Abstract
The RNA world hypothesis presumes that RNA will be competent for varied essential cellular functions. One such indispensable cell function is regulation of membrane permeability. Though this was not a known RNA activity, selection–amplification yielded RNAs that bound phosphatidylcholine:cholesterol liposomes. At least eight distinct, ≈95-mer sequences bind well to the outside of the lipid bilayer, though randomized sequences had no such activity. No distinct sequence motif for lipid binding was found. However, truncation of one such RNA shows that a smaller, 44-nucleotide irregular RNA hairpin is an active membrane binding domain. Bound RNA increases the permeability of liposomes to 22Na+. In addition, using voltage clamp technique, four individual RNAs increase the ion permeability of the plasma membrane of cultured human cells. The existence of multiple sequences that bind membranes and provoke permeability changes suggests that these may be elementary RNA functions that could be selected in vivo.
Cells communicate with their environments across phospholipid bilayer membranes. However, the permeability of such bilayers (relatively permeable to water, but virtually impermeable to polar molecules, even to small ions) is dramatically mismatched to cellular needs. Cellular life therefore absolutely requires facilitation of transport through phospholipid boundaries. The RNA-world hypothesis (1) posits ancestral cells in which RNA plays many of the roles taken by modern proteins. Might RNAs serve membrane functions?
We have approached this question, first, by isolating RNAs that bind to pure phospholipid membranes by using selection–amplification (2, 3). In this technique, novel RNA activities are isolated by selecting infrequent, perhaps unique active molecules from large pools of transcripts (≈1014 different molecules) with randomized sequences. Repetition of the selection (purification) is made possible by nucleic acid amplification (replication) applied to the partially purified pools, so that large cumulative purifications are possible after multiple cycles of selection–amplification. Ultimately, cDNA cloning and in vitro transcription make available single pure active RNAs for study. Second, we have reintroduced purified membrane-binding RNAs from selection–amplification into several membrane systems to measure their effects on permeability, as has long been done for channel proteins (4).
To act in membranes, RNA must interact with membrane constituents. Phospholipids, the main components of the biological membrane, are chemically tripartite. They consist of a polar head group, glycerol phosphate, and fatty acids. RNAs should easily interact with polar head groups, particularly cationic ones. Glycerol phosphate (which resembles the RNA backbone) also presents easily used hydrogen-bonding opportunities to an RNA. In contrast, fatty acids might be thought of as improbable RNA ligands. However, it has previously been shown that RNAs fold to form specifically shaped, hydrophobic sites that interact favorably with hydrocarbons [the valine and isoleucine side chains (5, 6)]. Earlier work also had hinted at weak, nonspecific, readily dissociated interactions between membranes and homogeneous oligonucleotides (7, 8). Taken together, these data suggest that RNA should be capable of binding and some degree of insertion into phospholipid bilayers. Such RNA binding might perturb membrane permeability, conceivably even changing it specifically.
EXPERIMENTAL PROCEDURES
Selection.
Four cycles of selection were performed in 50 mM Hepes (pH 7.0), 5 mM MgCl2, 1 mM CaCl2, and 50 mM NaCl. Then, the medium was changed to 50 mM Mes (pH 5.5), 20 mM MgCl2, 10 mM CaCl2, 100 mM NaCl, 40 mM KCl, 100 μM MnCl2, and 20 μM ZnCl2, which appeared to be more favorable for the selected activity. For selections, incubation of RNA [1,000 pmol (cycles 1–4) and 100 pmol (cycles 5–11)] with liposomes (20 μl of 20 mg/ml lipid) was performed at room temperature for 15 min, followed by gel filtration on Sephacryl S-1000 (Amersham Pharmacia). To decrease nonspecific sorption, the column was presaturated with both liposomes and nonspecific RNA. Fifty-microliter fractions were collected, and the first two fractions containing the leading edge of the liposome peak were pooled, chloroform extracted, precipitated and processed as described (9).
Initial RNA Pool.
