbi.1996.0409. [DOI] [PubMed] [Google Scholar]
- 15.Hope I A, Struhl K. Cell. 1986;46:885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- 16.O’Shea E K, Klemm J D, Kim P S. Science. 1991;254:529–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 17.Oas T G, McIntosh L P, O’Shea E K, Dahlquist F W, Kim P S. Biochemistry. 1990;29:2891–2894. doi: 10.1021/bi00464a001. [DOI] [PubMed] [Google Scholar]
- 18.Hill D E, Hope I A, Macke J P, Struhl K. Science. 1986;234:451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- 19.Gartenberg M R, Ampe C, Steitz T A, Crothers D M. Proc Natl Acad Sci USA. 1990;87:6034–6038. doi: 10.1073/pnas.87.16.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sellers J W, Vincent A C, Strhul K. Mol Cell Biol. 1990;10:5077–5086. doi: 10.1128/mcb.10.10.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sitlani A, Crothers D M. Proc Natl Acad Sci USA. 1996;95:1404–1409. doi: 10.1073/pnas.95.4.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paolella D M, Palmer C R, Schepartz A. Science. 1994;264:1130–1133. doi: 10.1126/science.8178171. [DOI] [PubMed] [Google Scholar]
- 23.Hockings S C, Kahn J D, Crothers D M. Proc Natl Acad Sci USA. 1998;95:1410–1415. doi: 10.1073/pnas.95.4.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellenberger T E, Brandl C J, Struhl K, Harrison S C. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 25.Keller W, König P, Richmond T J. J Mol Biol. 1995;254:657–667. doi: 10.1006/jmbi.1995.0645. [DOI] [PubMed] [Google Scholar]
- 26.Halcomb R L, Boyer S H, Danishefsky S J. Angew Chem Int Ed Engl. 1992;31:338–340. [Google Scholar]
- 27.Metallo S J, Schepartz A. Chem Biol. 1994;1:143–151. doi: 10.1016/1074-5521(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 28.Capranico G, Zunino F, Kohn K W, Pommier Y. Biochemistry. 1990;29:562–569. doi: 10.1021/bi00454a033. [DOI] [PubMed] [Google Scholar]
- 29.Hai T, Curran T. Proc Natl Acad Sci USA. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hai T, Liu F, Coukos W J, Green M R. Genes Dev. 1989;3:2883–2090. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- 31.Kahn J D, Crothers D M. Proc Natl Acad Sci USA. 1992;89:6343–6347. doi: 10.1073/pnas.89.14.6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paloma L G, Smith J A, Chazin W J, Nicolaou K C. J Am Chem Soc. 1994;116:3697–3708. [Google Scholar]
- 33.Ikemoto N, Kumar R A, Ling T T, Ellestad G A, Danishefsky S J, Patel D J. Proc Natl Acad Sci USA. 1995;92:10506–10510. doi: 10.1073/pnas.92.23.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zein N, Sinha A M, McGahren W J, Ellestad G A. Science. 1988;240:1198–1201. doi: 10.1126/science.3240341. [DOI] [PubMed] [Google Scholar]
- 35.Hawley R C, Kiessling L L, Schreiber S L. Proc Natl Acad Sci USA. 1989;86:1105–1109. doi: 10.1073/pnas.86.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mah S C, Townsend C A, Tullius T D. Biochemistry. 1994;33:614–621. doi: 10.1021/bi00168a029. [DOI] [PubMed] [Google Scholar]
- 37.Walker S, Landovitz R, Ding W D, Ellestad G A, Kahne D. Proc Natl Acad Sci USA. 1992;89:4608–4612. doi: 10.1073/pnas.89.10.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bailly C, Waring M J. J Am Chem Soc. 1995;117:7311–7316. [Google Scholar]
- 39.Hawley R C, Kiesseling L L, Schreiber S L. Proc Natl Acad Sci USA. 1989;86:1105–1109. doi: 10.1073/pnas.86.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker S, Valentine K G, Kahne D. J Am Chem Soc. 1990;112:6428–6429. [Google Scholar]
- 41.Walker S, Yang D, Kahne D, Gange D. J Am Chem Soc. 1991;113:4716–4717. [Google Scholar]
- 42.Krishnamurthy G, Ding W-D, O’Brien L, Ellestad G A. Tetrahedron. 1994;50:1341–1349. [Google Scholar]
- 43.Walker S, Andreotti A H, Kahne D. Tetrahedron. 1994;50:1351–1360. [Google Scholar]
- 44.Kumar R A, Ikemoto N, Patel D J. J Mol Biol. 1997;265:187–201. doi: 10.1006/jmbi.1996.0718. [DOI] [PubMed] [Google Scholar]
- 45.Oakley M G, Mrksich M, Dervan P B. Biochemistry. 1992;31:10969–10975. doi: 10.1021/bi00160a005. [DOI] [PubMed] [Google Scholar]
- 46.Li T, Zeng Z, Estevez V A, Baldenius K U, Nicolaou K C, Joyce G F. J Am Chem Soc. 1994;116:3709–3715. [Google Scholar]
- 47.Drak J, Iwasawa N, Danishefsky S, Crothers D M. Proc Natl Acad Sci USA. 1991;88:7464–7468. doi: 10.1073/pnas.88.17.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]