The initial RNA pool consisted of 95-nucleotide oligomer transcripts containing 50 central randomized positions, encoded within flanking PCR primer sequences containing a T7 promoter (5′CGGAAGCTTCTGCTACATGCAATGG-N50-CACGTGTAGTATCCTCTCCCTATAGTGAGTCGTATTAGAATTCGC-3′). Approximately 1015 molecules of 95-nucleotide RNA transcribed from 2 × 1014 independently synthesized DNA templates (10) were heated at 65°C for 3 min and were cooled to room temperature over 10 min.
Truncation of RNA Substrate.
Variants of isolate 13 were synthesized by in vitro transcription from either double-stranded DNA obtained by PCR (72- and 64-mer) or directly from deoxyoligomers (for 53-, 44-, and 40-mer) (10). Additional 5′ mutations were introduced to facilitate transcription.
RNase T1 and S1 Protection.
5′ labeled RNA bound to liposomes was isolated by gel filtration and was digested with RNase T1 (GIBCO/BRL) at concentrations ranging between 0.025 units/μl and 0.2 units/μl for 1 min at room temperature. RNase S1 (GIBCO/BRL) was used at concentrations ranging between 0.5 units/μl and 2 units/μl) for 20 min at room temperature. Reactions were stopped by several phenolchloroform extractions, and RNA was ethanol-precipitated and separated on 10% denaturing polyacrylamide gel. Gels were quantitated by using a Bio-Rad phosphorimaging system.
Liposome Preparation.
Unilamellar liposomes were prepared from phosphatidylcholine (1, 2-dioleoyl-sn-Glycero-3-Phosphocholine):cholesterol 4:1(Avanti Polar Lipids) at a final concentration 20 mg/ml, using the Avanti Mini-Extruder according to manufacturer’s protocol. This lipid was chosen for fluidity under all experimental conditions. Liposomes were stored at 4°C under argon for up to 2 weeks. Electron microscopic examination of the liposomes suggested that unilamellar vesicles were common and that the liposome population was relatively homogeneous in size, with diameters centered at 80 nm (11) (Tom Giddings, University of Colorado).
RNA Affinity To Free Choline and Liposomes.
RNA affinity to free choline was measured by using [14C]choline (Sigma) in an equilibrium gel-filtration assay (12). Kd values for RNA-liposome binding were estimated from Kd = kon/koff. The forward rate, kon, was estimated as the diffusion controlled rate (kon ≈ 108 M−1⋅s−1). koff was estimated from the survival of the complex during storage after gel chromatography; t1/2 ≥ 20 hr, koff ≤ 10−5⋅sec−1.
22Na Efflux Experiments.
Liposomes loaded with 22NaCl were prepared by the detergent removal method previously described (4). Dried mixtures of phosphatidylcholine and cholesterol (5:1) were dissolved in 20 mM Hepes (pH 7.0), 150 mM NaCl, 20 mM MgCl2, and 10 mM CaCl2 containing 10% Triton X-100 (Sigma). The lipid solution was diluted with the same buffer, containing 10 μCi 22NaCl/ml, bringing the final concentrations of Triton-X-100 and phosphatidylcholine to 1.75% (wt/vol) and 1.7% (wt/vol), respectively. To remove Triton X-100, the solution was incubated with BioBeads SM-2 (Bio-Rad). The amount of liposome-incorporated 22Na+ was measured by dilution of 20 μl of a vesicle suspension loaded with 22NaCl in 200 μl of 20 mM Hepes (pH 7.0) with sucrose (Fisher) varied to compensate the osmotic pressure. This was applied to a Dowex 50W-X8 column presaturated with 50 μl of liposomes. Liposomes were immediately eluted with 4 ml of Hepes-sucrose buffer. External 22Na+ is adsorbed to the Dowex resin; 22Na+ eluted from the column represents intravesicular 22Na+. RNA-stimulated 22Na+ efflux was measured by adding RNA to a 20-μl aliquot of a vesicle suspension loaded with 22NaCl (RNA concentration 5 μM; ≈100 molecules/liposome).
Patch Clamp Procedure.
Patch clamp recordings were made in HEK 293 cells (human epithelial kidney) from the American Type Culture Collection (accession no. CRL-1573.1; http://www.atcc.org) by using the whole cell patch configuration (13). The bath solution contained 119 mM NaCl, 11 mM MgCl2, 6.8 mM CaCl2, 10 mM glucose, and 10 mM Hepes (pH 7.35). The pipette solution contained 110 mM KCl, 5 mM K4/1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate, 5 mM K2ATP, 1 mM MgCl2, and 10 mM Hepes (pH 7.2).
RESULTS
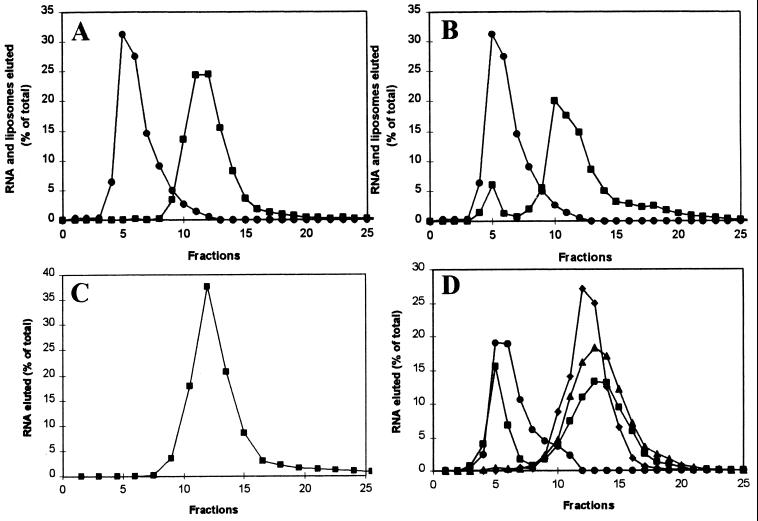
Selection of Liposome Binding RNA. To obtain RNA molecules capable of binding to lipid bilayers, we used a selection procedure based on gel-filtration of liposomes. A pool of 1015 RNA molecules each containing a randomized 50-nucleotide tract was incubated with phosphatidylcholine:cholesterol liposomes. Bound RNA was captured by fractionation of liposomes into the void volume of a Sephacryl S-1000 column. Initial randomized RNA had no detectable tendency to fractionate with liposomes (Fig. 1A), but ≈15% of total RNA migrated with liposomes after the eighth cycle of selection (Fig. 1B). Appearance of RNA in the void volume depended on RNA-liposome interactions; eighth cycle RNA alone eluted as a single included peak (Fig. 1C). Thus, the selected activity is not RNA aggregation. RNA-liposome binding requires both Mg2+ and Ca2+ ions (Fig. 1D). Binding was completely abolished by omission of either from the reaction buffer. Other divalent cations present during selection were dispensable.
Figure 1.
Elution of RNA and liposomes from a Sephacryl S-1000 column. (A) Randomized RNA (■) and liposomes (●). (B) RNA (■) and liposomes (●) after the eighth selection. (C) RNA after eighth selection (■), but without liposomes. (D) RNA and liposomes after the 11th selection (●) in the presence of 20 mM MgCl2 and 10 mM CaCl2 (■), only 20 mM MgCl2 (♦), and only 10 mM CaCl2 (▴).
Electron microscope images (11) of the liposome preparations in the presence and absence of selected RNA reveal no differences in size distribution. Thus, RNA binding to liposomes is not accompanied by detectable change in the liposomes, including fusion. Bound RNAs are accessible from the outside of the vesicles (see below), and therefore appear to have become attached to the exterior of preexisting liposomes.
Choline Involvement in RNA-Liposome Binding.
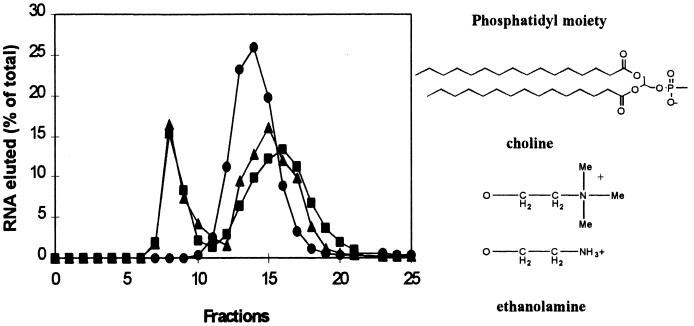
An evident liposomal surface target for RNA recognition is the cationic choline moiety of phosphatidylcholine. Indeed, 11th-round pool RNA did not detectably bind liposomes in the presence of the potential competitor, 5 mM choline (Fig. 2). In contrast, a similar small cationic control molecule, 5 mM ethanolamine, did not interfere with binding. From comparison of phosphatidylcholine and ethanolamine, choline methyl groups may contribute to RNA affinity. However, attempts to measure RNA binding to free choline were unsuccessful, indicating that Kd is probably ≥10−4 M. In contrast, the estimated Kd for RNA-liposome binding is 10−12–10−13 M (see Experimental Procedures). This apparent difference in affinity suggests that several cholines, or choline plus other bilayer features, comprise the RNA site.
Figure 2.
Elution of 11th selection RNA from the Sephacryl S-1000 column after incubation with liposomes (■) and in the presence of 5 mM choline (Sigma) (●) or of 5 mM ethanolamine (Sigma) (▴).
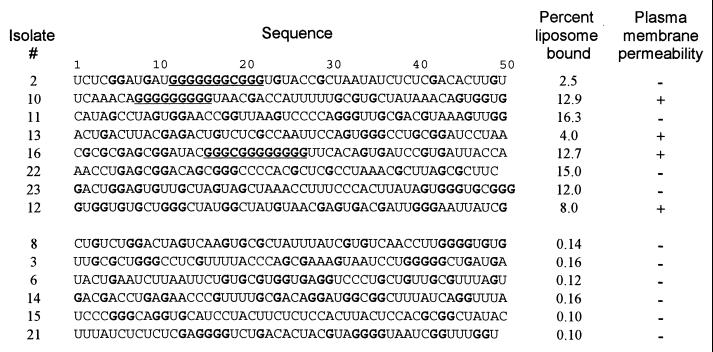
Liposome Binding Motif in RNA. After the 11th cycle of selection, ≈30% of RNA migrated with liposomes (Fig. 1D), and no increase in binding was detected during the last two rounds of selection. RNAs from pool 11 were cloned and characterized (Fig. 3). About 70% of individual pool RNA sequences bound to liposomes when tested as pure individual transcripts. RNA-liposome binding is linear over at least a 1,000-fold range of RNA concentrations (10 nM–10 μM). Thus, binding probably occurs via interaction of single RNA molecules with liposomes.
Figure 3.
RNA sequences after 11 cycles of selection. Gs are bold; G clusters are underlined. Liposome binding is percent of RNA coeluted with liposomes normalized to RNA eluted from the column [10 min of incubation at room temperature by using 100 pM RNA with 20 μl liposomes (20 mg/ml phosphatidylcholine and 3 mg/ml cholesterol)]. Permeability positive means induced current within 30 min after 2 μM RNA was added to HEK 293 cells.
Sequence comparison of the cloned RNAs did not reveal any common pattern among RNAs with affinity for liposomes except for frequent G-stretches. Oligo-G and -U stretches have been previously noted in RNAs that bind hydrophobic ligands and may indicate a hydrophobic patch in RNA (14). Nevertheless, long G tracts are not essential for bilayer affinity or permeation activity because they are absent from some RNAs showing both binding and permeability effects (Fig. 3; see below). Accordingly, there may be two or more types of binding structures and modes of binding. The absence of a specific sequence pattern for lipid bilayer recognition suggests that binding depends on RNA structures that are either small, degenerate, or multiple; thus RNA-bilayer interactions can be said to be readily made.
Characterization of Selected RNA-Phospholipid Bilayer Affinity.
RNA-liposome association is rapid. Binding of RNA is already maximal when RNA and liposomes are mixed and chromatographed on Sephacryl as rapidly as a manual experiment can be performed. RNA binding also is only slightly reversible. After rechromatography of isolate 13 RNA bound to liposomes, >80% of RNA remains associated after 12 hr at room temperature (not shown). Thus partial binding (Fig. 3) to liposomes is not necessarily attributable to weak association. Instead, such observations also can indicate alternative RNA conformations; this seems to be the case for isolate 13 RNA.
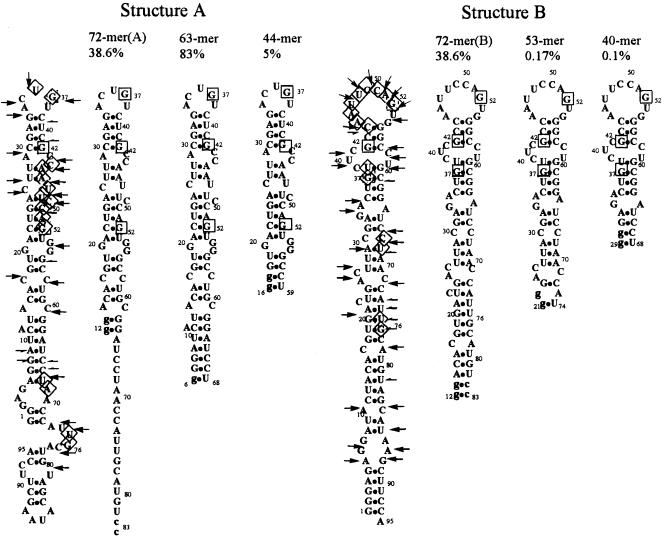
Only One Conformer of Isolate 13 Has Affinity to Liposomes.
Modeling of isolate 13 RNA predicted two equally stable alternative secondary structures (Fig. 4). Sites of RNA backbone flexibility (Pb2+ cleavage) (Fig. 4) are also consistent with existence of isolate 13 as an approximately equal mixture of two secondary structures (A and B). In further support of conformational variation, different folding conditions (fast or slow cooling) vary the fraction of bound isolate 13 RNA from 5 to 68% (not shown). Conceivably, such multiple conformations were forced by the selection. RNA binding to the membrane may require formation of a hydrophobic surface patch in the free molecule. Such a patch would have a high free energy of formation in H2O, and alternative conformations, inactive in membrane binding, may therefore be frequent.
Figure 4.
Truncation and binding by isolate 13 RNA. Structures A and B are two computed minimal free energy structures (15) for isolate 13 RNA. Sites of RNA backbone flexibility (Pb2+ cleavage) are indicated by arrows and half-arrows (16). Arrows are cleavage sites in agreement with the structure while half-arrows indicate sites supporting the alternative conformation. Guanines protected from RNase T1 digestion by liposomes are in squares. Nucleotides protected from nuclease S1 digestion on liposomes binding are in rhombs. Nucleotides mutated to improve transcription are in small letters. Numbers are the fraction of RNAs bound normalized to concurrent experiments with full-sized, optimally folded isolate 13 RNA.
Minimization of Liposome Binding RNA and Identification of the Active Region. Truncation of isolate 13 RNA suggests that only conformer A is active in binding (Fig. 4). First, 72-mer RNA, which can fold to both conformers, is partially active. Two truncated RNAs (52- and 40-mer), which cannot fold to yield a variant of A, but readily fold into derivatives of structure B, are inactive. On the other hand, truncated 64-mer RNA, designed to fold into structure A, is 84% active. Further RNA truncation maintaining type A structure resulted in the 44-mer RNA that retains 5% activity.
Fifteen nucleotides were protected from S1 nuclease digestion in liposome-bound RNA (Fig. 4). Such extensive protection is probably the sum of two different effects. First is real protection of internucleotide bonds in the active conformer A on binding (U36, G37). Second, we expect loss of S1 nuclease cuts unique to inactive conformer B in the liposome-bound fraction (nucleotides 44–52).
Three Gs (G37, G42, and G52) are also significantly protected from digestion by T1 RNase on liposome binding. Presently, we cannot distinguish protection of nucleotides by association with the membrane from protection because of RNA structural changes, and both may occur. In any case, these three T1 RNase-insensitive G residues are present in the active 44-mer, along with the S1 nuclease protections. Thus, the central, isolate 13-derived, 44-nucleotide hairpin is the smallest RNA known to have an active region for membrane affinity.
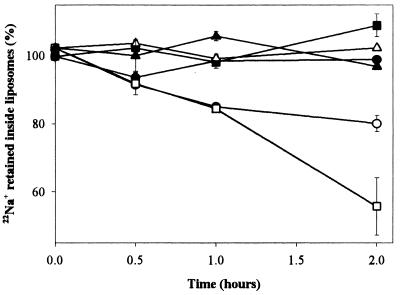
RNA-Induced Permeability in an Artificial Bilayer Measured by 22Na+-Efflux. Strong binding of RNA to liposomes with a very low off rate, and protections of several nucleotides near the hairpin loop, suggests some degree of RNA insertion into lipid membrane. This probably implies some local structural rearrangement in the phospholipid leaflet. A bound macromolecule surrounded by altered phospholipid could well constitute a site of altered membrane permeability. Therefore, permeability in artificial bilayers was evaluated by using RNA-induced 22Na+-efflux from the liposomes used for selection, paralleling a technique developed for protein channels. Liposomes were loaded with 150 mM 22NaCl, were incubated with RNA, and were applied to a Dowex column, which quantitatively adsorbs free Na+. Within liposomes, Na+ is shielded from the resin and is detected at the column void volume. Increased membrane permeability therefore decreases 22Na+ in column flow-through.
Although neither random RNA nor hydrolyzed active RNAs induced 22Na+-efflux, a mixture of 14 individual RNAs (Fig. 3) induced ≈45% efflux after 2 hr (Fig. 5). Pure isolate 13 RNA was also active in 22Na release. Such sodium efflux from the liposomes used in the initial selection strictly depended on the presence of Mg2+ and Ca2+. The common divalent requirement for both liposome binding and 22Na+ efflux supports an overlapping mechanism for both activities.
Figure 5.
Sodium permeability: RNA-induced 22Na+ efflux from phosphatidylcholine:cholesterol liposomes. Shown are no RNA (■), random RNA (●), isolate 13 (□), mixture of equal amounts of 14 individual RNAs (sequences in Fig. 3) (p), mixture of 14 individual RNAs in the absence of Mg2+ and Ca2+ (▴), and mixture of 14 RNAs treated with NaOH (20 mM NaOH, 30 min, 92°C; neutralized with 20 mM HCl) (▵). Each point is the mean of six samples derived from two independent experiments; bars show SEM.
Maximum 22Na+ efflux was achieved after 2 hr of RNA-liposome incubation (Fig. 5). Because RNA-liposome binding happens quickly and ion equilibration is expected to occur very rapidly after channel formation (in the millisecond time range), this technique measures the fraction of vesicles in which a channel has formed. Efflux is less than complete, perhaps because of the formation of internal membrane systems retaining 22Na+, but is inaccessible to externally applied macromolecules. The same limitation is observed with protein channels. For example, sodium channels from rat brain reconstituted into phosphatidylcholine liposomes release 35% of liposome-bound 22Na+ (17). In contrast to binding experiments, in which RNA appeared fully bound in <1 min, permeability effects assayed by 22Na+ release seem to require a subsequent slower process (Fig. 5).
Liposome Binding RNAs Alter Biological Membranes. RNA binding to an artificial bilayer makes it permeable to ions. Parallel biological effects were sought on intact cellular membranes. We used voltage clamp technique to evaluate RNAs applied to both external and internal faces of the human HEK 293 plasma membrane. HEK 293 is a cell line with low endogenous currents, frequently used to evaluate protein ion channel function.
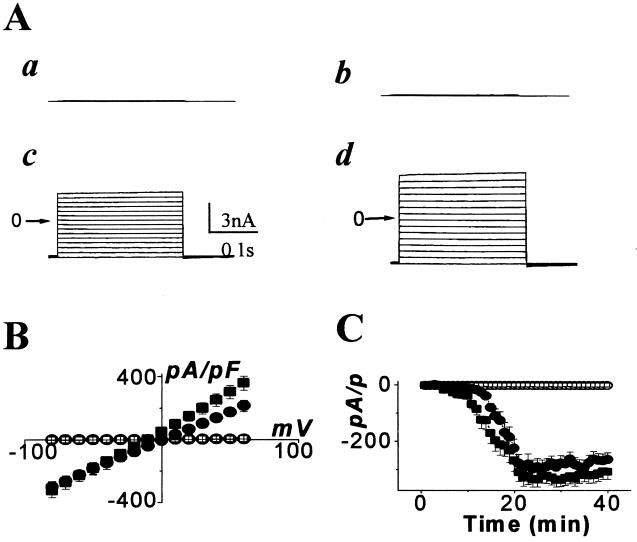
Randomized RNA sequences did not induce significant current (20 min at 5 μM) in whole cell patch mode. However, 11th-cycle pool RNA induced nanoampere-scale currents through the plasma membrane, with reversal potential around 0 mV (Fig. 6 A and B). An even larger effect was found with the mixture of 14 individual RNAs (Fig. 6 A and B). As for artificial bilayers, current through a cell membrane required high Mg2+ and Ca2+ (not shown). Accordingly, we expect an overlapping mechanism for RNA effects on both artificial and natural membranes.
Figure 6.
Induction of ionic currents by extracellular RNAs in HEK 293 cells. The membrane potential was first held at −80 mV. (A) Representative tracings recorded with 10 mV depolarizing steps from −80 to +60 mV for 300 msec in a control cell (a), 20 min after exposure to random RNA (b), RNA after 11 cycles of selection for liposome binding (c), and mixture of individual RNAs (d). (B) Averaged current–voltage relationships of steady state currents obtained in five control cells (□), 20 min after exposure to random RNA (□; n = 3), RNA after 11 cycles of selection for liposome binding (●; n = 5), and mixture of 14 individual RNAs (■; n = 6) under the same experimental conditions as in A. Currents are normalized to the membrane capacitance in picofarads in each cell (a cell ≅20 pF). (C) Time course of membrane current at −80 mV in 5 control cells (○), induced by random RNA (□; n = 3), RNA after 11 cycles of selection for liposome binding (●; n = 3), and mixture of 14 individual RNAs (■; n = 4). Data points were obtained at 1 min intervals. Bars indicate the SEM.
About half of individual pure liposome-binding RNAs displayed permeability effects (Fig. 3) when tested as transcripts applied to HEK 293 plasma membranes. Thus, the RNA structures required to increase membrane permeability are frequent among, but distinguishable from, the structures that allow membrane affinity. As in an artificial bilayer, RNA-induced permeability changes in a real plasma membrane are slow. The observed 5- to 10-min lag may indicate essential slow events: e.g., conformational change and/or assembly of active RNAs within the membrane (Fig. 6C).
Plasma membranes are chemically asymmetric. Choline-containing phospholipids are selectively sorted to the outer leaflet (18) whereas glycolipids (19) and glycoproteins (19), whose carbohydrates might exclude macromolecules like RNA, are exclusively outside (20). Thus, an internal membrane face comprises a distinct RNA target. Inside-out patches, in which a small area of membrane is captured on the patch electrode, allow application of RNA to a plasma membrane’s inner face. In fact, starting at 6 min after exposure of the internal membrane leaflet, isolate 13 RNA increases ionic permeability of an inside-out patch (not shown). Action from both inside and outside a human plasma membrane, taken together with activity on pure phospholipid bilayers, suggests that these RNAs will perturb many biological membranes.
DISCUSSION
We report two activities for multiple small RNA molecules. First, RNA is easily selected for binding to phospholipid bilayers. Selected RNA binding is relatively rapid and not readily reversed. Second, some bound RNAs can be shown to increase the ionic permeability of both artificial bilayers and natural plasma membranes.
However, membrane binding and permeability change are distinguishable processes. First, although binding is rapid, a slower process must occur before permeability effects can be detected. Second, the RNA structures required for the two processes can be distinguished. A specific set of RNA structures is clearly required to observe strong phospholipid membrane binding. This property cannot be measured in a randomized pool of sequences and requires selection of individual active RNAs. In further support of specific active structures, relatively small alterations of the selected RNA structure can disrupt binding. About half of RNAs selected for affinity to the phospholipid bilayer also increase permeability of biological plasma membranes. Thus, the permeability effects require a subset of RNA structures with phospholipid membrane affinity.
Surprisingly, neither binding nor permeability is obviously correlated with highly specific nucleotide sequences. Some evidence points to G tracts (sequence features previously correlated with hydrophobic ligands) and to the choline head group as regions of RNA-membrane interaction. Because the RNA membrane binding structure can be small (≤44 nucleotides) and is probably degenerate, many RNAs may have membrane affinity. Such interactions should be sought in vivo.
RNAs may partially insert and perturb both leaflets of the membrane or, alternatively, may interfere with membrane structure by interacting with the proximal phospholipid leaflet. Whether these RNAs have structure or function comparable to transmembrane proteins or use a mechanism unique to RNA is unknown and a matter for further experimentation. Nevertheless, even the present data suggest that RNA might have functioned as rudimentary channels and taken other membrane roles during cellularization in an RNA world.
Acknowledgments
We thank the members of our lab and also Kurt Beam, Dmitriy Kirpotin, and Alexey Wolfson for helpful discussions. This work was supported by National Institutes of Health Grants GM30881 and GM4080 to M.Y. and HL49330 to M.T.
References
- 1.Gilbert W. Nature (London) 1986;319:618. [Google Scholar]
- 2.Tuerk C, Gold L. Science. 1990;3:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 3.Ellington A D, Szostak J W. Nature (London) 1990;30:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 4.Talvenheimo J A, Tamkun M M, Catterall W A. J Biol Chem. 1982;257:11868–11871. [PubMed] [Google Scholar]
- 5.Majerfeld I, Yarus M. Nat Struct Biol. 1994;1:287–292. doi: 10.1038/nsb0594-287. [DOI] [PubMed] [Google Scholar]
- 6.Majerfeld I, Yarus M. RNA. 1998;4:471–478. [PMC free article] [PubMed] [Google Scholar]
- 7.Budker V G, Kazatchkov Y A, Naumova L P. FEBS Lett. 1978;95:143–6. doi: 10.1016/0014-5793(78)80070-2. [DOI] [PubMed] [Google Scholar]
- 8.Budker V G, Godovikov A A, Naumova L P, Slepneva I A. Nucleic Acids Res. 1980;8:2499–2515. doi: 10.1093/nar/8.11.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciesiolka J, Illangasekare M, Majerfeld I, Nickles T, Welch M, Yarus M, Zinnen S. Methods Enzymol. 1996;267:315–335. doi: 10.1016/s0076-6879(96)67021-9. [DOI] [PubMed] [Google Scholar]
- 10.Milligan J F, Uhlenbeck O C. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 11.Hope M J, Wong K F, Cullis P R. J Electron Microsc Tech. 1989;13:277–287. doi: 10.1002/jemt.1060130403. [DOI] [PubMed] [Google Scholar]
- 12.McConn J, Ku E, Himoe A, Brandt K G, Hess G P. J Biol Chem. 1971;246:2918–2925. [PubMed] [Google Scholar]
- 13.Uebele V N, England S K, Chaudhary A, Tamkun M M, Snyders D J. J Biol Chem. 1996;271:2406–2412. doi: 10.1074/jbc.271.5.2406. [DOI] [PubMed] [Google Scholar]
- 14.Yarus M. J Mol Evol. 1998;47:109–117. doi: 10.1007/pl00006357. [DOI] [PubMed] [Google Scholar]
- 15.Jaeger J A, Turner D H, Zuker M. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciesiolka J, Lorenz S, Erdmann V A. Eur J Biochem. 1992;204:575–81. doi: 10.1111/j.1432-1033.1992.tb16670.x. [DOI] [PubMed] [Google Scholar]
- 17.Tamkun M M, Talvenheimo J A, Catterall W A. J Biol Chem. 1984;259:1676–1688. [PubMed] [Google Scholar]
- 18.Bretscher M S. Science. 1973;181:622–629. doi: 10.1126/science.181.4100.622. [DOI] [PubMed] [Google Scholar]
- 19.Fishman P H, Brady R O. Science. 1976;194:906–915. doi: 10.1126/science.185697. [DOI] [PubMed] [Google Scholar]
- 20.Hirano H, Parkhouse B, Nicolson G L, Lennox E S, Singer S J. Proc Natl Acad Sci USA. 1972;69:2945–2949. doi: 10.1073/pnas.69.10.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]